Abstract
As the deluge of genomic DNA sequence grows the fraction of protein sequences that have been manually curated falls. In turn, as the number of laboratories with the ability to sequence genomes in a high-throughput manner grows, the informatics capability of those labs to accurately identify and annotate all genes within a genome may often be lacking. These issues have led to fears about transitive annotation errors making sequence databases less reliable. During the lifetime of the Pfam protein families database a number of protein families have been built, which were later identified as composed solely of spurious open reading frames (ORFs) either on the opposite strand or in a different, overlapping reading frame with respect to the true protein-coding or non-coding RNA gene. These families were deleted and are no longer available in Pfam. However, we realized that these may perform a useful function to identify new spurious ORFs. We have collected these families together in AntiFam along with additional custom-made families of spurious ORFs. This resource currently contains 23 families that identified 1310 spurious proteins in UniProtKB and a further 4119 spurious proteins in a collection of metagenomic sequences. UniProt has adopted AntiFam as a part of the UniProtKB quality control process and will investigate these spurious proteins for exclusion.
Introduction
Currently, the UniProtKB protein sequence database contains >17 million protein sequences (1). This wealth of data is helping us to understand biology at an ever increasing rate. A large fraction of these sequences can be grouped into a few thousand common protein families. Proteins within these families often share common functions that can allow information experimentally gleaned on one protein to be transferred to uncharacterized ones. This process of transitive annotation is essential to make sense of the rapidly growing amount of sequence data. There are concerns about transitive annotation not being robust and thus leading to numerous annotation errors (2). Although this phenomenon does occur it seems clear that high-quality manual curation of the protein sequence databases, the careful use of databases of protein families for annotation and feedback from users of protein databases have largely kept the gross errors in check. For example, incorrect protein function assignments from large-scale genome projects in general have not been transferred to hundreds or thousands of other proteins as feared. On the other hand, subtler misannotations such as assigning an incorrect but related enzymatic activity to a protein (for example phosphorylating the wrong substrate) occur. Due to the lack of experimental work on most proteins, it is quite difficult to judge the prevalence of this subtle misannotation. A recent estimate for six large enzyme superfamilies studied suggested a range of 5–63% of incorrect annotations (3).
A further source of error in the sequence databases is the prediction of spurious genes (4). Automatic gene prediction methods in prokaryotes are increasingly accurate, Glimmer3, for example, both improves start site prediction relative to Glimmer2 and reduces the high false-positive rate for high GC genomes. (5). However, given the large number of proteins being deposited in the sequence databases, it is still likely that many thousands of the included sequences are either wholly spurious or improperly extended, past their true start sites. As the capacity to manually curate gene predictions diminishes, it is essential to create new methods to identify spurious gene predictions. It has been noted that certain alternate reading frames seem more likely to give rise to long spurious open reading frames (ORFs) (6). Normark et al. (7) found that frames +3 and −1 were most likely to give rise to long spurious ORFs. Although alternative overlapping reading frames are used in viral genomes, there are relatively few confirmed cases found in prokaryotes or eukaryotes.
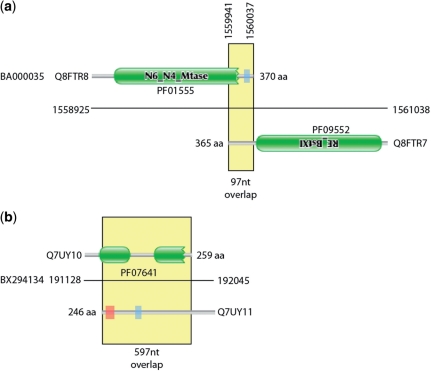
During the construction of the Pfam database of protein families (8), we have occasionally been alerted to the presence of families that were entirely composed of spuriously predicted ORFs. Once one gene has been spuriously predicted and put in the sequence database, there is a danger that future genome projects will annotate new protein-coding genes by similarity to the first spurious ORF. This can lead to entire families of spurious ORFs. In the worst-case scenario, these spurious families may even be annotated as having a function. This was the case pointed out by Tripp et al. (9) where a spuriously predicted gene on the opposite strand of ribosomal RNA had been given the incorrect function of cell wall hydrolase (PF10695). It may seem surprising that spuriously predicted ORFs would appear to have conservation like bona fide proteins. However, at the protein level the alignment of spurious ORFs can look like a normal protein alignment. In Figure 1, we show the multiple sequence alignment for former Pfam family PF10695, showing a protein-like conservation pattern. This conservation is actually due to the selective forces conserving the opposite strand rRNA sequence and structure. Once these errors are propagated to Pfam and other databases, then there is a danger that the error will be widely transferred and hence difficult to correct. Figure 2 shows contrasting examples of overlapping gene predictions. The first example (Figure 2a) shows a pair of proteins with correctly identified homology domains but with an uncharacteristically long tail-to-tail overlap. The second is an example of a hidden Markov model (HMM)-based domain definition identifying a region in a spurious gene call that overlaps a true gene (Figure 2b).
