Abstract
The breadth of information resources available to researchers on the Internet continues to expand, particularly in light of recently implemented data-sharing policies required by funding agencies. However, the nature of dense, multifaceted neuroscience data and the design of contemporary search engine systems makes efficient, reliable and relevant discovery of such information a significant challenge. This challenge is specifically pertinent for online databases, whose dynamic content is ‘hidden’ from search engines. The Neuroscience Information Framework (NIF; http://www.neuinfo.org) was funded by the NIH Blueprint for Neuroscience Research to address the problem of finding and utilizing neuroscience-relevant resources such as software tools, data sets, experimental animals and antibodies across the Internet. From the outset, NIF sought to provide an accounting of available resources, whereas developing technical solutions to finding, accessing and utilizing them. The curators therefore, are tasked with identifying and registering resources, examining data, writing configuration files to index and display data and keeping the contents current. In the initial phases of the project, all aspects of the registration and curation processes were manual. However, as the number of resources grew, manual curation became impractical. This report describes our experiences and successes with developing automated resource discovery and semiautomated type characterization with text-mining scripts that facilitate curation team efforts to discover, integrate and display new content. We also describe the DISCO framework, a suite of automated web services that significantly reduce manual curation efforts to periodically check for resource updates. Lastly, we discuss DOMEO, a semi-automated annotation tool that improves the discovery and curation of resources that are not necessarily website-based (i.e. reagents, software tools). Although the ultimate goal of automation was to reduce the workload of the curators, it has resulted in valuable analytic by-products that address accessibility, use and citation of resources that can now be shared with resource owners and the larger scientific community.
Database URL: http://neuinfo.org
Introduction
The Neuroscience Information Framework (NIF) is a rich and diverse system for discovering biological information of broad relevance to neuroscience. It was funded by the Blueprint for Neuroscience Research, a consortium of institutes that support neuroscience research at the National Institutes of Health, to improve the ability to find, access and utilize resources, defined here as data, tools, materials and services. Despite significant government investment and the availability of numerous on-line search engines, the biomedical research community remains largely unaware of resources created for their use. In addition, funding agencies need a system to provide an account of available resources to avoid duplication and identify areas in need. The necessity for new search strategies is particularly acute for online dynamic databases, the content of which is not well served by most web search engines for several reasons. One of the most important is that data in databases are typically served after the user fills in a set of forms that generate a dynamic web page, a function that search engines currently do not perform well. Although databases provide a very good set of tools for accessing, analyzing and manipulating data, even if the data are accessible to search engines, the meaning of their content may not be clear when out of the context of the database's site. For example, without a significant amount of experimental metadata, a string of numbers (i.e. ‘42’) found in a cryptic column titled ‘GExp’ may mean that a gene is expressed or not, because numerical data without context is utterly meaningless. The NIF addresses these issues by providing an overarching and practical framework for unified resource representation and access, designed to accommodate the diversity of current neuroscience resources.
The NIF has developed a search portal for information contained within distinct indices: the NIF Registry, the Data Federation and Literature. Through the NIF portal, each of these indices is simultaneously searched with an interface that organizes and represents the search results. The backbone of this substantial system is the NIF Registry, a semantically enhanced catalog of over 4500 biologically relevant web resources. The NIF Registry is a backbone of the NIF system because it is the entry point for any resource that is integrated with the NIF. The Data Federation is an extension of the registry, providing access to deep and continuously updated (see description of DISCO tools below) content of over 100 of those databases and data sets. The Data Federation organizes data according to domain-specific knowledge that the data represent, such as brain activation foci, nervous system connectivity and nervous system levels such as cellular or molecular levels. The literature databases are imported monthly from database dumps provided by the source journals (e.g. Journal of Visualized Experiments, JOVE), PubMed and PubMed Central.
