Abstract
Molecular docking is the most commonly used technique in the modern drug discovery process where computational approaches involving docking algorithms are used to dock small molecules into macromolecular target structures. Over the recent years several evaluation studies have been reported by independent scientists comparing the performance of the docking programs by using default ‘black box’ protocols supplied by the software companies. Such studies have to be considered carefully as the docking programs can be tweaked towards optimum performance by selecting the parameters suitable for the target of interest. In this study we address the problem of selecting an appropriate docking and scoring function combination (88 docking algorithm-scoring functions) for substrate specificity predictions for feruloyl esterases, an industrially relevant enzyme family. We also propose the ‘Key Interaction Score System’ (KISS), a more biochemically meaningful measure for evaluation of docking programs based on pose prediction accuracy.
A key objective of the commonly used molecular docking programs is to predict the correct placement of small molecules or ligands within the binding pocket of an enzyme or protein and the biological implications of this process. This knowledge is subsequently applied to identify novel ligands through virtual screening of compound libraries1,2. Several commercial and academic softwares are available for molecular modeling and docking studies. A bundle of studies on the evaluation of molecular docking programs and scoring functions have been published focusing on pose prediction (re-docking a compound with a known conformation and orientation into the target's active site followed by selection of the docking program that return poses below a preselected Root Mean Square Deviation value from the known conformation) and virtual screening (docking a decoy set of inactive compounds that has been mixed with compounds with known activity against the target in question followed by selection of the docking program based on enrichment studies)3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. A very surprising and interesting recent study by Cross et al (2009)21 on comparison of molecular docking programs for pose prediction and virtual screening accuracy showed that there is significant variability on the performance of docking programs based on the target enzyme or protein family. The findings of Cross et al change the paradigm of traditional or previous evaluation studies that used an array of diverse protein structures and standard datasets like DUD (Directory of Useful Decoys)22,23,24. Every molecular docking program or scoring function has a bias for particular physical properties of the target protein or enzyme of interest. It has been proposed that the differences in performance of the molecular docking programs could be attributed to the composition of the training sets used while developing particular docking programs that have different intended goals21. So, selection of a molecular docking program for a particular target needs careful consideration, as each program gives results of varying quality depending on the target. A recent trend is to select docking programs that suit your protein of interest25,26 while conclusions from previous evaluation studies should be exploited as a rough guide for selecting a docking program rather than sticking to the statements of expected performance based on diverse set of proteins or ligands.
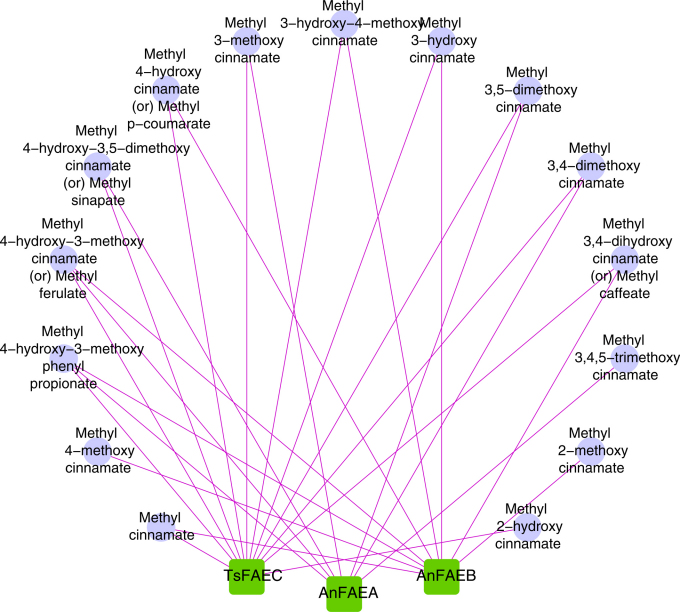
In this study we start anew in the evaluation and selection of molecular docking programs suitable for a specific target of interest. We address the problem of selecting an appropriate docking and scoring combination for substrate specificity predictions, specifically for the feruloyl esterase families, where each family possess both overlapping as well as unique specificity to the individual substrates (Fig. 1). The framework presented here is applicable to select software packages for docking studies for every enzyme or protein family. We recently proposed a novel classification system for feruloyl esterases (FAEs) that resulted in 12 families, which have the capability of acting on a large range of substrates for cleaving ester bonds and synthesizing high-added value molecules through esterification and transesterification reactions27. As mentioned above, there is some overlapping in the substrate-activity maps of the members of the various FAE families (FEFs) due to the flexibility of their residues in the FAE binding pocket. We therefore consider as the ultimate challenge for a docking program to correctly predict the ‘sensitive’ substrate specificity profile of the FAE families, which will position it superior among the others and more suitable for enzymes with high flexibility. We also propose an assessment measure, the Key Interaction Score System (KISS), to evaluate pose prediction accuracy. KISS carries both biological and chemical interaction information and it is presented and discussed in detail below.
Figure 1. Overlapping substrate specificities among the members (TsFAEC, AnFAEA and AnFAEB) of different FAE families; the diagram was created using Cytoscape version 2.841,42.
The enzymes TsFAEC, AnFAEA and AnFAEB were capable of hydrolyzing 12, 7 and 9 substrates respectively.
