Abstract
The present study measured variation in body weight using a combined analysis in an F2 intercross and an F34 advanced intercross line (AIL). Both crosses were derived from inbred LG/J and SM/J mice, which were selected for large and small body size prior to inbreeding. Body weight was measured at 62 (±5) days of age. Using an integrated GWAS and forward model selection approach, we identified 11 significant QTLs that affected body weight on ten different chromosomes. With these results we developed a full model that explained over 18% of the phenotypic variance. The median 1.5-LOD support interval was 5.55 Mb, which is a significant improvement over most prior body weight QTLs. We identified nonsynonymous coding SNPs between LG/J and SM/J mice in order to further narrow the list of candidate genes. Three of the genes with nonsynonymous coding SNPs (Rad23b, Stk33, and Anks1b) have been associated with adiposity, waist circumference, and body mass index in human GWAS, thus providing evidence that these genes may underlie our QTLs. Our results demonstrate that a relatively small number of loci contribute significantly to the phenotypic variance in body weight, which is in marked contrast to the situation in humans. This difference is likely to be the result of strong selective pressure and the simplified genetic architecture, both of which are important advantages of our system.
Introduction
Phenotypic variation in complex quantitative traits is attributed to combinations of genes, environmental factors, and their interactions with each other (Flint and Mackay 2009; Cheverud et al. 2010). Variation in body weight is linked to health disorders in both humans and agricultural animals (Bouchard 1991; Campfield and Smith 1999; Willet et al. 1999). Levi et al. (2010) describe numerous environmental influences contributing to the recent spike in body weight in humans, but strong evidence also indicates that body weight has a significant genetic component. Family and twin studies estimate that the heritable variation contributing to body weight ranges from 30 to 70% (Rankinen et al. 2006). More recently, genome-wide association studies (GWAS) have identified genetic variants that contribute to body weight, obesity, and body mass index (BMI) in human populations, yet each individual variant explains less than 0.05% of the heritable variation (Loos and Bouchard 2008; Stratigopoulos et al. 2008; Willer et al. 2009; Scherag et al. 2010; Speliotes et al. 2010). The discovery that these single-nucleotide polymorphisms (SNPs) account for only a tiny fraction of the genetic variation creates uncertainty regarding the ability of GWAS to identify the bulk of heritable variation for body weight. Reasons for this “missing heritability” are thought to be due in part to the presence of rare alleles, epistasis, and gene-by-environment interactions (Manolio et al. 2009).
Mouse models are complementary to human genetic studies and offer unique advantages, including the ability to control environmental variance, perform dangerous or invasive procedures, conduct well-defined crosses, functionally evaluate candidate genes in vivo or in vitro, and undertake rigorous mechanistic studies. Quantitative trait locus (QTL) studies have been successful in identifying chromosomal regions associated with body weight in mice, yet gene identification has remained elusive (Brockmann et al. 1998; Morris et al. 1999; Ishikawa and Namikawa 2004; Bennett et al. 2005; Neuschl et al. 2007). This is partially because QTL studies in mice have traditionally used recombinant inbred lines (RI), backcrosses, F2 intercrosses (F2), and similar strategies to identify QTLs that underlie phenotypic variability. Due to a lack of recombination, these techniques are able to identify only large genomic regions and are thus unsuitable for identifying the genes that underlie QTLs (Peters et al. 2007; Flint 2011).
This serious limitation can be addressed by using populations with greater numbers of accumulated recombinations, such as advanced intercross lines (AILs) (Darvasi and Soller 1995; Parker and Palmer 2011). AILs are produced by randomly mating many individuals beyond the F2 generation. These additional breeding generations produce additional recombinations, which allows for the more precise identification of QTL regions. Because AILs are derived from two inbred founders, they maintain the simplicity of more traditional crosses. The F2 and F34 AIL used in the present study are derived from the Large (LG/J) and Small (SM/J) inbred mouse lines originated from mice selected for large or small body size at 60 days of age, respectively (Goodale 1938; MacArthur 1944). After both lines were fully inbred, they displayed a 24-g difference in body weight at 60 days of age (Chai 1956). These strains have been studied extensively as genetic models of body weight and obesity-related traits (Cheverud et al. 2001; Ehrich et al. 2003, 2005a, b; Fawcett et al. 2008, 2010). The extreme phenotypic variability between these strains makes them ideally suited for the identification of genetic influences on body weight since they are expected to possess many segregating alleles that confer differences.
In the present study, we use the power of LG/J × SM/J F2 mice and the precision of LG/J × SM/J F34 AIL mice in conjunction with a forward model selection procedure to identify and fine-map loci associated with body weight. Knowledge about the genes identified in these regions may have many applications, including improvement of farm animal meat quality and breeding procedures or the development of treatments for growth- and obesity-related disorders in humans. In addition, from a technical perspective, the use of a highly recombinant mouse population in conjunction with our forward model selection QTL mapping procedure has broader applications for the analysis of complex traits.
