Abstract
The Comparative Toxicogenomics Database (CTD) is a public resource that promotes understanding about the effects of environmental chemicals on human health. CTD biocurators manually curate a triad of chemical–gene, chemical–disease and gene–disease relationships from the scientific literature. The CTD curation paradigm uses controlled vocabularies for chemicals, genes and diseases. To curate disease information, CTD first had to identify a source of controlled terms. Two resources seemed to be good candidates: the Online Mendelian Inheritance in Man (OMIM) and the ‘Diseases’ branch of the National Library of Medicine's Medical Subject Headers (MeSH). To maximize the advantages of both, CTD biocurators undertook a novel initiative to map the flat list of OMIM disease terms into the hierarchical nature of the MeSH vocabulary. The result is CTD’s ‘merged disease vocabulary’ (MEDIC), a unique resource that integrates OMIM terms, synonyms and identifiers with MeSH terms, synonyms, definitions, identifiers and hierarchical relationships. MEDIC is both a deep and broad vocabulary, composed of 9700 unique diseases described by more than 67 000 terms (including synonyms). It is freely available to download in various formats from CTD. While neither a true ontology nor a perfect solution, this vocabulary has nonetheless proved to be extremely successful and practical for our biocurators in generating over 2.5 million disease-associated toxicogenomic relationships in CTD. Other external databases have also begun to adopt MEDIC for their disease vocabulary. Here, we describe the construction, implementation, maintenance and use of MEDIC to raise awareness of this resource and to offer it as a putative scaffold in the formal construction of an official disease ontology.
Database URL: http://ctd.mdibl.org/voc.go?type=disease
Introduction
Many diseases are the product of the interactions between genes and the environment. An important component of the environment is chemical exposure. The Comparative Toxicogenomics Database (CTD; http://ctd.mdibl.org/) was developed to help researchers understand the connections between environmental chemicals and gene products, and their effects on human health (1–4).
CTD biocurators read the scientific literature and manually curate a triad of core data describing chemical–gene, chemical–disease and gene–disease relationships using an online curation application (5). CTD's curation paradigm uses controlled vocabularies to streamline curation, ensure consistency among biocurators, allow for quality control and to facilitate aggregation and analysis of information. The CTD Gene vocabulary is based on official gene symbols from NCBI Gene (6), and the CTD Chemical vocabulary is a subset of the ‘Chemicals and Drugs’ [D] branch of MeSH (7).
Finding a vocabulary for capturing disease data initially proved problematic. CTD had certain requirements for a disease vocabulary; it had to be robust, publicly available, relatively stable, regularly maintained, and, preferably, used as an annotation source by other sectors of the scientific community to facilitate interoperability. An ideal solution would have been an official Disease Ontology (8), similar to the highly successful Gene Ontology (GO) used for gene annotations (9). However, at the time of CTD's implementation of disease curation in 2006, the Disease Ontology (DO) project had yet to provide a stable, mature vocabulary. The requirement that the vocabulary be publicly available also eliminated well-known, restricted sources such as SNOMED-CT (Systematized Nomenclature of Medicine-Clinical Terms). The UMLS Metathesaurus, a multi-dimensional electronic version of different biomedical vocabularies, is freely available, but requires account creation, compliance with annual licensing terms, periodic reporting and subjection to restrictions and separate agreements on the use of some content (10). Since CTD is a small bioinformatics group, we needed a solution that would be more practical to manage and integrate with our curation paradigm and to make publicly available.
Two familiar resources looked promising: OMIM (11) and the MeSH ‘Diseases’ branch (7). Here, we describe our evaluation of these two sources, including the advantages and limitations of each with respect to the needs of CTD, and our decision to merge the two into a single artifact to capitalize on the advantages of both vocabularies. This resource is called MEDIC (MErged DIsease voCabulary), and we have used it successfully in our curation paradigm to describe over 2.5 million disease-associated toxicogenomic relationships at CTD (Table 1). We recognize and acknowledge that MEDIC is neither an ontology nor a perfect solution. Nonetheless, it has quickly filled a need in the database community, evidenced by it being adopted as a disease vocabulary by external groups such as the Rat Genome Database (12) and the Mouse Genome Database (13). We hope that the scientific community and ontology experts will develop a true disease ontology that either replaces or evolves from MEDIC's foundation. Until then, we introduce and offer MEDIC as a practical resource and scaffold for others to employ and build upon.
Table 1.
CTD disease data content (as of 5 October 2011)
| Disease data | Count |
|---|---|
| Direct chemical–disease interactions | 14 102 |
| Direct gene–disease interactionsa | 14 218 |
| Inferred chemical–disease relationships | 351 439 |
| Inferred gene–disease relationships | 1 906 178 |
| Inferred disease–GO relationships | 281 580 |
| Inferred disease–pathway relationships | 28 776 |
| Total | 2 596 293 |
aVia CTD biocurators and automatic integration of OMIM data.