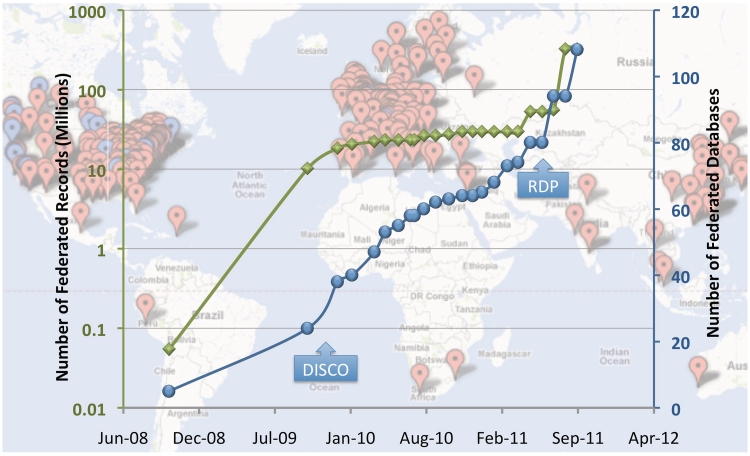
In the NIF Registry, each entry is manually curated according to a set of policies meticulously implemented and periodically reviewed (for a complete list, see http://confluence.crbs.ucsd.edu/display/NIF/Resources+and+Curation). Each resource is given a unique identifier in the form of a uniform resource identifier (URI) and a page within the NeuroLex, a semantic wiki also housing the NIF ontologies, where the resource can be aligned to additional ontological terms. Once represented in the NIF Registry, NIF curators collaborate with resource providers to completely expose their deep content via the NIF Data Federation, using a set of tools and services designed to work with most resource types as described by Gupta and colleagues (1). To our knowledge, the NIF Registry is the largest and most comprehensive catalog of biologically-relevant web resources available. This catalog is large because not only have the NIF curators worked efficiently, but also NIF has incorporated other existing registries such as the BioSiteMaps, who make their data publicly available, and implemented an automated resource discovery pipeline (RDP), described below. This data interchange between registries is greatly facilitated by the use of community ontologies that define a common set of descriptive terminologies [e.g. the Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC), the Biomedical Resource Ontology, Ontology for Biomedical Investigations and eagle-i]. NIF has worked with these communities often spearheading efforts to standardize resource representations so that these can be shared among catalogs. NIF Registry content is made available in several formats including RESTful web services and a queryable Resource Description Framework (RDF) graph. Figure 1 shows the current landscape of resources represented in the registry. Note that the NIF Registry has high level descriptions of over 4500 individual projects (databases, software tools, services; red and blue pointers on the map in the background of Figure 1) and of those, only 110 resources are registered with the Data Federation (blue pointers in Figure 1 background), which results in over 330 000 000 data records.
Figure 1.
NIF Resource Landscape. Background: each point on the map represents a global location that houses one or more resources registered with the NIF (via NeuroLex). Red points represent NIF registry entries and blue points represent databases and data sets incorporated into the data federation. Foreground: the blue line represents a plot of the number of federated data sources over time, and the green line represents the number of records in the NIF Data Federation over the same time (note these records come from only the blue dots and that the scale is logarithmic). The DISCO protocols and automated resource crawling were integrated into NIF system function in November 2009 and led to a growth of NIF holdings and in mid 2011, significant enhancements of the DISCO protocols allowing for enhanced automation of data ingestion, as well as the current resource discovery pipeline (RDP) were implemented.
Until recently, registration and integration into the NIF Registry and Data Federation was largely a manual process. Resources were identified by NIF curators through web and literature searches or through presentations at scientific conferences. A public nomination form also allows members of the community to recommend resources for inclusion. Despite the size and breadth of the NIF Registry, we recognized that the number of resources was much greater than could be identified through human input alone. Neuroscience is a broad and large field, with tentacles extending into all aspects of the life sciences, physical sciences and increasingly, social sciences. Thus, while the content is largely annotated and organized with respect to a neural focus, the resources themselves are drawn from all disciplines. From modest beginnings, e.g. Gardner et al. (2) and Gupta et al. (1), the NIF Registry and Data Federation have grown dramatically (Figure 1). As the NIF is built from independently maintained and developed resources, NIF also has had to grapple with how to make content understandable, and to keep content up-to-date and accurate. NIF curators and developers have delved deeply into the nature of available resources and content of experimental databases, identifying and overcoming technical barriers. In this report, we describe the development of automated resource discovery and curation techniques.
Methods and Results
The NIF curation process overview
The NIF project is maintained by two full-time curators and curatorial assistants who are responsible for identification, representation, updation and integration of resources within the NIF Registry and Data Federation. As mentioned in the introduction, the scope of NIF is neuroscience, but as neuroscience is broad, the types of resources included have expanded beyond those that are strictly neural in focus to include other relevant biomedical resources. In the registration pipeline, curators or resource owners are asked to provide a truly minimal set of information that represents the resource, mainly the name, uniform resource locator (URL), brief description, location (typically a university) and some keywords. The philosophy of the NIF is that automated tools do a much better job at providing detailed descriptions of resource content and keeping dynamic content up to date, but that human curation is necessary for accurate resource identification and consistent resource representation (a broad overview of this process is shown in Figure 2 and Table 1). Once identified, the NIF Data Federation and Resource update tools are employed to keep the content up to date. The steps in the curation process are shown in Table 1. Steps 1–3 are completed for all web resources represented in the public Registry. Steps 4–5 enable the inclusion of a database in the NIF Data Federation. Step 6 creates an enhanced index of literature that allows papers to be found based on information not necessarily found in the paper itself, e.g. the catalog number of an antibody reagent.