Results
Protein models and their substrate spectra
Detailed substrate specificity spectra is available only for three enzymes viz., feruloyl esterase A (AnFAEA) and feruloyl esterase B (AnFAEB) from Aspergillus niger, and feruloyl esterase C (TsFAEC) from Talaromyces stipitatus (their experimental kinetic data are given in Supplementary Table S1, see Section A in Supplementary Information). In our earlier study on the development of a FAE classification system27, pharmacophore models, based on key pharmacophore features of their substrate spectra, were proposed for those three FAEs and the respective sub-families that they belong to. While the three-dimensional crystal structure of AnFAEA has been resolved28, the crystal structures of the other two enzymes are not available yet. In the absence of any resolved X-ray or NMR structures, the three-dimensional atomic models for AnFAEB and TsFAEC were modeled from multiple threading alignments29 and iterative structural assembly simulations using the I-TASSER algorithm, an extension of the previous TASSER method30,31,32,33,34. Structure refinement of the modeled structures was carried out using the Discovery Studio software suite version 3.0 (Accelrys Inc, USA). Structural information and validation data (Supplementary Table S2) of the modeled FAEs are given in Section B of Supplementary Information. The coordinates of the model structures (see Supplementary Fig. S1) were submitted to the Protein Model DataBase (PMBD)35.
Evaluation of docking program-scoring function sets
Many evaluation studies have been performed using the default settings of the docking programs, which only provides a baseline performance of each program and lacks the insights of different options provided in the respective software. This is a point that should be considered carefully when claiming performance differences between the programs3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20. In the present study, docking programs were evaluated using the recommended optimized options in the respective software for a particular task, which eliminated the user bias to particular software or results. Additional support was received from the lead application scientific specialists (see Acknowledgments) of the respective software companies. This contribution also facilitated the elaboration of the observed variability in the results obtained by algorithms of the same program (e.g., Glide XP and Glide SP for docking functions in Schrödinger suite). Since new versions of docking programs are frequently released, these must be evaluated by the community almost in an annual base. To the best of our knowledge, this is not only the first evaluation study with the most recent versions (released in 2011) of popular state-of-the-art commercial docking suites, but probably also the most complete with 88 docking algorithm-scoring function sets (involving 24 docking algorithms and 24 scoring functions). As briefly discussed above, the evaluation or selection of the best docking program involves two major steps; first, to predict the pose of the ligand correctly when compared with the conformation in a co-crystallized protein or enzyme, and second, to predict binding affinities close to experimental observations.
Key Interaction Score System
The proposed Key Interaction Score System (KISS) is suggested as an improvement to the first step, namely pose selection, since the ability of a docking program to produce the correct binding mode is a prerequisite to later predict a set of reliable binding affinities. Even though the traditional approach of evaluating the docking programs using the RMSD (Root Mean Square Deviation) is commonly used, the main drawback is not taking into account the interactions between the ligand and the receptor. Hence, as an extension of the RMSD evaluation, we analyzed here whether the docked ligand pose reproduced the same interactions with the receptor as those observed in the cognate-ligand crystal structure. The cognate ligand crystal structure of the AnFAEA (PDB ID: 1UWC)28 was analyzed for key interactions (hydrogen bonds, polar and non-polar contacts, pi-interactions) of the ligand with the receptor. The most important point that should be remembered when comparing the interactions of the docked and crystal structure pose is that the crystal structures do not contain the coordinates for hydrogen, so hydrogens must be included before any comparison or simulation/docking process. The preprocessing of the protein structures is described in Section D of the Supplementary Information, while the observed differences in the interactions of unprocessed and processed crystal structures of 1UWC (as illustrated in Supplementary Fig. S2) only reinforce our assessment for the utility of this step before docking or simulation studies. For ranking the docking programs based on the KISS score, the hydrogen bond interactions in the ‘processed’ crystal structures were used as control systems. The function for calculating the KISS score is given below:
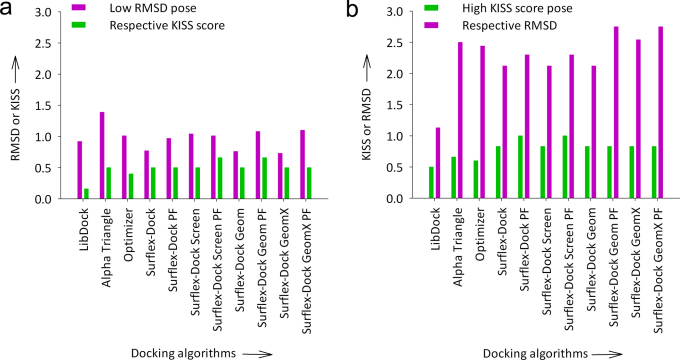
where, Ir = Number of reproduced hydrogen bond interactions by the docked pose. Ic = Total hydrogen bond interactions present in the binding pose of processed cognate ligand crystal structure. The hydrogen bond interactions between ligand and protein were explicitly taken into account when comparing the docked poses with the preprocessed cognate ligand crystal structures for calculating the KISS score. No cut-offs were used in analyzing the docked poses for calculating the KISS score. Imposing cut-offs would result in overweighting or underweighting of interactions or side chains or groups. Since no cut-offs were imposed, KISS score is extensible and could be included in various docking algorithms and scoring functions. A high KISS score can be achieved if the docked pose of the ligand reproduces the ‘same’ hydrogen bond interactions with the receptor seen in the crystal structures irrespective of low or high RMSD. Having a large RMSD between the experimental ligand pose and the computationally calculated pose by a docking program does not indicate a low quality of its force field implementation or scoring algorithm implemented, if the overall binding modes and interactions are reproduced the same way as seen in the crystal structure. Despite the general speculation that the lower the RMSD, the more likely the docked ligand will reproduce the interactions of the ligand in the crystal structure, this does not hold true for all cases. In this study we consider and compare both RMSD and KISS, even though more focus is given to the latter due to its biological significance. RMSD and KISS score are inversely correlated for the docking algorithms listed in Table 1. On the other hand, for approximately half of the docking algorithms in this study the lowest RMSD score does not correspond to the highest KISS score (Fig. 2a) and vice versa (Fig. 2b). For example in the case of pose selection studies with AnFAEA, even though a high RMSD of 2.5 Å was observed from the binding mode seen in the crystal structure, the docked pose 3 generated by the Alpha Triangle docking algorithm reached a KISS score of 0.66. Whereas, the best pose (pose rank 1) according to the low RMSD consideration (1.39 Å) generated by the same Alpha Triangle docking algorithm was considered to be less accurate as it showed a KISS score of 0.5 (see Supplementary Table S3 and Supplementary Fig. S3). Similar trend was observed for FA-1UWC docking with the Optimizer docking algorithm and the variants of the Surflex-Dock docking algorithm (Fig. 2).