Disease vocabularies
OMIM
OMIM is one of the most well-known and utilized resources for detailed information about human genetic diseases (11). We were initially drawn to OMIM because it is familiar to our users and its data are indexed with NCBI Gene records, providing a wealth of genetic disease terms that could be easily integrated into CTD via shared gene accession identifiers (IDs). OMIM, however, is a flat list of different concepts (phenotypes, genes, phenotypes without genes, genes with phenotypes, etc.), which does not provide connections between similar diseases. For example, a query at OMIM with ‘breast cancer’ retrieves ‘BREAST CANCER’ (OMIM:114480) annotated to 21 genes, as well as ‘BREAST-OVARIAN CANCER, FAMILIAL, SUSCEPTIBILITY TO, 3’ (OMIM:613399) annotated to one gene not currently associated with the ‘BREAST CANCER’ record. For CTD, we needed a way for our users to come to one umbrella term (e.g. breast neoplasms) and find information associated with individual and related diseases. While OMIM efficiently catalogs genetic diseases corresponding to mutations, CTD is also interested in environmental diseases, which are not necessarily associated with gene mutations, so we required a vocabulary that included non-genetic disorders as well.
We also needed a way to allow users to navigate between broad and specific disease levels. For example, instead of selecting data exclusively for ‘ALZHEIMER DISEASE’ (OMIM:104300), a CTD user might want a broader perspective for all neurodegenerative diseases, including Alzheimer, Parkinson and Lou Gehirg diseases. The flat OMIM structure does not provide a way to view aggregate information from such higher levels.
OMIM contains a mixture of different types of information, identifiable by a character prefix in front of the record ID. Since we wanted to avoid using OMIM gene pages as part of our disease vocabulary, we excluded in our initial mapping all OMIM records prefixed with an asterisk that identifies records for gene descriptions. We only collected records prefaced with a number sign (# phenotype description, molecular basis known), a percent sign (% phenotype description, molecular basis unknown), a plus sign (+ gene and phenotype combined) or no symbol (phenotype description, Mendelian basis not clearly established). We also excluded deleted OMIM records, identifiable by a caret symbol, as well as terms that seem to be more of a trait instead of a disease, such as ‘BLOOD GROUP, P SYSTEM’ (OMIM:111400).
To streamline its initial creation, MEDIC only included OMIM terms that were associated with an NCBI Gene accession ID. Since its inception, MEDIC is updated by including new OMIM records as they are assigned new gene annotations.
MeSH
MeSH is a controlled vocabulary thesaurus composed of over 26 000 primary terms that are used to index and annotate scientific abstracts in MEDLINE (7). Currently, the MeSH hierarchy is divided into 16 branches. The ‘Diseases’ [C] branch of MeSH, like other branches, is structured as a hierarchy that can be navigated between broad and specific terms (14). Hierarchies are extremely valuable in curation, as they allow associated data to be viewed at various levels of granularity, with data annotated to children of a branch to be aggregated at each higher level of the hierarchy. As an indexing source at PubMed, MeSH provides an efficient way to triage the literature for specific articles to be used in disease curation. However, MeSH does not include genes that are known to be associated with their disease terms, it is deficient in many detailed diseases (especially complex syndromes), and it contains some idiosyncrasies that present challenges to data navigation and analysis. For example, ‘Autistic Disorder’ (MESH:D001321) is not a child in the ‘Diseases’ [C] branch, but rather maps to the ‘Psychiatry and Psychology’ [F] branch. As such, CTD would need to include both the entire ‘Diseases’ [C] branch (and its supplementary concept terms) and the [F03] ‘Mental Disorders’ (MESH:D001523) sub-branch since our users would expect autism spectrum disorders (and other mental disorders) to be listed in a manner similar to other diseases.
MEDIC
For CTD's needs, we wanted to take advantage of both disease vocabularies: the familiarity and immediate genetic data offered by OMIM terms associated with NCBI Gene IDs, combined with the navigation utility and PubMed indexing feature of MeSH terms. An obvious solution was to create a merged vocabulary that integrated both OMIM and MeSH disease terms. In December 2006, two CTD biocurators spent three weeks manually reviewing, integrating and merging the appropriate OMIM disease terms (see above) into the MeSH disease hierarchy using a spreadsheet to form the basis of MEDIC.