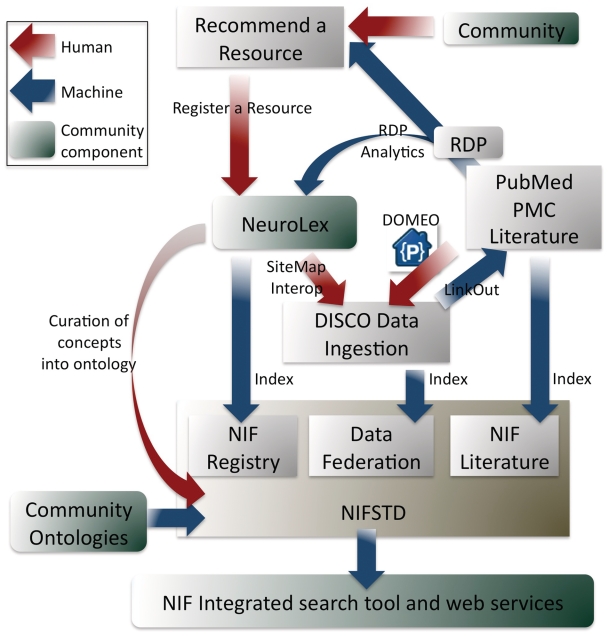
Figure 2.
A high level overview of the NIF system. This figure emphasizes where inputs and outputs of the NIF lie as a function of some of NIF's tools. Red arrows represent human steps, blue arrows represent automated steps and green boxes represent places in the system where community interactions are likely. The input of data is done using a suite of tools including NeuroLex (the first step for all data ingestion), DISCO (for deep data registration), LinkOut (linking data to PubMed, PMC PubMed Central literature), DOMEO (for literature annotation) and the RDP automated text-mining resource discovery pipeline that recognizes resources and recommends them to curators for possible inclusion in the NIF Registry. The creation of indices is informed by the ontology, as are the search tools and public web services. Note, all data moves through a process where it is recommended, registered to the NeuroLex, then included in the NIF Registry index and becomes available to DISCO tools for deeper content integration.
Table 1.
The NIF curation process
| NIF curation process | Details | Status | NIF system integration |
|---|---|---|---|
| (1) Recommend a resource | Is it relevant to neuroscience? yes—register with NeuroLex, receives wiki page and unique id | Semiautomated | NIF Registry—RDP |
| (2) Determine resource type | What type of resource is it? How is it accessible? What is its structure? Can users add data? Inputs? Outputs? | Semiautomated | NIF Registry—RDP |
| (3) Check periodically for resource validity and updates | Includes automated validity check of resource status (is the web site responding), and neuroscience content value (what is the neuroscience score from extracted text), as well as contacting resource providers to review their pages | Semiautomated | NIF Registry—RDP |
| (4) Write configuration files (Interop) | Display and index properly, integrate with existing resources (normalize), weights and keywords | Manual/curators or resource owners | NIF Data Federation—DISCO |
| (5) Check periodically for data validity and updates | Includes use of DISCO tool suite to automatically crawl database content, compare to the previous version and assign a date modified stamp, manual approval from curators of new data, and contacting resource providers to review their NIF representation | Semiautomated | NIF Data Federation—DISCO |
| (6) Annotate literature | Using the DOMEO tool, curators and community members can add data (e.g. unique identifiers of reagents or proteins) to literature that can be used to enhance search capabilities of NIF and other search engines. | Manual/curators or community | NIF Literature |
RDP: automated tools for resource identification and annotation
With the growth in numbers and the mercurial nature of web resources, it has become impractical for curators to manually identify, annotate and check resources regularly. Although we still employ web search engines and community input to identify potential new resources, the major source of potential resources remains the scientific literature. Thus, we have recently implemented text mining applications developed on top of the Textpresso text mining system (3) to help identify potential new resources from the literature, implement a rudimentary resource description, check for status and updates, determine if the resource is being cited elsewhere and suggest to curators when a resource may need review.