Figure 1.
Seed alignment for the AntiFam family derived from PF10695. Amino acids are colored by average similarity according to the BLOSUM62 amino acid substitution matrix from most similar (light blue) to less similar (gray). ‘S’ and ‘E’ in the first row stand for sequence start and sequence end, respectively. The final row features a consensus sequence. The alignment was displayed using the Belvu software (http://www.sanger.ac.uk/resources/software/seqtools/).
Figure 2.
Graphical representation of exemplar overlapping and spurious proteins. (a) shows two proteins from the Corynebacterium efficiens genome that encode components of a restriction system. The C-termini of the two proteins overlap by 97 nt. (b) Two highly overlapping predicted proteins from the Rhodopirellula baltica genome coded on opposite strands of DNA. The Q7UY10 protein contains two Pfam DUF1596 domains. There is no evidence that these are true expressed proteins. Green boxes represent regions matched by Pfam families, the red shaded areas represent transmembrane domains predicted by Phobius (10) and the blue shaded areas represent regions of low complexity (11).
AntiFam matches to predicted proteins in some cases will suggest that modifications to the extent of the coding region are needed rather than complete deletion of the protein from the sequence database. Most prfB genes, encoding the bacterial translation release factor 2, have a +1 programmed frameshift early in the coding region (12). The region downstream of the frameshift site is easily identified by gene finders, but unreconstructed extension 5′ to the frameshift results in translation of the wrong reading frame. AntiFam now includes model Spurious_ORF_21 to identify these improper treatments of the prfB gene.
Description of the resource
AntiFam is a freely available collection of multiple sequence alignments and profile HMMs. These models are designed to identify commonly recurring spuriously predicted ORFs. Some of the multiple sequence alignments used are taken from the Pfam database seed alignments for families identified as spurious ORFs. These alignments are kept as they appeared in the final release of Pfam before they were withdrawn (Table 1). Several additional custom families have been created to identify other commonly recurring spurious ORFs (Table 2). The profile HMMs have been constructed using the HMMER3 package with default parameters (13). The profile HMM library can be searched against any set of protein sequences using the ‘hmmsearch’ command. Due to the speed of the HMMER3 package, searching a sequence database such as UniProtKB will take a few minutes and searching a complete proteome will take seconds.
Table 1.
AntiFam entries derived from Pfam families
| Pfam accession number (identifier) | Last Pfam release present | Reason for deleting from Pfam | No. of matches in UniProt | No. of matches in metagenomics data seta |
|---|---|---|---|---|
| PF07612 (DUF1575) | 15.0 | Proteins may not be expressed. Evidence for homology to known protein on opposite strand | 3 | 0 |
| PF07616 (DUF1578) | 15.0 | Proteins may not be expressed. Evidence for homology to known protein on opposite strand | 6 | 6 |
| PF07630 (DUF1591) | 15.0 | Proteins may not be expressed. Evidence for homology to known protein on opposite strand | 6 | 0 |
| PF07633 (DUF1594) | 15.0 | Proteins may not be expressed. Evidence for homology to known protein on opposite strand | 5 | 0 |
| PF11370 (DUF3170) | 25.0 | Probable shadow ORF of Clp protease | 16 | 7 |
| PF11194 (DUF2825) | 25.0 | Probable CRISPRb repeat regions | 159 | 18 |
| PF11664 (DUF3264) | 25.0 | Probable CRISPR repeat regions | 21 | 13 |
| PF10695 (Cw-hydrolase) | 25.0 | Antisense to rRNA (9) | 225 | 1,654 |
| PF10919 (DUF2699) | 26.0 | Shadow ORF of PF00665 (integrase core domain 1) | 25 | 11 |
| PF07641 (DUF1596) | 26.0 | Dubious genome annotation. Family comprises only three sequences from Rhodopirellula baltica, two overlapping | 3 | 0 |
The final two columns show the number of matches of each AntiFam entry to UniProtKB and to a metagenomic data set.