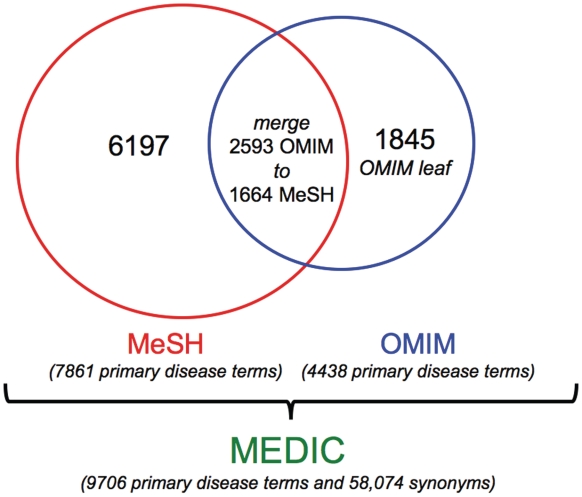
MEDIC is updated on a monthly basis, and is freely available to download in a variety of formats from CTD (Figure 1). As of October 2011, MEDIC contains 9706 unique diseases (plus 58 074 disease synonyms), composed of 6197 primary MeSH terms and IDs, 1845 primary OMIM terms and IDs (made leaves of MeSH terms) and 1664 MeSH terms that contain 2593 OMIM terms merged to them (Figure 2).
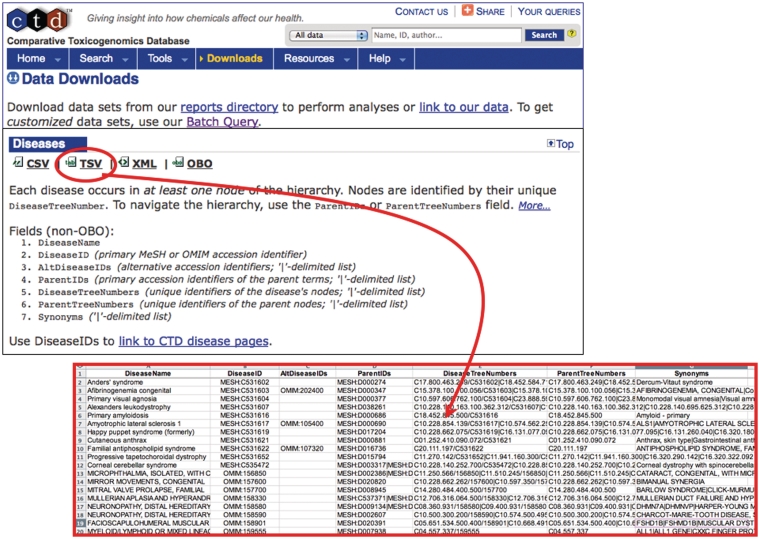
Figure 1.
MEDIC is freely available from CTD. To obtain the most recent version of MEDIC, use the ‘Downloads’ menu tab. The vocabulary can be downloaded in various formats including CSV, TSV (red circle and inset), XML and OBO. We encourage other databases that use MEDIC to provide a direct link from their disease page to CTD’s equivalent disease page to promote interoperability between databases.
Figure 2.
Components of MEDIC. As of October 2011, MEDIC contained 9706 unique disease primary terms and 58 074 synonyms. It includes 6197 MeSH primary terms, 1845 OMIM primary terms (as leaf nodes) and 1664 MeSH primary terms (that have 2593 OMIM primary terms merged to them).
By combining the primary terms, synonyms and IDs from both OMIM and MeSH into a single resource, MEDIC becomes a flexible solution that can be mapped to other disease vocabularies or ontologies. For example, the current version of the DO also includes some terms, synonyms and IDs from OMIM, MeSH and SNOWMED-CT, allowing groups that use the DO to migrate to MEDIC via term and ID mapping. Vice versa, groups that start out by initially adopting MEDIC will have the flexibility to migrate to a more robust DO or other disease vocabulary in the future by similar term and ID mapping. Data management tools such as the interactive Ontology Lookup Service could help streamline and enhance the cross-platform analysis and mapping of these shared vocabularies (15).
MEDIC mapping guidelines
The MeSH disease hierarchy is used as the backbone of MEDIC, with OMIM terms either merged to a MeSH term or added as a leaf (child) to one or more MeSH terms. Where the same disease is represented in OMIM and MeSH, the OMIM name, synonyms and ID all become synonyms of the equivalent MeSH term. This fusion gives our users more power to query diseases at CTD. OMIM primary terms and synonyms are kept in their capitalized format on CTD web display, thereby allowing biocurators and users to readily distinguish between OMIM and MeSH terms.