The automated suite consists of three pipelines: one main and two auxiliaries. The main pipeline performs monthly scans of the PubMedCentral (PMC) open access archive, which contained 358 561 articles as of October 2011. First, the tool detects all URLs within the full text of PMC papers (currently, the URL total is 281 960). Second, URLs are cleaned by removing duplicates and comparing them with URLs in the NIF registry (152 254 total URLs). Third, URLs are verified to ensure they link to active web pages and, if so, descriptive information is extracted from the homepage or the ‘About Us’ section. This text is then parsed with the NIF annotation web service, using the NIF standard ontology (NIFSTD) ontology, covering the broad domains of neuroscience (4) to annotate neuroscience-relevant terms and assign a rudimentary relevance score to each resource. Each term that matches an ontology concept is given a score of 1, and any term that does not match receives a 0. The sum over all terms provides a rough estimate for neuroscience-relevance and is used to suggest the resources inclusion into the NIF Registry and/or Data Federation. Furthermore, terms that match the resource ontology module of NIFSTD, such as ‘database’ or ‘software tool’ are annotated and extracted as suggestions for resource type. For further identification of resource type, the algorithm counts terms that are contained within the resource module of the NIFSTD ontologies (NIF Investigation; http://ontology.neuinfo.org/NIF/DigitalEntities/NIF-Investigation.owl). The more hits within a particular subset of the resource ontology, e.g. software, database branches, the stronger the likelihood that the resource type has been correctly identified. Finally, all URLs are divided into top-level domains such as ‘.org’ (∼5.7 K), ‘.edu’ (∼1.4 K), ‘.com’ (∼5.7 K) lists and each subset is ranked by the score. This last feature was important so that curators could focus their efforts in one domain, for instance the university-specific sites only.
Although this matching system is relatively simple, curators report that resources found with a score over 20 have a high degree (>70%) of validity for inclusion into NIF (Table 2). To date, more than 200 resources (a small fraction of the 4500 total) have been added to the NIF Registry based on suggestions from this automated pipeline. Of the resources identified by the pipeline, most ‘hits’ cover the software domain, with the software package R garnering the largest number of citations, but mature databases such as WormBase also gather hundreds of mentions. Additionally, as of October 2011, there are approximately 800 resources that have scored over 20 and are awaiting curation. As resources are curated, they are removed from the list on subsequent crawls, but new resources are also added so we do not anticipate a significant decrease to the backlog in the near future.
Table 2.
Example of evaluation of a potential resource found in PMC archive
| RDP actions | Example result |
|---|---|
| Use pattern matching to find in full text | URL: http://nbase.biology.gatech.edu |
| Use script to download the page and extract the page title | Name: NBase, Neisseria Meningitidis Online Database |
| Submit extracted text to public NIFSTD web service (tags indicate type of the annotated words) | Partial page content:
|
| Extract all nlx_res_tags and count | Possible Type for the entire page: Databases=3 Data=2 Funding=1 |
The web service discussed above is linked to and documented in NIF's developers’ section http://neuinfo.org/developers.
The two auxiliary pipelines consist of modules of the main pipeline, performed on a weekly basis. In order to determine if a resource has changed or been removed, the validity checker examines all web pages in the NIF Registry and creates a log of invalid URLs. NIF curators are made aware of potentially invalid URLs and how many weeks the page has been down in order to review the status of the resource. It is NIF's curation policy to maintain resource descriptions even when the resources themselves are no longer on line as a record of the resource, but the fact that these are invalid for a longer period is noted in the resource description. The second auxiliary process is used to monitor changes to the homepage of a resource. Using the NIF annotation web service described above, this pipeline compares the number of ontology terms found on the ‘Homepage’ or on the ‘About us’ page from week to week. A list of resources is updated and sorted by variability, i.e. the difference in ontology score from Week 1 to Week 2, alerting NIF curators that the resource has potentially received significant changes and should be reviewed. This is presented as a rank ordered list.
While the main pipeline is primarily for NIF curators, it can also be used to provide important metrics for online scientific resources. As the automated pipeline scans literature for URLs, names and other unique identifiers of resources, it is able to report where and when a resource has been cited, regardless of whether a literature citation to the resource was included, information currently not available to resource providers through existing scientific websites (i.e. Web of Science, PubMed). This information is particularly valuable to resource providers as it allows them to determine the utilization and impact of their resource. In the near future, we plan to add these tables to our DISCO database, and display these analytics on each resource's description page on the NIF NeuroLex wiki (described further below). We anticipate that providing additional citation information to web-based science projects will enhance our understanding of the use of online or software resources by members of the scientific community in a way that has not been accurately captured by traditional citation indices.