Materials and methods
Subjects
All procedures were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC) in accordance with NIH guidelines. Details regarding the mice and genotypes used in the present study have been described previously (Cheng et al. 2010; Lionikas et al. 2010; Samocha et al. 2010). We obtained inbred male SM/J and female LG/J mice from the Jackson Laboratory (Bar Harbor, ME). These mice were used to produce LG/J × SM/J F1 mice, which were then bred to create the F2 generation (n = 488, 239 females and 249 males).
In addition, we obtained 140 F33 breeders from the laboratory of Dr. James Cheverud (Washington University, St. Louis, MO). The F33 mice were outbred, with more than 50 families having been maintained per generation since their inception. Breeding was random except that siblings were not mated with one another. We chose to study the SM/J and LG/J strains mainly because they were selected to have high and low body weight and so were expected to segregate many relevant loci, and because of the availability of an F33 AIL, which represents almost 10 years of breeding. Records from Dr. Cheverud’s lab allowed us to construct a pedigree for each F33 mouse that traced back to the original inbred founders. From these 140 F33 mice, 119 were successfully bred to create an F34 generation (n = 701, 343 females and 358 males) in which phenotypes were measured. We produced only one F34 litter per breeding pair. Breeding pairs were rotated after each litter in order to avoid producing large numbers of full sibs; however, the phenotyped (F34) generation inevitably contained many sibs, half-sibs, and cousins as well as more distant and complex relationships. All F2 and F34 mice were housed in standard laboratory conditions with a 12:12-h light cycle and ad libitum access to standard lab chow and water.
Phenotyping
Mice were weighed when they were approximately 2 months old (mean age = 62 days, SD = 5 days) at the same time of day during the light phase of the day using a Fisher Scout II scale; weights were rounded to the nearest 0.1 g. These measurements were taken as part of a behavioral study investigating methamphetamine sensitivity (Cheng et al. 2010), but were obtained before any drug was administered. Additional data collected from these mice after the measurement of body weight have also been published (Lionikas et al. 2010; Samocha et al. 2010). Our study uses a novel forward selection technique and investigates body weight QTLs, which were not studied in any of the previous papers.
Genotyping
Genotyping was performed as previously described (Cheng et al. 2010). Briefly, 162 evenly spaced SNPs were used as markers in the F2 mice (Petkov et al. 2004). For the F34 mice, we designed a custom SNP array that assayed SNPs using the Illumina Infinium Platform (http://www.illumina.com). SNPs were chosen to provide uniform coverage of the mouse genome and contained ~4,000 markers that were polymorphic between LG/J and SM/J strains. A full list of these SNPs is available at the JAX Phenome website under the name “Chicago1” (http://phenome.jax.org/db/q?rtn=projects/detailsandsym=Chicago1). We performed genome-wide association analysis in the combined population of the F2 and F34 intercrosses using the R package QTLRel, which is available from CRAN (http://cran.r-project.org/web/packages/QTLRel/index.html). This software allowed us to account for the complex relationships (e.g., sibling, half-sibling, cousins) among the F34 mice by using a mixed model as described previously (Cheng et al. 2010). Because of well-known effects of sex on body weight, we explored genetic models where sex was included as an additive covariate.
Multiple QTL mapping and model selection
The initial genome scan identified QTLs on nine chromosomes that contributed to variation in body weight, with multiple peaks on the same chromosome. After the initial scan, we fit our model with all of the identified QTLs, sequentially performed a series of tests, and removed QTLs that were not significant given other QTLs in the model. For model selection, we used Akaike’s (1974) information criterion (AIC):
where log(M) is the log-likelihood of model M under consideration, |M| is the number of parameters in the model M, and k = 2. Markers were tested using a 2-degree-of-freedom test which models both additive and dominant QTL effects. Instead of using the classical AIC in which k = 2, we chose k to be half the 0.05 genome-wide threshold. This extended AIC is called BICδ by Broman (2002) and posits that the chance of selecting any QTL by the criterion will be 0.05 if there is actually no QTL.
We wanted to select the multi-QTL model that produced the minimum AIC value. Because the number of loci tested was large, it was impractical to search through the whole model space to select the optimal model. Therefore, we adopted two well-known model search strategies, forward selection and backward elimination. First, we performed a forward selection, starting with the model that included no QTL. Next, a genome scan was performed and the locus that resulted in the smallest AIC was added to the model. A second genome scan was conducted while including the previously selected QTL in the model. This procedure was repeated until no loci had sufficiently large AIC to be added to the model. The forward selection method identified 11 QTLs, two of which were on chromosome 6. These results were broadly similar to the results obtained when we performed a single QTL scan; however, the forward selection procedure identified a QTL on chromosome 14 that was not significant when using a single QTL scan, which demonstrates the advantage of our approach.