We used the following guidelines in our manual mapping of OMIM terms to MeSH terms in the initial construction of MEDIC. In our analysis, we considered a number of factors, including: the semantic similarity of the OMIM disease term to a MeSH term as determined by the biocurator (e.g. OMIM ‘LUNG CANCER’ is similar to MeSH ‘Lung Neoplasms’), OMIM synonyms, the disorders described in the OMIM report, its accompanying cited literature and the MeSH terms annotated to its cited literature.
An OMIM primary term is either merged directly to the most appropriate MeSH term or else is made a leaf (child) of one or more MeSH terms. Example: ‘LUNG CANCER’ (OMIM:211980) is merged to ‘Lung Neoplasms’ (MESH:D008175), while ‘MYELOPROLIFERATIVE DISORDER, CHRONIC, WITH EOSINOPHILIA’ (OMIM:131440) is made a leaf of two terms: ‘Myeloproliferative Disorders’ (MESH:D009196) and ‘Eosinophilia’ (MESH:D004802). An individual OMIM term cannot be both merged to and made a leaf of MeSH terms. An OMIM term cannot be made the leaf of another OMIM term.
If an OMIM disease term uses the word ‘susceptibility’ in its name, then that term is merged to the MeSH disease term that is concordant with the core name of the OMIM term. Example: ‘ASTHMA, SUSCEPTIBILITY TO’ (OMIM:600807) is merged to ‘Asthma’ (MESH:D001249). However, if the OMIM ‘susceptibility’ term is a complex of different diseases that do not match a single MeSH term, the OMIM term should be added as a leaf beneath all the appropriate MeSH terms. Example: ‘BREAST-OVARIAN CANCER, FAMILIAL, SUSCEPTIBILITY TO, 2’ (OMIM:612555) is added as a leaf node beneath both ‘Breast Neoplasms’ (MESH:D01943) and ‘Ovarian Neoplasms’ (MESH:D010051).
If an OMIM primary term uses a phrase describing heritability (e.g. ‘hereditary’, ‘autosomal’, ‘X-linked’, etc.), then the term is added as a leaf beneath the most appropriate MeSH term(s). Example: ‘DEAFNESS, AUTOSOMAL DOMINANT 12’ (OMIM:601543) is added beneath ‘Deafness’ (MESH:D003638).
If an OMIM primary term uses a numeral, then it is merged to the concordant MeSH term. Example: ‘SCHIZOPHRENIA 12’ (OMIM:608543) is merged to ‘Schizophrenia’ (MESH:D012559).
If an OMIM primary term uses the word ‘type’, then the term is added as a leaf beneath the most appropriate MeSH term(s). Example: ‘SYNDACTYLY, TYPE 1’ (OMIM:185900) is added beneath ‘Syndactyly’ (MESH:D013576).
For OMIM primary terms that describe syndromes, the biocurator first checks to see if that same syndrome exists in MeSH, and if it does, then the OMIM term is merged to the MeSH term. Example: ‘CHROMOSOME 5q DELETION SYNDROME’ (OMIM:153550) is merged to ‘5q- syndrome’ (MESH:C535323). If the OMIM syndrome is not in MeSH, then the OMIM term will become a leaf beneath one or more MeSH terms. Example: ‘ALOPECIA-MENTAL RETARDATION SYNDROME 2’ (OMIM:610422) is a leaf to both ‘Alopecia’ (MESH:D000505) and Intellectual Disability’ (MESH:D008607).
Updating and maintaining MEDIC
MEDIC is updated by CTD on a monthly basis. Since both OMIM and MeSH are constantly refining their own respective databases, it is inevitable that MEDIC will fall out of synchronization from time to time. To ensure the continued completeness and high quality of MEDIC, we implemented a two-tiered quality control process.
Completeness
From CTD’s perspective, the completeness of the MEDIC vocabulary is defined by its ability to capture OMIM-to-gene associations. To that end, we run a quarterly process that reads through the latest OMIM ‘mim2gene’ file and attempts to identify diseases that do not currently exist in MEDIC either as a discrete or merged term. All OMIM diseases are candidates for inclusion, with the exception of OMIM entries that are designated as no longer existing (i.e. carat prefix) and those designated as genes of known sequence (i.e. asterisk prefix). As the process reads through the ‘mim2gene’ file, if an OMIM disease is encountered that is not accounted for in MEDIC (and is considered valid for inclusion in CTD as defined above), it is checked against a list of OMIM terms that CTD has been unable to match to a MeSH term in the past (e.g. traits such as ‘BLOOD GROUP, P SYSTEM’). If the disease is not contained in the unmatched list, it is included in a report for CTD biocurators to review as the basis for entry of new terms into MEDIC.