NIF data federation and DISCO interoperability tools
Once a resource has been discovered and annotated, the next step in the curation process is establishing integration and interoperability of the resource's content within the NIF Data Federation (1). The NIF Data Federation provides access to the deep content of databases via the NIF search portal. The current NIF Federation searches over 330 million records, contained within 110 independent databases at the time of writing, though new databases are continually added (Figure 1). When multiple databases are identified that cover largely the same content, NIF defines a ‘horizontal view’ that integrates these different sources into a single virtual database. For example, the current NIF connectivity database comprises six independent data sets [namely Brain Architecture Management System (BAMS), Collations of Connectivity Data on the Macaque Brain (CoCoMac), BrainMaps, ConnectomeWiki, temporal-lobecom and the UCLA multimodal connectivity database] organized under a common view. This virtual database lists two connected brain regions per row, and supporting information such as projection strength, species, technique used to determine that the two are connected, reference(s) for each statement and the link back to the original database (see http://neuinfo.org/nif/nifgwt.html?query=%22Hippocampus%22&category=Data%20Type:Connectivity). Similarly, the AntibodyRegistry incorporates data from individual vendors like UCDavis' NeuroMab, antibody literature-linking databases such as the Journal of Comparative Neurology's database and several aggregators of mainly commercial antibodies such as BioCompare. NIF also provides a set of tools to promote interoperability among resources, to facilitate linking between resources such as databases and literature citations, or to make these resources available via other cataloging efforts, e.g. Biositemaps.
To facilitate data sharing between a variety of different resources on the Internet, we have implemented and further developed DISCO, an extensible web resource DISCOvery, registration and interoperation framework (5, 6). DISCO provides an XML-based format to describe different types of information to be harvested by automated aggregator systems such as NIF. The DISCO Framework provides a set of tools and automated services configured by resource providers that instruct the NIF system how and when to harvest information content. DISCO currently includes six types of information that can be exposed via the following DISCO services:
SiteMap: used to describe high-level information about a resource,
Terminology: a glossary of the terms used by a resource,
Interoperation: a logical description of how to access data provided by a resource for the purpose of interoperation with other resources,
Schema: used to describe the database schema of a resource,
LinkOut: used by a resource to create data links that extend Entrez NCBI's information about publications and data entries (e.g. neurons and genes) and
News: used by a resource to publicize special issues and activities.
To process the DISCO content, NIF has a specialized DISCO system capable of harvesting the data from resources implementing DISCO. In addition, we have recently developed a DISCO Dashboard to help track, manage and interoperate the shared content of these resources on NIF. Figure 3 shows an overview of DISCO and a portion of the DISCO Dashboard Web interface listing several NIF-registered resources and the various DISCO services utilized by each of those resources. NIF uses the data harvested by DISCO to support other technologies including ontological mappings, horizontal integration, and data update alerts to help develop an evolvable global scientific portal architecture in support of neuroscience research.
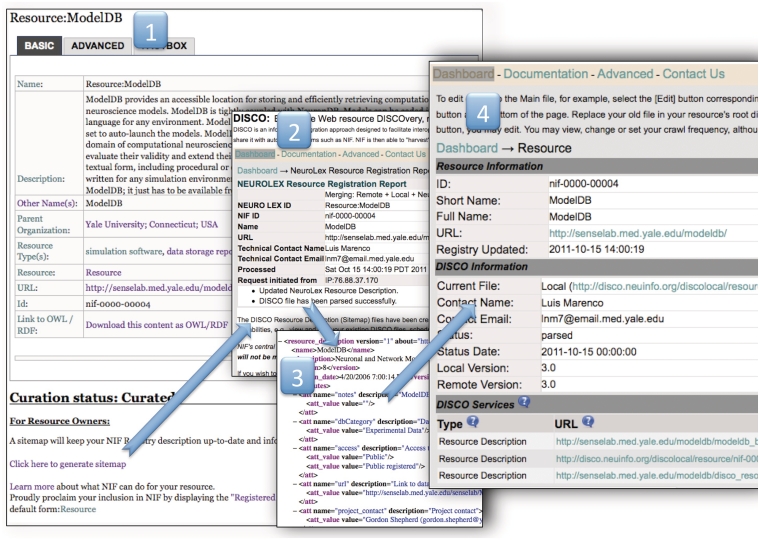
Figure 3.
The NIF Registration Pipeline. The NIF registration pipeline starts at a wiki page for each resource (i). This step shows an example public wiki page for the ModelDB resource. Anyone can nominate a resource, the curators will standardize the entry, the resource owner can change the description by simply hitting the edit button and adding information to the form and the owner can sign up to watch the page so that when any changes are made, he/she is notified. When the description is adequate, the curators will change the curation status to ‘curated’ and the ‘click here to generate sitemap’ link becomes visible. This link activates the DISCO system to generate a sitemap file using the text from the stable version of the resource in the NeuroLex wiki (ii). The event tracking system is activated, generating an email to the resource-provider tracking group in NIF, and instructions prompt the user to download the DISCO interop file (iii) and place it into the root directory of the resource. When this is complete, the DISCO dashboard updates and a new page is generated for the resource (iv) that allows the curators or the resource owner to regenerate, or edit the files that were created, schedule a crawl frequency and add additional files allowing for deeper interoperation with NIF such as including data in the Data Federation.