Table 1. List of docking algorithms, where the lowest RMSD corresponds to the highest KISS score for re-docking the cognate ligand on the crystal structure of the AnFAEA (PDB ID: IUSW).
| Docking algorithm | Low RMSD pose (Å) | KISS score |
|---|---|---|
| C-DOCKER | 0.99 | 0.66 |
| Flexible Docking | 0.82 | 0.33 |
| Glide SP | 0.62 | 0.66 |
| Glide XP | 1.21 | 0.66 |
| Glide HTVS | 5.24 | 0 |
| Schrodinger's IFD | 0.62 | 0.66 |
| Triangle Matcher | 0.39 | 0.66 |
| Alpha PMI | 5.66 | 0 |
| Proxy Triangle | 0.39 | 0.66 |
| FlexX TM | 0.25 | 0.66 |
| FlexX SIS | 0.34 | 0.66 |
| Simplex Evolution | 0.49 | 1 |
| Iterated Simplex | 0.47 | 0.8 |
Figure 2. Docking algorithms, where there is no correlation between RMSD and KISS score during cognate ligand docking accuracy studies on the crystal structure of the AnFAEA (PDB ID: IUSW).
(a) Lowest RMSD poses and their respective KISS scores. (b) Docked poses with highest KISS score and their respective RMSD.
In many of the docked poses generated from all the docking algorithms it was observed that the ligand establishes additional interactions with the amino acid residues of the binding pocket. Even though those poses increase the number of ligand-receptor interactions, they were considered as incorrect due to lack of the original key interactions seen in the crystal structures. From the examples discussed above, it is evident that having a low RMSD between the docked and the crystallographic pose does not necessarily mean that the ligand can actually form similar interactions or similar binding modes and that a high RMSD value does not indicate a vice versa situation. Hence, when evaluating docking programs it is also essential to look into all of scoring poses carefully. The high flexibility of the ligand/substrate and the flexibility of the binding pocket residues of FAEs27 increase the chances of high variability between the experimental and docked poses; although the same interactions were reproduced by docking programs that showed a KISS score of 1. It should also be noted that the degree of implementation of ligand and receptor flexibility varies widely between the docking algorithms. When we evaluated the docking algorithms for pose prediction accuracy just based on RMSD between the computationally docked pose and the pose in the crystal structure, FlexX TM, FlexX SIS, Triangle Matcher and Proxy Triangle were ranked superior in generating low RMSD (<0.4 Å) value poses; but, those poses were able to score a KISS value of only 0.66. Further rank order of docking algorithms that generated poses with the RMSD range between 0.4–1.4 Å was: Glide SP = Schrodinger's IFD > Surflex-Dock GeomX = Surflex-Dock Geom = Surflex-Dock > Flexible Docking = LibDock = Surflex-Dock PF = C-DOCKER > Surflex-Dock Screen PF = Optimizer > Surflex-Dock Screen > Surflex-Dock Geom PF > Surflex-Dock GeomX PF > Glide XP > Alpha Triangle. The weakest docking algorithms are Glide HTVS and Alpha PMI that generated poses with RMSD values greater than 5Å. Evaluation of the docking programs based on the KISS score of the docked poses revealed that Surflex-Dock PF, Surflex-Dock Screen PF and Simplex Evolution docking algorithms as the best with a KISS score of 1, which means that these three programs were able to produce the ligand-receptor interactions in the docked pose similar to the interactions observed in the processed cognate-ligand crystal structure. The other variants of Surflex-Dock algorithm viz., Surflex-Dock Screen, Surflex-Dock Geom, Surflex-Dock GeomX, Surflex-Dock Geom PF and Surflex-Dock GeomX PF were also able to generate high KISS score (0.83) poses. Hence, we concluded that Surflex-Dock in the SYBYL-X v1.3 suite is the best for pose prediction accuracy in the case of FAEs despite the higher values of RMSD compared to other software platforms. This shows the inadequacy of the energy terms or the interaction terms of the docking algorithm or the scoring function, which were not able to correctly identify the best conformation pose. Automatic calculation of KISS scores, considering the ligand-receptor interactions in crystal structure as a reference, by the software programs can lead to significant alterations in the evaluation of pose selections.