aThe metagenomic set of sequences is the same as that used by Pfam (14).
bCRISPR, Clustered Regularly Interspaced Short Palindromic Repeats.
Table 2.
AntiFam entries derived from custom multiple sequence alignment
| Identifier | Type of spurious family | No. of matches in UniProt | No. of matches in metagenomics data seta |
|---|---|---|---|
| Spurious_ORF_10 | Translated bacterial tRNA, tRNA01 | 196 | 795 |
| Spurious_ORF_11 | Translated bacterial tRNA, tRNA02 | 89 | 170 |
| Spurious_ORF_12 | Translated bacterial tRNA, tRNA03 | 143 | 408 |
| Spurious_ORF_13 | Translated bacterial tRNA, tRNA04 | 77 | 671 |
| Spurious_ORF_14 | Translated bacterial tRNA, tRNA05 | 156 | 191 |
| Spurious_ORF_15 | Translated bacterial tRNA, tRNA06 | 31 | 63 |
| Spurious_ORF_16 | Translated bacterial tRNA, tRNA07 | 40 | 17 |
| Spurious_ORF_17 | Translated bacterial tRNA, tRNA08 | 5 | 10 |
| Spurious_ORF_18 | Translated bacterial tRNA, tRNA09 | 4 | 39 |
| Spurious_ORF_19 | Translated bacterial tRNA, tRNA10 | 7 | 12 |
| Spurious_ORF_20 | Translated bacterial tRNA, tRNA11 | 43 | 28 |
| Spurious_ORF_21 | PrfB frameshift | 24 | 5 |
| Spurious_ORF_22 | From a lncRNA, LINC00174 | 26 | 1 |
aThe metagenomic set of sequences is the same as that used by Pfam (14).
AntiFam is primarily a tool that is aimed at bioinformaticians to be used as part of genome annotation projects. Therefore, we have not implemented a standalone website for viewing entries in AntiFam. The AntiFam alignments and profile HMMs can be downloaded from the following URL: ftp://ftp.sanger.ac.uk/pub/databases/Pfam/AntiFam/
Of the 1310 proteins identified in UniProtKB as probably being spurious the large majority were from TrEMBL, the unreviewed part of UniProtKB. This means that no annotator had been involved in the creation of the entries. They had been automatically created from the records in the European Nucleotide Archive, GenBank or DNA Data Bank of Japan (DDBJ). These protein entries are in the process of being checked for removal from UniProtKB. One spurious protein found in the reviewed Swiss-Prot section of UniProtKB was Y114_CHLMU (Q9PLI5) that is an uncharacterized protein from Chlamydia muridarum. This belonged to the previously mentioned spurious Cw-hydrolase family and was removed in UniProt release 2011_10. An additional 13 spurious proteins in the reviewed portion of UniProtKB are also identified, of which 8 are due to non-coding RNA translations:
O67358.1 Aquifex aeolicus Trigger factor contains frameshift extension;
P19773.1 Mycobacterium tuberculosis protein matching DUF2699;
P47080.1 yeast protein YJL007C product of a dubious gene prediction;
P92540.1 Arabidopsis protein;
Q04100.1 yeast protein YDR445C product of dubious gene prediction and partly overlaps YDR444W;
Q52M62.3 human product of a dubious coding sequence (CDS) prediction. Probable non-coding RNA;
Q6ZQT7.1 human product of a dubious CDS prediction. Probable non-coding RNA;
Q6ZRM9.1 human product of a dubious CDS prediction. Probable non-coding RNA;
Q75L30.1 human product of a dubious CDS prediction. Probable non-coding RNA;
Q9CJR2.1 Pasteurella multocida tRNA-derived match;
Q9CMD0.1 P. multocida tRNA-derived match;
Q9CMX0.1 P. multocida tRNA-derived match; and
Q9CMZ6.1 P. multocida tRNA-derived match.