High quality
The most recent MeSH and OMIM vocabularies are loaded from their respective databases to CTD each month. To ensure that MEDIC is synchronized with any changes in these vocabularies, CTD biocurators are notified of all disease name changes (whether by MeSH or OMIM) for all mapped terms. This notification is determined by computationally comparing the disease names that were used when the OMIM–MeSH mappings were originally made to the name of the disease in the most recent monthly download. The biocurators research the definitions of the terms in this list to determine if the semantics of the disease (and therefore potentially its association in MEDIC) have changed. Changes in accessions and/or dropped terms are also checked to ensure that they are properly addressed each month.
We have not yet resolved all quality control issues, including, for example, when OMIM changes the character prefix for an OMIM ID. This change can sometimes result in a phenotype report now becoming a gene page (identifiable by an asterisk), something we exclude from MEDIC. We are working on ways to identify and resolve such records in MEDIC. Even with its limitations, however, MEDIC has been a practical vocabulary to implement at CTD in the absence of a more formal, stable, and mature disease ontology.
Implementing MEDIC at CTD
Curating to MEDIC
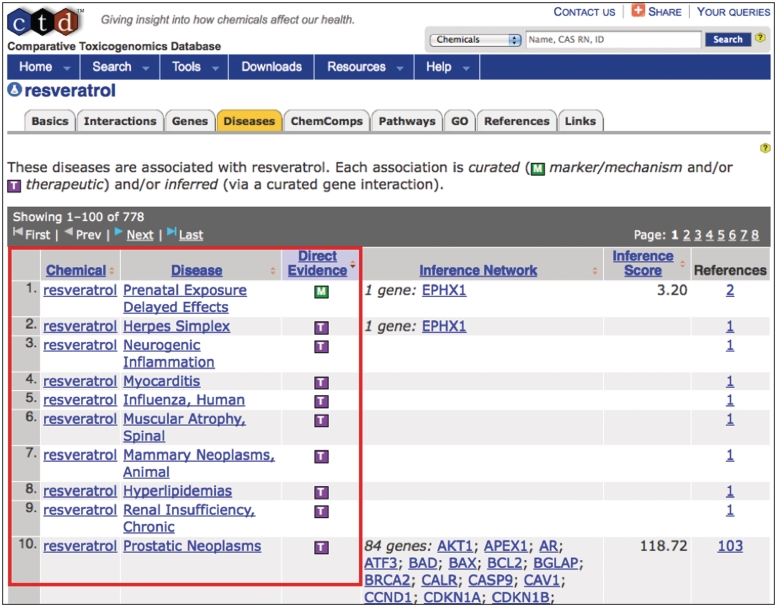
As part of the curation process at CTD, biocurators manually curate chemical–disease and gene–disease relationships from the literature (4–5). Chemicals and genes can be associated to a disease via two types of interactions. The chemical/gene can act as a biomarker or play a molecular role in the disease process (an M-type relationship), or the chemical/gene can be a known or putative therapeutic for the disease (a T-type relationship). CTD biocurators have successfully used MEDIC as a vocabulary to curate disease relationships from the scientific literature for 5471 genes and 2701 chemicals. For example, the chemical resveratrol has a curated relationship to over 50 different diseases from MEDIC (Figure 3). Users can seamlessly explore all of these interactions from the perspective of any of the appropriate chemical, gene, or disease pages in CTD.
Figure 3.
Curating to MEDIC. CTD biocurators use MEDIC as their disease vocabulary when curating chemical–disease and gene–disease data. The ‘Diseases’ tab (orange) on CTD's chemical page for resveratrol displays the curated relationships between the chemical and over 50 diseases (red box, partial screenshot). The green M icon indicates resveratrol is a marker for or plays a molecular role in the disease; the purple T icon indicates the chemical is a real or putative therapeutic for the disease. Every disease term is hyperlinked to its own disease page, allowing users to seamlessly explore chemical-gene-disease networks.