Enrichment and Rank-ordering studies
The docking programs for enrichment of docked poses according to the experimental substrate spectra of the three FAEs described before was evaluated together with the ability of the scoring functions to rank-order the docked poses according to the experimental binding affinities observed. Generally, docking programs include both a docking algorithm for the analysis of different ligand confirmations and a scoring function that should ideally be able to rank the ligands according to the experimental binding affinity. The scoring functions that have been developed still remain as weak predictors of binding affinity and are in need of significant improvements16. Assigning the lowest energy score to the correct binding pose has proved to be a major challenging task for the scoring functions, which is the major reason for the inability to rank-order the compounds. The binding affinity of a ligand also depends upon the collective interactions with binding pocket residues of the receptor, which makes the rank-ordering task more challenging for scoring functions. In addition, the cooperative effects of interactions have only been considered recently, whereas the development of target-dependent scoring functions has also been suggested36,37,38. With the above points in mind, we evaluated the scoring functions both for enrichment and for rank-ordering of ligands specific to FAEs. Unfortunately, the Km values (the measure of affinity) of the FAEs, used in our evaluation, among different substrates are quite close (Supplementary Table S1), which poses a major challenge for docking algorithms or scoring schemes to rank-order the substrates. So the identification of active substrates by the docking algorithms or scoring schemes was set as a realistic aim for assessment.
Even though reviewing of different assessment methods for evaluating docking programs is out of the scope of the present work, they are briefly discussed here due to their importance in the evaluation process. The standard tool for measuring docking enrichment is the enrichment factor, which is simply the ratio of the number of actives retrieved in a specified top x% of the database to the number of actives expected at random. The only advantage of this methodology is simplicity and can be used easily in large virtual screening studies. But, it has several disadvantages. Enrichment factors are highly sensitive to the ratio of actives and decoys and it is hard to compare results obtained using different ligand sets or to evaluate different programs. Most importantly, a decision needs to be made as to where to set the cut-point in the database, which is not always obvious. Another metric that has been used for enrichment studies is ROC (received operating characteristic) curve, which although is independent of the active-decoy ratio, has disadvantages when comparing ROC curves of different data sets. For example, ROC curves of different shapes can have the same Area under Curve (AUC) value and the complexity further increases when evaluating the ROC curves of different docking programs for different protein families21. Matthews Correlation Coefficient (MCC) is a metric used in many fields of engineering and medicine and it is now being adopted for enrichment studies39. Thus MCC was used in this work to evaluate the randomness of the enrichment.
The positive prediction accuracy or sensitivity Sn = TA/(TA+FI) and negative prediction accuracy or specificity Sp = TI/(TI+FA) are also introduced. The overall accuracy is defined as Oq = (TA+TI)/(TA+FI+TI+FA). The different terms are: True Active TA (correctly predicted active substrates), False Inactive FI (active substrates incorrectly predicted as inactive), True Inactive TI (correctly predicted inactive substrates), and False Active FA (inactive substrates incorrectly predicted as active).
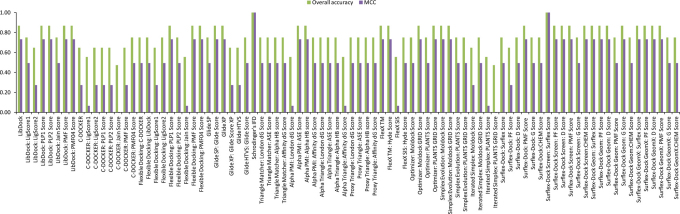
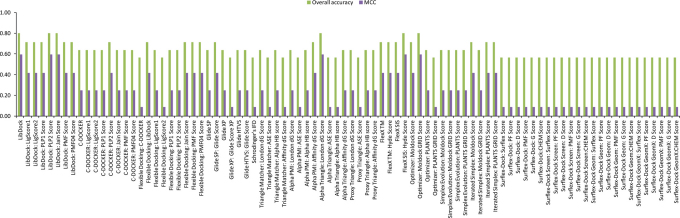
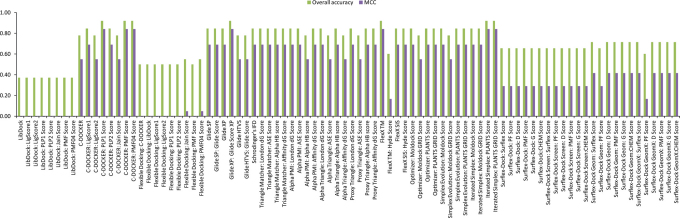
Different programs exhibited large performance differences in enrichment studies of the three FAEs that we have examined (see Fig. 3, Fig. 4 and Fig. 5). The three FAEs, members of different FAE families27, present high diversity in their binding sites (see Fig. 6A, 6B and 6C) and types of ligands. Several factors like binding pocket environment (ex: hydrophobicity), volume of the binding pocket and number of rotatable bonds that deal with the flexibility of the binding pocket play significant role on the performance of the docking algorithms/scoring functions. So, which docking program should we choose when dealing with enzymes with sensitive substrate profile like FAEs? The answer should be given individually for the three aspects viz., pose prediction, enrichment and rank-ordering. In the case of pose prediction accuracy, we could safely say that the Surflex-Dock suite (SYBYL-X v1.3 software package) is accurate in terms of the KISS score, but still there is room for improvement for its algorithms in terms of generating low RMSD poses. Whereas, Simplex evolution algorithm (MVD v4.3.0 software package) performed well in both aspects of pose prediction accuracy (Table 1). In the case of enrichment studies for the AnFAEA, Schrödinger's