The usage of DISCO has grown significantly over the past 3 years, with 642 resources (out of the total 4500 NIF Registry entries) hosting at least one of the DISCO files as of October 2011. For example, ModelDB, a database of computational models, currently employs four DISCO files hosted at the root directory of ModelDB. The sitemap DISCO file tells NIF and other ‘robots’ that there is a resource at the root URL with specific characteristics, enabling robots, spiders and crawlers to maintain a good description of the resource, similar to other site map files. The Interop file allows NIF to harvest some of the data from ModelDB according to the specification given by the ModelDB group. The LinkOut file creates a set of annotations to be ingested by PubMed, for use by the PubMed Link Out services. Rather than each resource provider implementing these services separately, the NIF LinkOut Broker submits these on behalf of the ModelDB database. Although in this example, the database owner controls the files, this is not strictly required as NIF staff can both write and host these files. In recent months, the DISCO team implemented a data life cycle tracking system, where each chunk of data is compared with a previous version, and NIF logs any changes including how many elements are present in a database, which ones have been updated, which have been added and which have been deleted. These enhancements, currently unpublished, will be described fully in a separate publication.
Integrated resource registration pipeline
In the past year, we have worked to smooth the resource registration pipeline and integrate it more fully with the NIF website. With the release of NIF 3.0 in the fall of 2010, we linked the resource registry with the NeuroLex wiki (http://neurolex.org), so that each resource entered into the NIF Registry received its own category page. The NeuroLex is built on the MediaWiki platform, extended significantly such that it can properly display ontological concepts and resources. To facilitate a consistent representation of the resource in the Wiki, we created a wiki form that is automatically displayed when someone categorizes a page as a type of resource. When the NIF curators or someone other than the resource owner adds a resource, the NIF curators contact the resource owners to validate the descriptive information in the registry description. During the past 2 years, over 800 resource owners have been contacted and over 500 have either approved the entries or sent changes back to NIF making this a successful effort and ensuring the quality of the NIF Registry. NIF focused initially on contacting databases and software tool providers that had active communities. Thus, this should not be taken to be a random statistical sample.
The use of wiki allows resources to be linked easily to the entities within the NIF ontologies, but also allows each resource owner to take ‘ownership’ of the wiki page that houses the full description of their resource. For example, resource owners can link their descriptions to their Twitter or RSS feeds. We added an automatic function to the Wiki page for resource owners to generate a DISCO sitemap from the wiki description with a single click, making the generation of these files relatively simple (Figure 3). However, the DISCO functions may only be utilized by a resource owner because they require access to the resource root directories. As NIF seeks to promote interoperability of resources in general, the DISCO protocol also recognizes other sitemap formats, e.g. Biositemaps, so that resource providers do not have to maintain multiple site map files.
Annotation of resources in publications
The automated pipelines and interoperability tools described above are designed to help reduce barriers between resource representations in different forms, e.g. in the Web versus the literature. However, in the course of text mining the biomedical literature for biomedical resources, it became apparent that while some resources can be found and characterized at least semiautomatically, many are not easily extracted accurately by text mining engines, particularly reagents and other materials. One such case, where text mining often fails is description of antibody reagents, which often lack information such as catalog numbers and the other descriptive components (protein recognized, target species, raised in species, clone id) are scattered through several sentences or paragraphs. For cases where the automated pipeline fails, we developed a plug-in to DOMEO, a semantic literature annotation tool described below that facilitates the identification of resources inside of published papers by curators and subsequent export of these data for use in the NIF system.
As the test case for developing this plug-in, we used the problem of identifying antibodies within neuroscience-relevant papers. Because of the anatomical, cellular, subcellular and molecular heterogeneity and complexity of the nervous system, neuroscientists rely heavily on antibodies for unraveling the spatial organization of signaling networks. As part of the NIF data federation, we created the AntibodyRegistry, a large database of over 890 000 individual antibody records, covering almost 80% of the commercial antibodies used in Journal of Comparative Neurology articles over the last 5 years. Each antibody is given a unique ID that can be used to track the antibody across different vendors or for identifying uniquely the many noncommercial antibodies used and shared by individual investigators. The AntibodyRegistry contains links between published articles with reagent-specific descriptors, including vendor catalog identifiers. Vendor-specific catalog information can then be accessed transparently on the Web via the PubMed LinkOut broker function, described above. The registry database is searchable from the NIF Portal and individual records can be added and edited by users through a public interface.