Identification of problematic Pfam families
In addition to the families reported by Pfam users, we tried to identify if further spurious families existed. The large majority of proteins in the TrEMBL portion of UniProtKB come from translations found in entries in the European Nucleotide Archive, GenBank or DDBJ. Thus, we scanned TrEMBL entries to identify UniProtKB entries that overlapped with each other in the nucleotide entry. We confined our scan to the prokaryotic entries because the nature of overlaps is relatively simple compared to the complex patterns of interlacing and nesting found in eukaryotic gene structures. The scan identified 73 853 proteins that were found to be overlapping. This list of proteins was then used to identify further Pfam families that contained numerous overlapping genes. We ordered the Pfam families by the fraction of overlapping proteins found within it. This list can be found in Supplementary Table S1. Using this measure means that large well-known families that are likely to have many overlaps by chance are not at the top of the list.
Future plans
The first release of AntiFam contains only a modest number of families. However, we see a number of ways to increase this in the future. The first of these is to increase the number of non-coding RNA-based families. We currently have only one ribosomal RNA-based family and we can add many further families. We can identify proteins related to ribosomal RNAs initially using tblastn, which compares a protein to a nucleotide sequence considering all six reading frames. In addition, we could also consider comparing a large database of RNA sequences to the protein sequence databases to identify further potentially spurious proteins. To date, we have only been able to investigate the Pfam families with the highest fraction of overlapping proteins. But in the coming months, we will investigate this list more thoroughly to identify if any further Pfam families should be deleted and added to AntiFam.
Conclusions
The first release of AntiFam contains 23 families derived from Pfam as well as a small number of non-coding RNAs that were erroneously translated into protein sequences. We expect that this number will grow in the future and we have several ideas to help us to achieve this. This should increase the power of AntiFam to reduce the number of spurious ORFs finding their way into the sequence databases. We hope that AntiFam will become an indispensible tool for quality control in metagenomic and genomic studies. We are particularly keen for biocurators and experimental biologists to remain vigilant and alert us to new cases of spurious ORFs so that we can add them to this resource.
Supplementary Data
Supplementary data are available at Database Online.
Funding
Wellcome Trust (grant number WT077044/Z/05/Z); National Human Genome Research Institute (grant number R01 HG004881). Funding for open access charge: Wellcome Trust (grant number WT077044/Z/05/Z).
Conflict of interest. None declared.
Acknowledgements
We are grateful to James Tripp from University of California Santa Cruz, who took the time to alert us to one of these spurious families.
References
- 1.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011 doi: 10.1093/database/bar009. bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner SE. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- 3.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bork P, Bairoch A. Go hunting in sequence databases but watch out for the traps. Trends Genet. 1996;12:425–427. doi: 10.1016/0168-9525(96)60040-7. [DOI] [PubMed] [Google Scholar]
- 5.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veloso F, Riadi G, Aliaga D, et al. Large-scale, multi-genome analysis of alternate open reading frames in bacteria and archaea. Omics. 2005;9:91–105. doi: 10.1089/omi.2005.9.91. [DOI] [PubMed] [Google Scholar]
- 7.Normark S, Bergstrom S, Edlund T, et al. Overlapping genes. Annu. Rev. Genet. 1983;17:499–525. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 8.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2011;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tripp HJ, Hewson I, Boyarsky S, et al. Misannotations of rRNA can now generate 90% false positive protein matches in metatranscriptomic studies. Nucleic Acids Res. 2011;39:8792–8802. doi: 10.1093/nar/gkr576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 12.Craigen WJ, Cook RG, Tate WP, Caskey CT. Bacterial peptide chain release factors: conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl Acad. Sci. USA. 1985;82:3616–3620. doi: 10.1073/pnas.82.11.3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 14.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]