IFD algorithm and Surflex-Dock suite (Surflex-Dock Screen: Surflex Score) are accurate with an MCC value of 1 (Fig. 3). The other variants of Surflex-Dock algorithm also performed well in enrichment studies with an MCC value of 0.73. Even though Accelrys LibDock algorithm failed completely in enrichment studies for the TsFAEC (Fig. 5), it performed reasonably with an MCC value of 0.6 in enrichment studies for the AnFAEB (Fig. 4). Whereas, Accelrys C-Docker algorithm failed for the cases of AnFAEA (Fig. 3) and AnFAEB (Fig. 4), it performed well in the enrichment studies for TsFAEC (Fig. 5). Full rank list of the 88 docking algorithm-scoring sets for enrichment studies of all three FAEs is given in Supplementary Table S4 of the supporting information. As expected, weak correlations were obtained when comparing the rank-ordering of the active substrates by all the 88 docking algorithm-scoring function sets with the experimentally derived binding affinities. This may be due to the fact that the scoring functions calculate the final score as the additive value of contacts between the ligand/substrate and the receptor. For example, a large substrate that has similar binding affinity with a companion small substrate has the possibilities to create more contacts with the residues of the binding pocket (when compared to the interaction possibilities of the small substrate), which may lead to overestimation of its affinity by the scoring function. Within the obtained sensitivity values for rank-ordering of active substrates, the only algorithm that was top ranked as the best for all the three enzymes (AnFAEA, AnFAEB and TsFAEC) was Accelrys Flexible docking algorithm and its scoring functions PMF04, PMF and PLP1 with Sn values of 0.43, 0.22 and 0.17, respectively (see Supplementary Fig. S4, Supplementary Fig. S5 and Supplementary Fig. S6).
Figure 3. Evaluation of docking algorithm-scoring function sets for AnFAEA substrate enrichment studies.
The final assessment was done based on Matthews Correlation Coefficient (MCC).
Figure 4. Evaluation of docking algorithm-scoring function sets for AnFAEB substrate enrichment studies.
The final assessment was done based on Matthews Correlation Coefficient (MCC).
Figure 5. Evaluation of docking algorithm-scoring function sets for TsFAEC substrate enrichment studies.
The final assessment was done based on Matthews Correlation Coefficient (MCC).
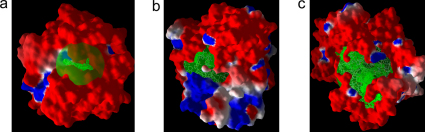
Figure 6. Electrostatic surface diagrams of the three FAEs used in enrichment and rank-ordering studies.
The binding cavity is depicted in gren mesh and the volume of binding pocket is depicted as green trasparent sphere. It is evident from the diagrams that the volume of binding cavity of TsFAEC is very large that allows high degree of freedom for flexible docking algorithms for posing the ligands. (a) AnFAEA. (b) AnFAEB. (c) TsFAEC.
Is the observation of only ligand-receptor interactions enough to identify actives and inactives? The answer is ‘yes’ only if the information regarding the residues involved in key interactions between ligand and receptor is available. This information can be deduced by observing the top scoring docked poses of both active and inactive substrates. When the top scoring poses obtained during enrichment studies for AnFAEA by Schrödinger's Glide SP algorithm (see Supplementary Fig. S7) were analyzed, it was observed that all the active substrates were able to form hydrogen bond interactions with Thr 68 and Leu 134 amino acid residues of the binding pocket, whereas the inactive substrates were not able to do so. If this interaction information can be further applied as a constraint for docking, we may obtain 100% accuracy in the enrichment. Rank-ordering of the substrates based on either the Glide SP score (see Supplementary Table S5) or Glide docking energy (see Supplementary Table S6) alone could not identify the actives. When the key interaction information (hydrogen binding with Thr 68 and Leu 134) was combined with the Glide SP score which ensures that the unfavourable but energetically accessible protonation and tautomeric states are penalized accordingly, we could identify the actives and the rank-ordering of the substrates correlates with the experimental data (see Supplementary Table S7). As evident from Supplementary Table S7, the combination of the Key Interaction System and Glide SP score not only overcomes the problem of false positives and false negatives but also rank the substrates according to experimental binding affinity (Km). Extraction of interaction information is not possible without the availability of minimum experimental data, which is not straightforward for all proteins. At the very least, these receptor-ligand complexes can be visually inspected for the key interactions by modellers and medicinal chemists to obtain a qualitative idea of the KISS score. As for now, visualization of the binding modes of the receptor/ligand in question can help to choose the correct pose. The most important measure of the effectiveness of the KISS system will come from its automation by docking programs and further its actual use in structure-based drug design projects in the biotechnology and pharmaceutical industry.