In order to populate the links between antibodies and publications, we implemented a text mining assisted (semiautomatic) curation process inside of the DOMEO tool, because manual curation of antibody information from publications is tedious, but in most cases, quite necessary. When catalog numbers are listed in articles and those catalog numbers match the AntibodyRegistry, text-mining software detects and records the match, aligned with a specific offset in the article's text. Curators then evaluate the results and confirm or reject the entries. When catalog numbers are not present, information such as the clone number and the supplier can help the curator selecting one or more possible registry candidates.
To appropriately identify references to antibodies within text, we have developed a special plug-in for the DOMEO tool (7) a web application for producing stand-off annotation of online documents and document fragments. DOMEO supports manual and semiautomatic annotation. The manual annotation process consists of users highlighting a span of text in an online HTML/XHTML/XML document and attaching an annotation body to it. This body can be unstructured—simple free text or structured—according to data or ontological models. The semiautomatic annotation process allows NIF to leverage literature-mining software for producing annotation and eventually collect user feedback. The annotation can be exported in RDF format according to the Annotation Ontology (AO) (8).
The DOMEO list of features can be extended through the definition of software plug-ins. The NIF antibody plug in allows visual annotation of the content of a document with an antibody taken form the AntibodyRegistry. More precisely, the tool allows curators to highlight a span of text in an online document, search the AntibodyRegistry for the right entry and link it to the selected document fragment. Also, it is possible to specify which methods have been applied, which organisms have been studied through annotation with the NIFSTD ontologies. A comment field for free-text may also be attached to the annotation item. The RDF export of the antibody annotation is used to augment the information provided by the NIF framework—for both antibodies and publications—as well as to automatically populate the PubMed entries in the LinkOut section.
Discussion
Unlike traditional library catalogs or collections, the Internet presents unique challenges to the organization and representation of information resources. The Web is a dynamic medium, with websites, databases and wikis changing dramatically from year to year, month to month, and even day to day. New resources emerge continually from sources around the world; stewardship and authorship are not fixed. Despite the availability of curators and community contributors, the fluidity of the medium can easily overwhelm most aggregation sites, particularly those created for science, which relies on comprehensive, up-to-date and accurate information. Accurate and comprehensive information is necessary not only for scientists who seek to exploit resources to enhance their research but to assure that resource providers and funding administrators have the appropriate information available for making decisions about resource development and support.
In this article, we present several approaches to aid curators in expanding, enhancing and managing NIF’s information resources, chiefly the NIF Registry and the NIF Data Federation. As the largest and one of the most mature aggregation sites of its kind for neuroscience, NIF provides unique insights into the current resource landscape and its attendant curatorial challenges. Based on this experience, we can categorically state that despite the ready access and astounding capabilities of search engines like Google, identifying appropriate resources continues to be a challenge for most researchers.
There has been a significant investment in databases and tools for biological science, and frequent calls for more of them e.g. Akil et al. (9), but few calls to the biological community to adopt practices and frameworks for making their resources more easily discoverable. Resources are referenced in diverse sources, from web pages, databases, literature and personal conversations with colleagues and this makes for a haphazard mechanism for resource discovery. Although these mechanisms are effective for small communities, they are parochial for the totality of resources available, leading to fragmentation in the resource ecosystem. Thus, as we experienced when ingesting the self generated resource descriptions of the Biositemap project (10), a large number of the descriptions were incomplete and incomprehensible from the point of view of the NIF (see NIF blog at http://blog.neuinfo.org/index.php/essays/professional-vs-self-curation). The curation and resource representation strategy of the NIF utilizes a hybrid approach, relying on both automated tools, professional curators and community-based tools. Thus, the initial resource representation may be identified through our automated pipeline, the representation created by the NIF curators, and the final editing and approval performed by resource owner via the wiki. It has been our experience that when owners are provided with this base information, they are able to provide updates and corrections that are consistent with the NIF annotation standards, yet allows the resource owners to take ‘ownership’ of their resource descriptions. Through the implementation of the automated resource discovery and annotation pipeline, this process has been greatly streamlined. Via this pipeline, NIF is also providing information which resources are being used in various communities, although currently only for the open access literature, including articles deposited within PubMed Central. The question of whether this process will ever be fully automated or managed completely by the resource owners, at least in the near future, remains open. We believe, however, that as resource owners and funding agencies become more focused on resource discovery, practices may emerge that make this scenario more likely.
Resources retrieved by the automated discovery pipeline are now being used by NIF curators and are complementing the existing curation pipeline very well. In addition to suggesting resources to curators, information about those resources such as up-time and citation rates seem to be a very good fit for enhancing resource descriptions from a relatively static text-based description to a more dynamic and analytical view of the resource landscape.