After generating this model, we further refined the locations of these 11 QTLs by moving them to nearby locations that reduced the AIC. This was done by use of a coordinate descent algorithm (Nocedal and Wright 1999). For this we cyclically moved each of the 11 identified QTLs around its linkage region while keeping the locations of other QTLs constant. The location of each QTL was updated with the location that provided the smallest AIC; this procedure is similar to one previously described by Zeng et al. (1999).
Finally, we performed backward elimination to see if any QTL should be excluded from the model. The rationale is that the contribution of a QTL depends on other QTLs in the model and forward selection can result in extraneous QTLs (Broman 2002). This procedure did not remove any of the QTLs. We defined the confidence interval for each QTL as the 1.5-LOD dropoff on either side of the peak. This interval was expressed in physical map position (Mb) by using the genotyped SNP that was at or beyond the 1.5-LOD support interval.
Model testing
We used the following model to identify QTLs; for the ith individual
where xi represents covariates; ai,k = 1, 0, or −1 if the genotype at the kth QTL is AA, Aa, or aa; di,k = 1 or 0 if the genotype at the kth QTL is heterozygous or homozygous; β’s are the corresponding effects; gi is the polygenic effect; and εi is the residual effect. Furthermore, assume (g1, g2, …, gn) ~ N(0, Σ) and (ε1, ε2, …, ε3) ~ N(0, Iσ2). The model that we arrived at predicted body weight (yi) as follows:
where xsex = 1 or 0 if the ith individual is male or female and xage is its age in days.
Sequence analysis
Sequences for LG/J and SM/J inbred mice were provided by Dr. Jim Cheverud from The Genome Sequencing Center at Washington University (http://genome.wustl.edu) and are described in detail elsewhere (Norgard et al. 2011). Briefly, the sequencing data identified over 4 million autosomal polymorphisms between LG/J and SM/J inbred mice, and all SNPs used in the study are available at dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/). We used these data to identify nonsynonymous coding SNPs within our QTL regions.
Results
Phenotypic analysis
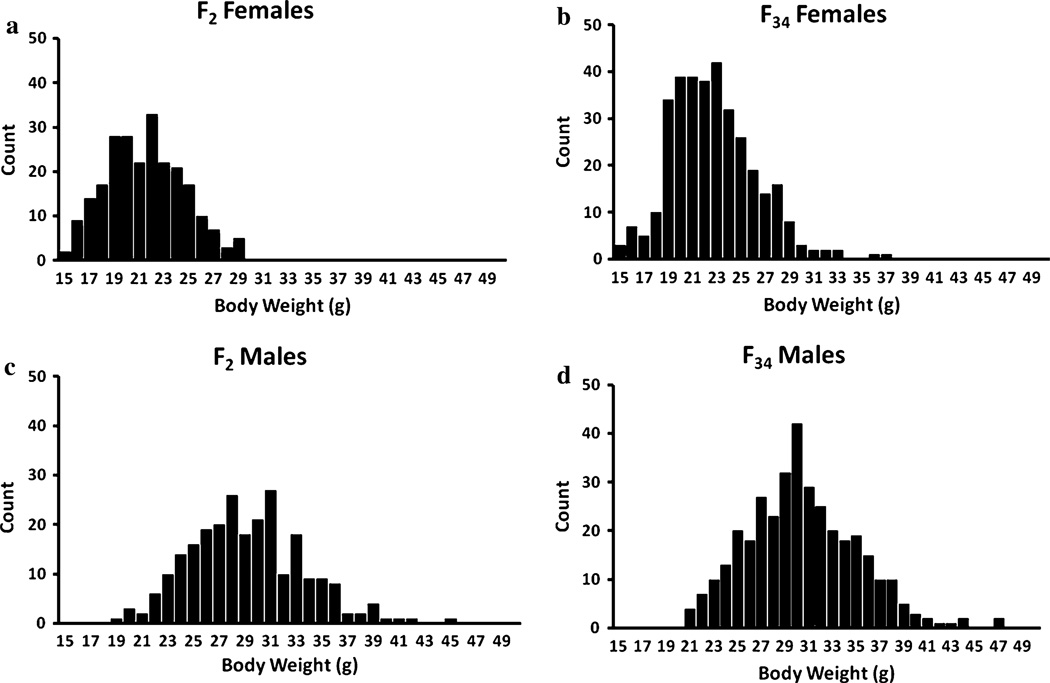
On average, F2 mice weighed 25.01 g (SD = 5.48 g). F34 mice weighed an average of 26.21 g (SD = 5.67 g); these differences were not significant. As expected, there was a highly significant effect of sex on body weight (p < 0.0001). Figure 1 displays the distribution of body weight, split by generation and sex.