Displaying and navigating MEDIC
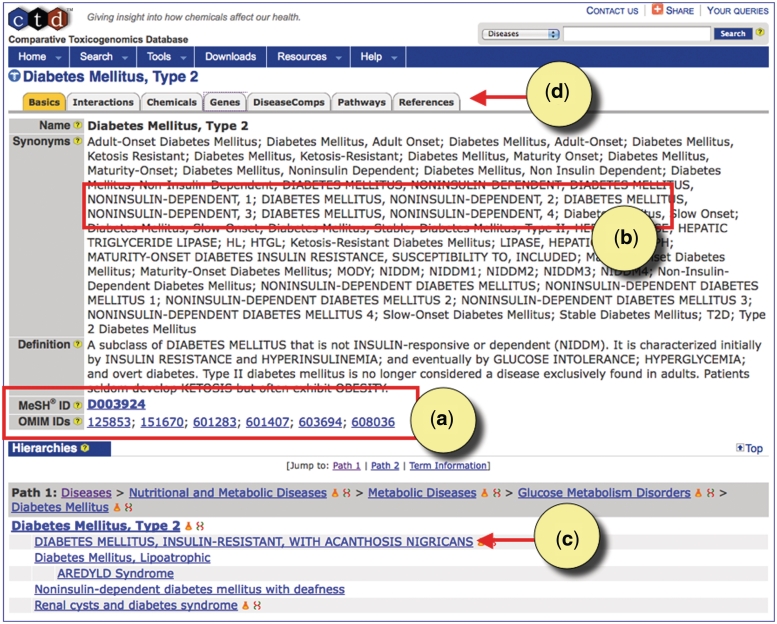
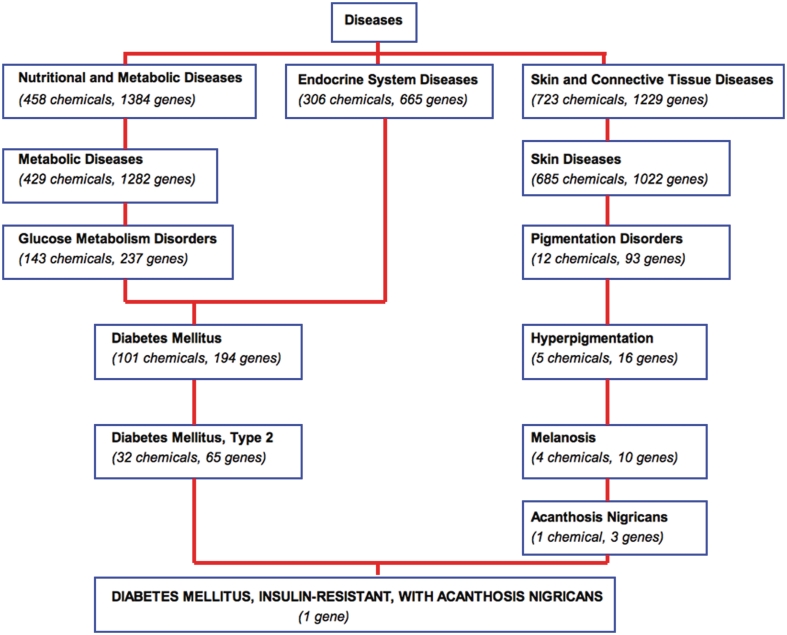
Every MEDIC primary term is displayed as a disease page in CTD. Users looking for information about type 2 diabetes will find the disease page anchored to the MeSH term ‘Diabetes Mellitus, Type 2’ (MESH:D003924) with similar OMIM terms having been merged to the page (Figure 4a), such as ‘DIABETES MELLITUS, NONINSULIN-DEPENDENT’ (OMIM:125853) and ‘DIABETES MELLITUS, NONINSULIN-DEPENDENT, 1’ (OMIM:601283). All the OMIM synonyms have been merged to the MeSH synonyms for this disease, and are recognizable by their capitalization (Figure 4b). Another OMIM term was added as a leaf beneath this disease, as can be seen in the hierarchy paths displayed at the bottom of the page (Figure 4c); here, ‘DIABETES MELLITUS, INSULIN-RESISTANT, WITH ACANTHOSIS NIGRICANS’ (OMIM:610549) is a leaf to both ‘Diabetes Mellitus, Type 2’and ‘Acanthosis Nigricans’ in MEDIC. CTD-curated data for type 2 diabetes can be found under the appropriate data-tabs at the top (Figure 4d).
Figure 4.
CTD's disease page for type 2 diabetes. (a) The disease page is anchored to the MeSH term ‘Diabetes Mellitus, Type 2’ (MESH: D003924). Equivalent OMIM diseases are merged to the MeSH page in MEDIC. All accession IDs are hyperlinked to their respective databases. (b) Merged OMIM terms and synonyms are easily recognizable by their capitalization. (c) OMIM terms can be leaf nodes beneath MeSH terms, and users can see the hierarchy in which the terms fall by following the Paths. (d) CTD-curated data for type 2 diabetes can be seen by clicking on the appropriate data-tabs.
The hierarchical nature of MEDIC (provided by the MeSH backbone) allows users the flexibility to navigate up and down the vocabulary to explore and discover chemicals and genes annotated to those diseases both by CTD biocurators and the automatic incorporation of OMIM genetic data (Figure 5). Thus, users looking for all genes related to type 2 diabetes would also find data for ‘DIABETES MELLITUS, INSULIN-RESISTANT, WITH ACANTHOSIS NIGRICANS’ listed as a disease leaf (Figure 5). If a user wanted to take a more broad view, they can navigate to a parent term (e.g. ‘Glucose Metabolism Disorders’) to see even more associated data. This ability to navigate to more generic levels should facilitate meta-analyses about chemical–gene–disease networks for broad concepts, such as ‘Neurodegenerative Diseases’ (MESH:D019636) or ‘Autoimmune Diseases’ (MESH:D001327).