Discussion
If the docking algorithms and scoring functions kiss different proteins in various ranges, as we have shown in this study, how can the researchers decide which docking program to use? Can we rely on the bundle of evaluation studies that has been published? In general, the docking program evaluation studies have been performed on several 3D structures and the researchers publish the average values (for example, average RMSD of docked poses by a particular program; average enrichment values), which we should see with magnifying lens. The very straightforward solution to this major question, as proposed with this study, is to choose the program that performs well with the protein/target of interest (of course, some experimental data are needed to make the evaluation possible). Comparison of molecular docking programs for pose prediction and enrichment showed that there is significant variability on the performance of docking programs based on the target protein. So docking program that performs well with the protein/target of interest should be chosen. The proposed KISS score provides a biochemical meaning in the selection of docking programs.
The KISS system has the ability to identify the beneficial docking poses (with high KISS score) irrespective of the RMSD value. RMSD is strictly a measure of fit based on the proportion of atoms aligned with the crystallographic pose, whereas the KISS system also considers docked poses with badly aligned atoms if they were able to form the same hydrogen bond interactions observed in the crystallographic pose. The KISS system thus reduces the problem of flexibility arising from the large number of poses or conformers. The KISS system considers a docked pose with very low RMSD as incorrect, if it has a KISS score of zero. Studies on evaluation of docking programs based on pose selection are problematic by the fact that docking poses are penalized and considered incorrect from 2Å to an infinitely poor RMSD16,40. Such a crude RMSD cutoff cannot rescue correct poses with high RMSD. The docking poses (false positives) with good RMSD but forming different interactions with the protein than those observed in crystallographic structure can be filtered by combining KISS and RMSD. KISS provides a biochemical dimension in the selection of docking poses and can be integrated with any of the docking programs that use RMSD as the measure for ranking the docked poses. We believe that the KISS system penalizes false negatives or false positives due to the fact that it introduces a biochemical measure that ranks high beneficial poses with high RMSD. Though KISS may not solve all the issues with the current docking algorithms and scoring functions, combining with RMSD will avoid discarding realistic poses.
Even though the work reported here mainly focused on selecting the best docking program for use in screening of compounds for FAEs, it also addressed the following questions. (A) How can pose selection studies be made biologically meaningful? (B) Can we rely completely on RMSD based studies to select a docking program? (C) Does RMSD and KISS score co-ordinate each other? (D) Does pose selection and enrichment/rank-ordering goes hand-in-hand? So, it's now the reader's turn to carefully select the docking program that is specific for his/her target structure of interest; the framework is readymade in this article.
Methods
Docking software suites
Docking small molecules (ligands) into larger protein molecules (receptors) is a complex and difficult task and requires several protocols/algorithms to help with docking. In general the calculations of receptor-ligand interactions involves two steps, first an algorithm is used to place various confirmations (if the algorithm allows) of the ligand molecules into the binding pocket of an enzyme or target structure, and second the binding energies of the docked molecules are calculated. The first process is referred as ‘docking’ and the second process is referred as ‘scoring’. Most of the docking programs developed perform both the processes. A large variety of docking algorithms and scoring functions exists and were used in this study; the detailed description of each algorithm/scoring function is beyond the scope of this paper, hence the reader is therefore referred to respective publications given in the brief description of the algorithms and scoring schemes used (see Section C of the Supplementary Information). Preprocessing of protein and ligand structures was done according to the protocols recommended in the respective docking programs. 3D coordinates of substrates structures that have been created in our previous work27 were used in this study.
Discovery Studio v3.0: Discovery studio version 3.0 is an integrated modeling and simulation solution for both small molecule and biotherapeutics-based research; and the latest version 3.0 used in this study has been released in December 2010 (Accelrys Inc, USA). It is built on the Pipeline Pilot Enterprise Server™ operating platform, allowing seamless integration of protein modeling, pharmacophore analysis, and structure-based design, as well as third-party applications (e.g., Catalyst, MODELER, CHARMm, etc).
Schrödinger Suite - Maestro v9.2: Maestro version 9.2 is the graphical user interface (GUI) for the latest versions of Schrödinger's suite computational programs released in April 2011 (Schrödinger LLC, USA): CombiGlide version 2.7, ConfGen version 2.3, Desmond version 3.0, Epik version 2.2, Glide version 5.7, Impact version 5.7, Jaguar version 7.8, Liaison version 5.7, LigPrep version 2.5, MacroModel version 9.9, Phase version 3.3, Prime version 3.0, PrimeX version 1.8, QikProp version 3.4, QSite version 5.7, SiteMap version 2.5, Strike version 2.0, and WaterMap.
Molecular Operating Environment (MOE) v2010.10: MOE version 2010.10 is fully integrated drug discovery software package released in November 2010 (Chemical Computing Group, Canada).
LeadIT v2.0.1: LeadIT vesion 2.0.1 is an interactive graphical user interface which embeds both docking and fragment-based design tools, FlexX and ReCore respectively, released in March 2011 (BioSolveIT GmbH, Germany).
Molegro Virtual Docker v4.3.0: Molegro Virtual Docker (MVD) version 4.3.0 is an integrated platform for predicting protein - ligand interactions43, released in February 2011 (Molegro ApS, Denmark). MVD handles all aspects of the docking process from preparation of the molecules to determination of the potential binding sites of the target protein, and prediction of the binding modes of the ligands.