Fig. 1.
Histogram of body weight in F2 and F34 mice. a The body weight distribution for F2 female mice. b The weight distribution in F34 females. c The weight distribution in F2 males. d The weight distribution in F34 males
QTL analysis
We performed multiple-QTL model selection using forward selection and backward elimination on the integrated LG/J × SM/J F2 and LG/J × SM/J F34 populations. The resulting full model consisted of 11 QTLs on chromosomes 1, 2, 4, 6 (2 loci), 7, 8, 9, 10, 11, and 14 (Supplementary Fig. 1). Table 1 displays the confidence intervals, peak LOD scores, number of genes in the interval, and percent variance explained for each QTL. The 1.5-LOD support interval for body weight QTLs ranged between 1.82 and 18.79 Mb, with a median interval length of 5.55 Mb. The number of annotated genes within these intervals ranged from 5 to 109, with a median of 39 genes.
Table 1.
1.5-LOD support intervals for the QTLs for body weight
| QTL | Chr | CI (Mb) | Width (Mb) | Peak (Mb) | LOD | Genes in interval |
% Var |
|---|---|---|---|---|---|---|---|
| BodWt1 | 1 | 133.16–137.59 | 4.44 | 136.186097 | 8.10 | 86 | 1.2 |
| BodWt2 | 2 | 153.44–156.37 | 2.93 | 154.430807 | 7.70 | 67 | 1.1 |
| BodWt4 | 4 | 53.93–55.82 | 1.89 | 54.622596 | 14.04 | 5 | 1.9 |
| BodWt6a | 6 | 76.09–81.85 | 5.76 | 77.070185 | 13.59 | 11 | 2.1 |
| BodWt6b | 6 | 142.67–144.50 | 1.82 | 143.980441 | 12.56 | 6 | 2.1 |
| BodWt7 | 7 | 114.79–120.60 | 5.81 | 118.259509 | 15.56 | 92 | 2.2 |
| BodWt8 | 8 | 28.87–47.66 | 18.79 | 47.658428 | 11.73 | 99 | 3.1 |
| BodWt9 | 9 | 47.86–50.19 | 2.33 | 50.090963 | 9.25 | 20 | 1.2 |
| BodWt10 | 10 | 87.27–92.82 | 5.55 | 88.687559 | 8.89 | 39 | 1.3 |
| BodWt11 | 11 | 90.65–97.67 | 7.02 | 94.39318 | 7.91 | 109 | 1.1 |
| BodWt14 | 14 | 87.93–102.70 | 14.77 | 93.996854 | 6.50 | 27 | 0.9 |
The width of the interval and peak marker location are given in Mb position based on Build 37 of the mouse chromosome. LOD score, number of genes in the interval, and percent variance explained are also provided
Sequence analysis
By comparing the sequences of LG/J and SM/J mice, we identified nonsynonymous coding SNPs in the 1.5-LOD support intervals of the QTLs and narrowed the list of candidate genes (Supplementary Table 3). The number of genes with nonsynonymous coding SNPs within each interval ranged from 0 to 52, with a median of 4 genes. Five of the QTL intervals contained fewer than 5 genes with nonsynonymous coding SNPs. Three of the genes that we identified (Rad23b, Stk33, and Anks1b) have been associated with adiposity and BMI in human GWAS studies (Willer et al. 2009; Croteau-Chonka et al. 2010).
Discussion
We performed genome-wide mapping of QTLs affecting body weight in an F2 and F34 AIL population of mice derived from strains selected for high and low body weight. We used a forward selection and backward elimination method to identify 11 QTLs affecting body weight. This procedure identified one additional QTL that was not significant when standard single QTL mapping was performed. As expected, all LG/J alleles were found to be associated with higher body weight. The integration of the F2 and F34 populations provided good power (F2) and good resolution (F34); the median resolution was 5.55 Mb, with four loci <3 Mb.
Traditionally, F2 intercrosses are used to identify QTLs underlying phenotypic variation, and fine-mapping is carried out as a second step using congenic strains or other fine-mapping tools. This time- and labor-intensive effort at subsequent dissection and gene identification is often derailed by the discovery that a single QTL of large effect is in fact caused by multiple loci of small effect located in the same chromosomal region (Legare et al. 2000; Mott et al. 2000; Cheng et al. 2010; Shao et al. 2010). An AIL is an improvement over these traditional methods because it merges identification and fine-mapping into a single step, which can often discriminate between loci that are due to single versus multiple alleles (Darvasi and Soller 1995). One drawback is that the power to detect QTLs in AILs is lower than in F2 populations. This is because the AILs have greater amounts of recombination than the F2, so more tests are performed and a corresponding higher threshold is needed to control type I errors. The integration of the F2 and F34 AIL populations combines the detection power of the F2 with the precision of the F34 AIL.