Figure 5.
Navigating MEDIC and its curated data. A bird's eye view of a section of MEDIC provides users with the ability to navigate and explore disease terms, relationships, and their associated CTD data. The disease ‘DIABETES MELLITUS, INSULIN-RESISTANT, WITH ACANTHOSIS NIGRICANS’ (OMIM:610549) is a leaf of ‘Diabetes Mellitus, Type 2’ (MESH:D003924) and ‘Acanthosis Nigricans’ (MESH:D000052). Chemicals and genes annotated to each MEDIC term are cumulated as the user navigates up to more broad concepts.
Using MEDIC for DiseaseComps
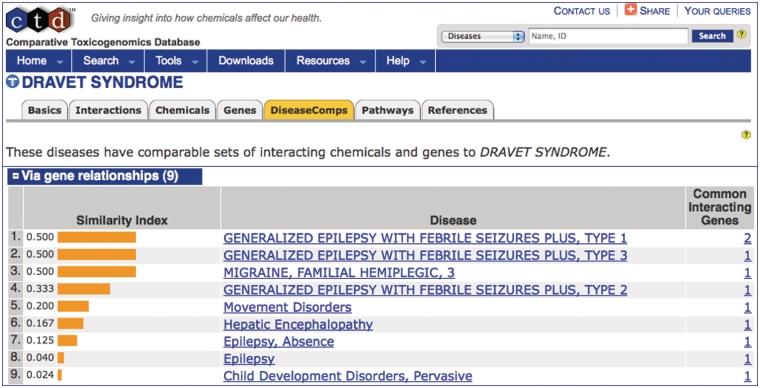
CTD provides unique metrics called GeneComps and ChemComps that find comparable genes and chemicals, respectively, based upon their shared toxicogenomics interactions and calculates a similarity index following the statistical method of the Jaccard score (16). We recently introduced DiseaseComps that now identify and rank similar diseases based upon their common molecular profiles as well (17). The use of MEDIC as a disease vocabulary for DiseaseComps helps provide insight to unfamiliar disorders. For example, the term ‘DRAVET SYNDROME’ (OMIM:607208) offers little insight to exactly what the disease is. However, DiseaseComps automatically finds similar diseases that share the same affected genes in ‘DRAVET SYNDROME’ (Figure 6). DiseaseComps ranks a mixture of both MeSH and OMIM terms (recognizable by its capitalization) based upon their similarity index to provide additional insight about ‘DRAVET SYNDROME’; here, it is seen that the disease shares genes with disorders involving epilepsy, migraines and hepatic encephalopathy.
Figure 6.
DiseaseComps use MEDIC. The DiseaseComps tab (orange) ranks diseases similar to ‘DRAVET SYNDROME’ based upon shared genes. DiseaseComps, which employs MEDIC as its disease vocabulary, ranks a mixture of both MeSH and OMIM terms (recognizable by its capitalization) based upon their similarity index. ‘DRAVET SYNDROME’ is discovered to share genes with epilepsy, migraines and hepatic encephalopathy.
Future directions
MEDIC is a practical disease vocabulary implemented at CTD. We envision it as an interim solution until a true ontology is developed; however, with rigorous work, it is possible that MEDIC itself could expedite development of a robust ontology by providing a foundation of disease terms and relationships on which to build. We will continue using MEDIC until a better resource is presented.
In early 2012, CTD will greatly expand its curated content, in part, as a result of a collaborative project that involved the curation within 10 months of over 50 000 toxicology publications selected for four disease areas (cardiovascular, renal, hepatic and neurological disorders). This project successfully used MEDIC as its annotation source, and resulted in curating more than 5300 chemicals and 6400 genes to over 2700 disease terms from MEDIC.
MEDIC currently contains 9700 unique disease terms (and 57 000 synonyms). To group similar diseases and make it easier to view associated annotations, we are developing a MEDIC-Slim vocabulary that will contain between 25 and 35 high-level terms. MEDIC-Slim can be used to help cluster similar diseases, which will aid visualization strategies at CTD.