SYBYL-X v1.3: SYBYL-X version 1.3 is a Small Molecule Modeling and Simulation package released in May 2011 (Tripos – A certara company, USA), provides industry proven tools for small molecule modeling and simulation, allowing researchers to perform critical tasks such as hit or lead expansion, lead or scaffold hopping, and to consider critical molecular properties or predicted ADME and physical properties early in the discovery process.
Author Contributions
DGU and NS performed the study. DGU and GPA designed the study and wrote the manuscript. NS and LO assisted in writing and editing the manuscript. DGU, NS, LO and GPA analyzed the data and reviewed the manuscript.
Supplementary Material
Supplementary Information
Supplementary Information
Acknowledgments
We thank Fabio Boni (Accelrys Inc, USA), Renate Baker (Accelrys Inc, USA), Annettte Höglund (Schrödinger LLC, USA), Kevin Turpeinen (Schrödinger LLC, USA), Claudia Leschinski (BioSolveIT GmbH, Germany), Dr. Rene Thomsen (Molegro ApS, Denmark), Filipa Vasconcelos (Tripos-A certara company, USA), Veronique Monjardet (Tripos-A certara company, USA), Inga Randoll (Tripos-A certara company, USA) and Boursereau Yohan (Tripos-A certara company, USA) for their support through providing software evaluation licenses. We are thankful to Dr. Gary Tresadern (Research Informatics, Janssen Research & Development, Pharmaceutical Companies of Johnson & Johnson, Spain), Dr. Anil Kumar and Sugandha Sharma (G.B. Pant University, India) and for valuable help and advice on docking studies with the MOE software. We gratefully acknowledge Dr. Katalin Nadassy (Accelrys Inc), Dr. Tien Luu (Lead Scientific Specialist, Accelrys Inc), Dr. Jeff Saunders (Applications Scientist, Schrödinger LLC), Dr. Hege S. Beard (Applications Scientist, Schrödinger LLC), Dr. Christian Lemmen (CEO, BioSolveIT GmbH), Dr. Rene Thomsen (CEO, Molegro ApS), Dr. Ulrike Uhrig (Application Scientist Manager & Discovery Software Trainer, Tripos-A Certara Company), Dr. Fabian Bös (Application Scientist, Tripos - A Certara Company), Prof. Leif A. Eriksson (National University of Ireland), Viraja Palwai (Örebro University), Dr. So Ha Lee and Ahmed Z. Abdelazem (Korea Institute of Science and Technology) for discussions and scientific support. Funding support for this research was provided by The Swedish Research Council (Vetenskapsrådet Grant no: 2008–2955).
References
- Toledo-Sherman L. M. & Chen D. High-throughput virtual screening for drug discovery in parallel. Curr Opin Drug Discov Devel 5, 414–421 (2002). [PubMed] [Google Scholar]
- Kitchen D. B., Decornez H., Furr J. R. & Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 3, 935–949 (2004). [DOI] [PubMed] [Google Scholar]
- Bissantz C., Folkers G. & Rognan D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J Med Chem 43, 4759–4767 (2000). [DOI] [PubMed] [Google Scholar]
- Stahl M. & Rarey M. Detailed analysis of scoring functions for virtual screening. J Med Chem 44, 1035–1042 (2001). [DOI] [PubMed] [Google Scholar]
- Wang R., Lu Y. & Wang S. Comparative evaluation of 11 scoring functions for molecular docking. J Med Chem 46, 2287–2303 (2003). [DOI] [PubMed] [Google Scholar]
- Bursulaya B. D., Totrov M., Abagyan R. & Brooks C. L. Comparative study of several algorithms for flexible ligand docking. J Comput Aid Mol Des 17, 755–763 (2003). [DOI] [PubMed] [Google Scholar]
- Schulz-Gasch T. & Stahl M. Binding site characteristics in structure-based virtual screening: evaluation of current docking tools. J Mol Model 9, 47–57 (2003). [DOI] [PubMed] [Google Scholar]
- Xing L., Hodgkin E., Liu Q. & Sedlock D. Evaluation and application of multiple scoring functions for a virtual screening experiment. J Comput Aided Mol Des 18, 333–344 (2004). [DOI] [PubMed] [Google Scholar]
- Perola E., Walters W. P. & Charifson P. S. A detailed comparison of current docking and scoring methods on systems of pharmaceutical relevance. Proteins 56, 235–249 (2004). [DOI] [PubMed] [Google Scholar]
- Kellenberger E., Rodrigo J., Muller P. & Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins 57, 225–242 (2004). [DOI] [PubMed] [Google Scholar]
- Kontoyianni M., McClellan L. M. & Sokol G. S. Evaluation of docking performance: comparative data on docking algorithms. J Med Chem 47, 558–565 (2004). [DOI] [PubMed] [Google Scholar]
- Ferrara P., Gohlke H., Price D. J., Klebe G. & Brooks C. L. 3rd Assessing scoring functions for protein-ligand interactions. J Med Chem 47, 3032–3047 (2004). [DOI] [PubMed] [Google Scholar]
- Kontoyianni M., Sokol G. S. & McClellan L. M. Evaluation of library ranking efficacy in virtual screening. J Comput Chem 26, 11–22 (2005). [DOI] [PubMed] [Google Scholar]
- Cummings M. D., DesJarlais R. L., Gibbs A. C., Mohan V. & Jaeger E. P. Comparison of automated docking programs as virtual screening tools. J Med Chem 48, 962–976 (2005). [DOI] [PubMed] [Google Scholar]