After identifying 11 QTLs associated with body weight that accounted for approximately 18% of the phenotypic variance, we then used the obesity gene map (http://www.obesitygenes.org), the mouse genome informatics database (http://www.informatics.jax.org/), and the published literature on body weight phenotypes to identify approximately 60 other body weight QTLs that shared overlapping confidence intervals with our regions of interest (Supplementary Table 1). While it is likely that some of the QTLs identified in our study are the same as those identified by other researchers, in most cases we have mapped QTLs with far greater precision than earlier body weight studies, which have a median QTL interval of 31.6 Mb. Interestingly, our QTLs also overlapped regions in other mouse populations associated with phenotypes such as obesity, bone mineral density, limb length, and organ weights (Supplementary Table 2). This may indicate the influence of a single gene on multiple phenotypic traits (pleiotropy), or may simply reflect the fact that body weight is a composite trait consisting of the weights of muscle and fat compounds as well as those of organs, bones, and body fluids (Brockmann et al. 1998). For example, Cheverud et al. (2010) reported that 20% of the weight of an animal in the LG/J × SM/J population is dissectible fat, and in some cases this proportion can approach 50%. While many of our body weight QTLs overlapped with regions associated with organ weights and fat percentages, only 1 of the 11 body weight QTLs overlapped with the muscle weight QTLs identified by Lionikas et al. (2010) in the same population of mice, suggesting that these two phenotypes may have different underlying genetic influences. Still, a limitation of our study design was that we considered only total body weight rather than the weight of specific components that may be driving the observed variation in body weight.
Using sequence data generously provided by Dr. Cheverud, we were able to examine more closely the candidate genes within the QTL intervals and search for polymorphic SNPs between strains (Supplementary Table 3). Three of the QTLs (BodWt4, BodWt7, and BodWt10) contained genes (Rad23b, Stk33, and Anks1b, respectively) with nonsynonymous coding SNPs that have been associated with adiposity, waist circumference, and BMI in human GWAS studies (Willer et al. 2009; Croteau-Chonka et al 2010). A knockout mouse exists for Rad23b that displays disruptions in adipose tissue, endocrine and exocrine glands, growth, size, and metabolism (Ng et al. 2002). The GWAS finding, the existence of a knockout mouse, and the fact that Rad23b is the only gene in the BodWt4 QTL interval with a nonsynonymous coding SNP makes it an especially promising candidate gene for follow-up studies. However, it is important to note that the polymorphisms underlying the observed trait variance may be due to differences in gene expression rather than in protein-coding genes. Indeed, one of our QTLs, BodWt6a, did not contain any genes with nonsynonymous coding SNPs in exonic regions. It is possible that this QTL and the others are due to SNPs in promoter or enhancer regions that gave rise to expression QTLs (eQTLs) resulting in differences in gene expression that underlie body weight QTLs. Loci identified by human GWAS are enriched for eQTLs, suggesting that the latter may cause the former (Nicolae et al 2010). Availability of genome-wide eQTL data in the LG/J and SM/J strains will greatly aid in the identification of specific genes underlying these QTLs.
Our study has several important limitations. First, because we have used a cross between two inbred strains, we are studying the alleles that segregate between them and not the total universe of alleles that segregate among other laboratory or wild mice. We did observe significant overlap in the QTLs we identified in our population with QTLs identified in other populations of mice (Supplementary Table 1), which is consistent with the idea that laboratory mice are segregating a relatively limited number of alleles (Yang et al. 2007). Additionally, we considered only body weight, but not body size or composition, and we examined only one developmental time point and one diet condition. Other studies have provided evidence that different genetic loci may affect body weight at different developmental stages, sexes, and diets (Cheverud et al. 2010; Lawson et al. 2011). Despite these limitations, the QTLs we identified showed significant overlap with QTLs identified by other researchers at a variety of ages, ranging from 21 to 252 days (Supplementary Table 1), as well as with the results of human GWAS studies (Willer et al. 2009; Croteau-Chonka et al 2010). Finally, we did not consider parent-of-origin effects, which are known to be important for body weight in these strains (Cheverud et al. 2010).
In conclusion, we have mapped a large number of body weight QTLs using a novel multiple-QTL mapping procedure and forward selection model in an AIL. This has allowed us to determine which QTLs contribute significantly to variation in body weight given the existence of other QTLs in the model. Some of the QTLs we identified correspond to regions identified by other researchers, yet in the majority of cases, we have narrowed the confidence intervals quite significantly compared to previous studies. In our study we have observed a relatively simple genetic architecture, where a significant fraction of phenotypic variation can be explained by a small number of loci; this is in contrast to efforts to identify similar loci in humans and reflects a strength of our approach. The use of a forward model selection procedure allowed us to identify an additional locus compared to a single-QTL analysis. Furthermore, the combination of high-resolution mapping and sequence data offers a powerful approach and permitted identification of several candidate genes that may underlie differences in body weight. In summary, AILs allow GWAS to be performed in a situation where all alleles are common and where uniform environmental conditions can be maintained, which limits the interactions between genes and environment. These advantages allowed us to map QTLs with a modest sample size and identify small regions that warrant further molecular evaluation.