Summary
CTD's merged disease vocabulary MEDIC provides a practical solution to a need not yet sufficiently fulfilled by the scientific community. It merges and combines the best of two disease sources: the freely available genetic data and disease description of OMIM combined with the hierarchical structure of MeSH. We acknowledge that this artifact is neither an ontology nor a perfect solution. Nonetheless, our initiative has been well received by other groups as a useful compromise. As with many other databases, we eagerly await a more robust, stable, mature, and maintained disease ontology. In the interim, we invite others to explore and use CTD's MEDIC as either a potential solution or a scaffold on which to build.
To date, MEDIC has been successfully implemented at CTD in curating more than 28,000 disease interactions describing the relationship between 2700 chemicals and 5400 genes to over 4600 disease terms. The hierarchical structure of MEDIC allows users to explore associated CTD data at different levels for meta-analysis.
MEDIC is freely available, and can be viewed and navigated on the web (with all of its associated CTD curated content) at: http://ctd.mdibl.org/voc.go?type=disease. MEDIC is updated monthly, and the most recent version can be downloaded in CSV, TSV, XML or OBO format from: http://ctd.mdibl.org/downloads/#alldiseases. We ask external databases that use MEDIC to cite CTD as a source and provide a direct link from their disease page to CTD's disease page, to promote global data integration.
Citing and linking to CTD
To cite CTD, please see: http://ctd.mdibl.org/about/publications/#citing. Currently, over 26 external databases link to or present CTD data on their own websites. If you are interested in establishing links to CTD data, please notify us (http://ctd.mdibl.org/help/contact.go) and follow these instructions: http://ctd.mdibl.org/help/linking.jsp.
Funding
National Institute of Environmental Health Sciences and the National Library of Medicine (R01ES014065 and R01ES014065-04S1); the National Center for Research Resources (grant number P20RR016463). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: NIEHS and NLM grants (R01ES014065 and R01ES014065-0451).
Conflict of interest. None declared.
Acknowledgements
We thank Ben King and Roy McMorran for continual CTD refinement, improvement and maintenance. We are also indebted to our dedicated team of professional biocurators for the implementation of MEDIC in their curation: Drs Cynthia Murphy, Cynthia Saraceni-Richards, Susan Mockus, Robin Johnson, Heather Keating, Jean Lay, Kelley Lennon-Hopkins and Daniela Sciaky.
References
- 1.Davis AP, King BL, Mockus S, et al. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. 2011;39:D1067–D1072. doi: 10.1093/nar/gkq813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gohlke JM, Thomas R, Zhang Y, et al. Genetic and environmental pathways to complex diseases. BMC Syst. Biol. 2009;3:46. doi: 10.1186/1752-0509-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis AP, Murphy CG, Saraceni-Richards CA, et al. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 2009;37:D786–D792. doi: 10.1093/nar/gkn580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis AP, Murphy CG, Rosenstein MC, et al. The Comparative Toxicogenomics Database facilitates identification and understanding of chemical-gene-disease associations: arsenic as a case study. BMC Med. Genomics. 2008;1:48. doi: 10.1186/1755-8794-1-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis AP, Wiegers TC, Murphy CG, et al. The curation paradigm and application tool used for manual curation of the scientific literature at the Comparative Toxicogenomics Database. Database. 2011 doi: 10.1093/database/bar034. 20 September 2011 [Epub ahead of print; doi:10.1093/database/bar034] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coletti MH, Bleich HL. Medical subject headings used to search the biomedical literature. J. Am. Med. Inform. Assoc. 2001;8:317–323. doi: 10.1136/jamia.2001.0080317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osborne JD, Flatow J, Holko M, et al. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl. 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 12.Shimoyama M, Smith JR, Hayman T, et al. RGD: a comparative genomics platform. Hum. Genomics. 2011;5:124–129. doi: 10.1186/1479-7364-5-2-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blake JA, Bult CJ, Kadin JA, et al. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson SJ, Johnston D, Humphreys BL. Relationships in medical subject headings. In: Bean CA, Green R, editors. Relationships in the Organization of Knowledge. New York: Kluwer Academic Publishers; 2001. pp. 171–184. [Google Scholar]
- 15.Cote RG, Jones P, Apweiler R, et al. The Ontology Lookup Service, a lightweight cross-platform tool for controlled vocabulary queries. BMC Bioinformatics. 2006;7:97. doi: 10.1186/1471-2105-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis AP, Murphy CG, Saraceni-Richards CA, et al. GeneComps and ChemComps: a new CTD metric to identify genes and chemicals with shared toxicogenomic profiles. Bioinformation. 2009;4:173–174. doi: 10.6026/97320630004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis AP, Rosenstein MC, Wiegers TC, et al. DiseaseComps: a metric that discovers similar diseases based upon common toxicogenomic profiles at CTD. Bioinformation. 2011;7:154–156. doi: 10.6026/97320630007154. [DOI] [PMC free article] [PubMed] [Google Scholar]