- Yang J. M., Chen Y. F., Shen T. W., Kristal B. S. & Hsu D. F. Consensus scoring criteria for improving enrichment in virtual screening. J Chem Inf Model 45, 1134–1146 (2005). [DOI] [PubMed] [Google Scholar]
- Warren G. L. et al. A critical assessment of docking programs and scoring functions. J Med Chem 49, 5912–5931 (2006). [DOI] [PubMed] [Google Scholar]
- Chen H., Lyne P. D., Giordanetto F., Lovell T. & Li J. On evaluating molecular-docking methods for pose prediction and enrichment factors. J Chem Inf Model 46, 401–415 (2006). [DOI] [PubMed] [Google Scholar]
- Onodera K., Satou K. & Hirota H. Evaluations of molecular docking programs for virtual screening. J Chem Inf Model 47, 1609–1618 (2007). [DOI] [PubMed] [Google Scholar]
- McGaughey G. B. et al. Comparison of topological, shape, and docking methods in virtual screening. J Chem Inf Model 47, 1504–1519 (2007). [DOI] [PubMed] [Google Scholar]
- Kellenberger E., Foata N. & Rognan D. Ranking targets in structure-based virtual screening of three-dimensional protein libraries: methods and problems. J Chem Inf Model 48, 1014–1025 (2008). [DOI] [PubMed] [Google Scholar]
- Cross J. B. et al. Comparison of Several Molecular Docking Programs: Pose Prediction and Virtual Screening Accuracy. J Chem Inf Model 49, 1455–1474 (2009). [DOI] [PubMed] [Google Scholar]
- von Korff M., Freyss J. & Sander T. Comparison of ligand- and structure-based virtual screening on the DUD data set. J Chem Inf Model 49, 209–231 (2009). [DOI] [PubMed] [Google Scholar]
- Huang N., Shoichet B. K. & Irwin J. J. Benchmarking sets for molecular docking. J Med Chem 49, 6789–6801 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J. J. Community benchmarks for virtual screening. J Comput Aided Mol Des 22, 193–199 (2008). [DOI] [PubMed] [Google Scholar]
- Hevener K. E. et al. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J Chem Inf Model 49, 444–460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeragraf M. et al. How reliable are current docking approaches for structure-based drug design? Lessons from aldose reductase. Angew Chem Int Edit 46, 3575–3578 (2007). [DOI] [PubMed] [Google Scholar]
- Udatha D. B., Kouskoumvekaki I., Olsson L. & Panagiotou G. The interplay of descriptor-based computational analysis with pharmacophore modeling builds the basis for a novel classification scheme for feruloyl esterases. Biotechnology advances 29, 94–110 (2011). [DOI] [PubMed] [Google Scholar]
- McAuley K. E., Svendsen A., Patkar S. A. & Wilson K. S. Structure of a feruloyl esterase from Aspergillus niger. Acta crystallographica. Section D, Biological crystallography 60, 878–887 (2004). [DOI] [PubMed] [Google Scholar]
- Wu S. & Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35, 3375–3382 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan S., Zhang W. & Hobrath J. V. Recent advances in structure-based virtual screening of G-protein coupled receptors. AAPS J 11, 178–185 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins-Structure Function and Bioinformatics 77, 100–113 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3 D structure prediction. Bmc Bioinformatics 9,40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins-Structure Function and Bioinformatics 69, 108–117 (2007). [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A. & Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrignano T., De Meo P. D., Cozzetto D., Talamo I. G. & Tramontano A. The PMDB Protein Model Database. Nucleic Acids Res 34, D306–309 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. H., Stephens E., O'Brien D. P. & Zhou M. Understanding noncovalent interactions: ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angewandte Chemie 43, 6596–6616 (2004). [DOI] [PubMed] [Google Scholar]
- Muley L. et al. Enhancement of hydrophobic interactions and hydrogen bond strength by cooperativity: synthesis, modeling, and molecular dynamics simulations of a congeneric series of thrombin inhibitors. Journal of medicinal chemistry 53, 2126–2135 (2010). [DOI] [PubMed] [Google Scholar]
- Lafleur K., Huang D., Zhou T., Caflisch A. & Nevado C. Structure-based optimization of potent and selective inhibitors of the tyrosine kinase erythropoietin producing human hepatocellular carcinoma receptor B4 (EphB4). Journal of medicinal chemistry 52, 6433–6446 (2009). [DOI] [PubMed] [Google Scholar]
- Ong S. A. K., Lin H. H., Chen Y. Z., Li Z. R. & Cao Z. W. Efficacy of different protein descriptors in predicting protein functional families. Bmc Bioinformatics 8, 300 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A. N. Bias, reporting, and sharing: computational evaluations of docking methods. J Comput Aid Mol Des 22, 201–212 (2008). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot M. E., Ono K., Ruscheinski J., Wang P. L. & Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R. & Christensen M. H. MolDock: a new technique for high-accuracy molecular docking. Journal of medicinal chemistry 49, 3315–3321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information