Supplementary Material
Acknowledgments
We thank Dr. James Cheverud and Dr. Heather Lawson for generously providing the F33 mice used to create the F34 and for the sequence data in the LG/J and SM/J mice. This work was supported by a grant from the Schweppe Foundation and by NIH grants MH079103, DA07255, DA024845, and DA021336.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00335-011-9349-z) contains supplementary material, which is available to authorized users.
Contributor Information
Clarissa C. Parker, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA
Riyan Cheng, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA.
Greta Sokoloff, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA.
Jackie E. Lim, Departments of Pharmacology and Cancer Biology, Duke University School of Medicine, Durham, NC 27710, USA
Andrew D. Skol, Department of Medicine, Section for Genetic Medicine, University of Chicago, Chicago, IL 60637, USA
Mark Abney, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA.
Abraham A. Palmer, Email: aap@uchicago.edu, Department of Human Genetics, University of Chicago, 920 E 58th St., CLSC-507D, Chicago, IL 60637, USA; Departments of Psychiatry and Behavioral Neuroscience, University of Chicago, Chicago, IL 60637, USA.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. [Google Scholar]
- Bennett B, Carosone-Link PJ, Lu L, Chesler EJ, Johnson TE. Genetics of body weight in the LXS recombinant inbred mouse strains. Mamm Genome. 2005;16:764–774. doi: 10.1007/s00335-005-0002-6. [DOI] [PubMed] [Google Scholar]
- Bouchard C. Current understanding of the etiology of obesity: genetic and nongenetic factors. Am J Clin Nutr. 1991;53:1561S–1565S. doi: 10.1093/ajcn/53.6.1561S. [DOI] [PubMed] [Google Scholar]
- Brockmann GA, Haley CS, Renne U, Knott SA, Schwerin M. Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics. 1998;150:369–381. doi: 10.1093/genetics/150.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW. A model selection approach for identification of quantitative trait loci in experimental crosses. J R Stat Soc B. 2002;64:641–650. doi: 10.1534/genetics.108.094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campfield LA, Smith FJ. The pathogenesis of obesity. Bailleres Best Pract Res Clin Ednocrinol Metab. 1999;13:13–30. doi: 10.1053/beem.1999.0004. [DOI] [PubMed] [Google Scholar]
- Chai C. Analysis of quantitative inheritance of body size in mice. II. Gene action and segregation. Genetics. 1956;41:167–178. doi: 10.1093/genetics/41.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185:1033–1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Vaughn TT, Pletscher LS, Peripato AC, Adams ES, Erikson CF, King-Ellison KJ. Genetic architecture of adiposity in the cross of LG/J and SM/J inbred mice. Mamm Genome. 2001;12:3–12. doi: 10.1007/s003350010218. [DOI] [PubMed] [Google Scholar]
- Cheverud JM, Lawson HA, Fawcett GL, Wang B, Pletscher LS, Fox AR, Maxwell TJ, Ehrich TH, Kenney-Hunt JP, Wolf JB, Semenkovich CF. Diet-dependent genetic and genomic imprinting effects on obesity in mice. Obesity. 2010 Silver Spring;19(1):160–170. doi: 10.1038/oby.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau-Chonka DC, Marvelle AF, Lange EM, Lee NR, Adair LS, Lange LA, Mohlke KL. Genome-wide association study of anthropometric traits and evidence of interactions with age and study year in Filipino women. Obesity. 2010 Silver Spring;19(5):1019–1027. doi: 10.1038/oby.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich TH, Kenney JP, Vaughn TT, Pletscher LS, Cheverud JM. Diet, obesity, and hyperglycemia in LG/J and SM/J mice. Obes Res. 2003;11:1400–1410. doi: 10.1038/oby.2003.189. [DOI] [PubMed] [Google Scholar]
- Ehrich TH, Kenney-Hunt JP, Pletscher LS, Cheverud JM. Genetic variation and correlation of dietary response in an advanced intercross mouse line produced from two divergent growth lines. Genet Res. 2005a;85:211–222. doi: 10.1017/S0016672305007603. [DOI] [PubMed] [Google Scholar]
- Ehrich TH, Hrbek T, Kenney-Hunt JP, Pletscher LS, Wang B, Semenkovich CF, Cheverud JM. Fine-mapping gene-by-diet interactions on chromosome 13 in a LG/J × SM/J murine model of obesity. Diabetes. 2005b;54:1863–1872. doi: 10.2337/diabetes.54.6.1863. [DOI] [PubMed] [Google Scholar]
- Fawcett GL, Roseman CC, Jarvis JP, Wang B, Wolf JB, Cheverud JM. Genetic architecture of adiposity and organ weight using combined generation QTL analysis. Obesity. 2008;16:1861–1868. doi: 10.1038/oby.2008.300. [DOI] [PubMed] [Google Scholar]
- Fawcett GL, Jarvis JP, Roseman CC, Wang B, Wolf JB, Cheverud JM. Fine-mapping of obesity-related quantitative trait loci in an F9/10 advanced intercross line. Obesity. 2010;18:1383–1392. doi: 10.1038/oby.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. Mapping quantitative traits and strategies to find quantitative trait genes. Methods. 2011;53(2):163–174. doi: 10.1016/j.ymeth.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale H. A study of the inheritance of body weight in the albino mouse by selection. J Hered. 1938;29:101–112. [Google Scholar]
- Ishikawa A, Namikawa T. Mapping major quantitative trait loci for postnatal growth in an intersubspecific backcross between C57BL/6J and Philippine wild mice by using principle component analysis. Genes Genet Syst. 2004;79:27–39. doi: 10.1266/ggs.79.27. [DOI] [PubMed] [Google Scholar]
- Lawson HA, Lee A, Fawcett GL, Wang B, Pletscher LS, Maxwell TJ, Ehrich TH, Kenney-Hunt JP, Wolf JB, Semenkovich CF, Cheverud JM. The importance of context to the genetic architecture of diabetes-related traits is revealed in a genome-wide scan of a LG/J × SM/J murine model. Mamm Genome. 2011;22:197–208. doi: 10.1007/s00335-010-9313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS, 2nd, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- Levi J, Vinter S, St. Laurent R, Segal LM. F as in Fat: How Obesity Threatens America’s Future [online], Trust for America’s Health and Robert Wood Johnson Foundation. [Accessed 25 April 2011];2010 http://healthyamericans.org/reports/obesity2010/Obesity2010Report.pdf.
- Lionikas A, Cheng R, Lim JE, Palmer AA, Blizard DA. Fine-mapping of muscle weight QTL in LG/J and SM/J intercrosses. Physiol Genomics. 2010;42A:33–38. doi: 10.1152/physiolgenomics.00100.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Bouchard C. FTO: the first gene contributing to common forms of human obesity. Obes Rev. 2008;9:246–250. doi: 10.1111/j.1467-789X.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- MacArthur J. Genetics of body size and related characters. I. Selection of small and large races of the laboratory mouse. Am Nat. 1944;78:142–157. [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KH, Ishikawa A, Keightley PD. Quantitative trait loci for growth traits in C57BL/6J × DBA/2J mice. Mamm Genome. 1999;10:225–228. doi: 10.1007/s003359900977. [DOI] [PubMed] [Google Scholar]
- Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine-mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA. 2000;97:12649–12654. doi: 10.1073/pnas.230304397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuschl C, Brockmann GA, Knott SA. Multiple-trait QTL mapping for body and organ weights in a cross between NMRI8 and DBA/2 mice. Genet Res. 2007;89:47–59. doi: 10.1017/S001667230700852X. [DOI] [PubMed] [Google Scholar]
- Ng JM, Vrieling H, Sugasawa K, Ooms MP, Grootegoed JA, Vreeburg JT, Visser P, Beems RB, Gorgels TG, Hanaoka F, Hoeijmakers JH, van der Horst GT. Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Mol Cell Biol. 2002;22:1233–1245. doi: 10.1128/MCB.22.4.1233-1245.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLos Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocedal J, Wright SJ. Numerical optimization. New York: Springer; 1999. [Google Scholar]
- Norgard EA, Lawson HA, Pletscher LS, Wang B, Brooks VR, Wolf JB, Cheverud JM. Genetic factors and diet affect long-bone length in the F(34) LG, SM advanced intercross. Mamm Genome. 2011;22(3–4):178–196. doi: 10.1007/s00335-010-9311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Palmer AA. Dark matter: are mice the solution to missing heritability? Front Gene. 2011;2:32. doi: 10.3389/fgene.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters LL, Robledo RF, Bult CJ, Churchill GA, Paigen BJ, Svenson KL. The mouse as a model for human biology: a resource guide for complex trait analysis. Nat Rev Genet. 2007;8:58–69. doi: 10.1038/nrg2025. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Cassell MA, Sargent EE, Donnelly CJ, Robinson P, Crew V, Asquith S, Haar RV, Wiles MV. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse. Genomics. 2004;83:902–911. doi: 10.1016/j.ygeno.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity. 2006 Silver Spring;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Lim JE, Cheng R, Sokoloff G, Palmer AA. Fine mapping of QTL for prepulse inhibition in LG/J and SM/J mice using F2 and advanced intercross lines. Genes Brain Behav. 2010;9:759–767. doi: 10.1111/j.1601-183X.2010.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, Muller TD, Grallert H, Wichmann HE, Balkau B, Heude B, Jarvelin MR, Hartikainen AL, Levy-Marchal C, Weill J, Delplanque J, Korner A, Kiess W, Kovacs P, Rayner NW, Prokopenko I, McCarthy MI, Schafer H, Jarick I, Boeing H, Fisher E, Reinehr T, Heinrich J, Rzehak P, Berdel D, Borte M, Biebermann H, Krude H, Rosskopf D, Rimmbach C, Rief W, Fromme T, Klingenspor M, Schurmann A, Schulz N, Nothen MM, Muhleisen TW, Erbel R, Jockel KH, Moebus S, Boes T, Illig T, Froguel P, Hebebrand J, Meyre D. Two new loci for bodyweight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in french and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Sinasac DS, Burrage LC, Hodges CA, Supelak PJ, Palmert MR, Moreno C, Cowley AW, Jr, Jacob HJ, Nadeau JH. Analyzing complex traits with congenic strains. Mamm Genome. 2010;21:276–286. doi: 10.1007/s00335-010-9267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan J, Mägi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segrè AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpeläinen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proencça C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grässler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jørgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, König IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaløy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimäki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O#x02019;Donnell CJ, O’Rahilly S, Ong KK, Oostra B, Paré G, Parker AN, Perola M, Pichler I, Pietiläinen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstråle M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tönjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kähönen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Smith GD, Erdmann J, Froguel P, Grönberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ MAGIC; Procardis Consortium. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratigopoulos G, Padilla SL, LeDuc CA, Watson E, Hattersley AT, McCarthy MI, Zeltser LM, Chung WK, Leibel RL. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, Lettre G, Lim N, Lyon HN, McCarroll SA, Papadakis K, Qi L, Randall JC, Roccasecca RM, Sanna S, Scheet P, Weedon MN, Wheeler E, Zhao JH, Jacobs LC, Prokopenko I, Soranzo N, Tanaka T, Timpson NJ, Almgren P, Bennett A, Bergman RN, Bingham SA, Bonnycastle LL, Brown M, Burtt NP, Chines P, Coin L, Collins FS, Connell JM, Cooper C, Smith GD, Dennison EM, Deodhar P, Elliott P, Erdos MR, Estrada K, Evans DM, Gianniny L, Gieger C, Gillson CJ, Guiducci C, Hackett R, Hadley D, Hall AS, Havulinna AS, Hebebrand J, Hofman A, Isomaa B, Jacobs KB, Johnson T, Jousilahti P, Jovanovic Z, Khaw KT, Kraft P, Kuokkanen M, Kuusisto J, Laitinen J, Lakatta EG, Luan J, Luben RN, Mangino M, McArdle WL, Meitinger T, Mulas A, Munroe PB, Narisu N, Ness AR, Northstone K, O’Rahilly S, Purmann C, Rees MG, Ridderstråle M, Ring SM, Rivadeneira F, Ruokonen A, Sandhu MS, Saramies J, Scott LJ, Scuteri A, Silander K, Sims MA, Song K, Stephens J, Stevens S, Stringham HM, Tung YC, Valle TT, Van Duijn CM, Vimaleswaran KS, Vollenweider P, Waeber G, Wallace C, Watanabe RM, Water-worth DM, Watkins N, Witteman JC, Zeggini E, Zhai G, Zillikens MC, Altshuler D, Caulfield MJ, Chanock SJ, Farooqi IS, Ferrucci L, Guralnik JM, Hattersley AT, Hu FB, Jarvelin MR, Laakso M, Mooser V, Ong KK, Ouwehand WH, Salomaa V, Samani NJ, Spector TD, Tuomi T, Tuomilehto J, Uda M, Uitterlinden AG, Wareham NJ, Deloukas P, Frayling TM, Groop LC, Hayes RB, Hunter DJ, Mohlke KL, Peltonen L, Schlessinger D, Strachan DP, Wichmann HE, McCarthy MI, Boehnke M, Barroso I, Abecasis GR, Hirschhorn JN Wellcome Trust Case Control Consortium; Genetic Investigation of Anthropometric Traits Consortium. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet WC, Dietz WH, Colditz GA. Guidelines for healthy weight. N Engl J Med. 1999;341:427–434. doi: 10.1056/NEJM199908053410607. [DOI] [PubMed] [Google Scholar]
- Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. NatGenet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- Zeng ZB, Kao CH, Basten CJ. Estimating the genetic architecture of quantitative traits. Genet Res. 1999;74:279–289. doi: 10.1017/s0016672399004255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.