Several other projects that rely exclusively on site-specific curators to add their data including eagle-i (11) and eBIRT (Biomedical Interactive Resource Tool; http://ebirt.emory.edu) have contributed significantly to describing resources in an interchangeable format. This interchange of information, among site-specific projects, and projects focusing on different communities such as NITRC (software), Biomedical Resource Ontology (BRO; web resources) and Biocatalogue (web services) although imperfect in many ways, are important steps towards fully describing the resource landscape, a goal that is too broad for any one community or project to undertake. Toward that end, we applaud the efforts of the international society for biocuration, and more specifically the BioDBCore and BioSharing projects, which appear to have involved some journal editors and publishers to bring resource representation to the forefront (12, 13). These efforts would be facilitated by maintaining a URI for each resource, so that resource registries can be easily integrated and so that identification of resources across different representations can be tracked. The NIF Resource Registry provides resource URI's via the NeuroLex wiki through a simple public registration form.
NIF also takes as its charge, resource promotion and interoperation, in addition to cataloging their existence. The NIF project has tried to implement solutions to reduce the current state of resource fragmentation to provide benefits not only to the researcher looking for appropriate data or tools but to the resource provider. Through our resource registration, curation and update tools, the NIF registry provides a quick and relatively painless method for resource providers to make their resource known and expose their data through the NIF portal. Through the NIF interoperability tools, resource providers can link their content and tools to other resources including the literature, alert the NIF and other aggregator sites when they make a change to their content, and monitor the impact of their resource through tracking literature citations and traffic through NIF. The wiki pages should also soon host access statistics from the NIF system; again tracking how often NIF users access the resource. It is likely that many more resource owners will use the NIF registry and related tools, if they can obtain useful information about the use and adoption of their resources. By porting the NIF Registry to a wiki-based platform and by providing a set of resource-tracking tools, we hope to make the NIF Registry a more vibrant system that supports resource providers in publicizing their work and facilitates resource discovery by users.
Funding
This work was supported by and has been funded in whole or in part through the NIH Blueprint for Neuroscience Research with Federal funds from the National Institute on Drug Abuse, National Institutes of Health, Department of Health and Human Services [Contract Number HHSN271200577531C]. P.W.S. is an Investigator with the Howard Hughes Medical Institute.
Conflict of interest. None declared.
Acknowledgements
We thank Mrs Andrea Stagg and many assistant curators for their hard work on the NIF Registry.
Appendix
Table A1.
List of abbreviations
References
- 1.Gupta A, Bug W, Marenco L, et al. Federated access to heterogeneous information resources in the Neuroscience Information Framework (NIF) Neuroinformatics. 2008;6:205–217. doi: 10.1007/s12021-008-9033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gardner D, Akil H, Ascoli GA, et al. The Neuroscience Information Framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6:149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller HM, Rangarajan A, Teal TK, et al. Textpresso for neuroscience: Searching the full text of thousands of neuroscience research papers. Neuroinformatics. 2008;6:195–204. doi: 10.1007/s12021-008-9031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bug WJ, Ascoli GA, Grethe JS, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6:175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marenco L, Wang R, Shepherd GM, et al. The NIF DISCO Framework: facilitating automated integration of neuroscience content on the web. Neuroinformatics. 2010;8:101–112. doi: 10.1007/s12021-010-9068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marenco L, Ascoli GA, Martone ME, et al. The NIF LinkOut broker: a web resource to facilitate federated data integration using NCBI identifiers. Neuroinformatics. 2010;6:219–227. doi: 10.1007/s12021-008-9025-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciccarese P, Ocana M, Clark T. DOMEO: a web-based tool for semantic annotation of online documents. Paper at Bio-Ontologies. 2011;2011 [Google Scholar]
- 8.Ciccarese P, Ocana M, Castro LJG, et al. An open annotation ontology for science on web 3.0. J. Biomed. Semantics. 2011;2(Suppl. 2):S4. doi: 10.1186/2041-1480-2-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akil H, Martone ME, Van Essen DC. Challenges and opportunities in mining neuroscience data. Science. 2011;331:708–712. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenenbaum JD, Whetzel PL, Anderson K, et al. The Biomedical Resource Ontology (BRO) to enable resource discovery in clinical and translational research. J. Biomed. Inform. 2011;44:137–145. doi: 10.1016/j.jbi.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torniai C, Brush M, Vasilevsky N, et al. Proceedings of the International Conference on Biomedical Ontology. Buffalo, NY: 2011. Developing an application ontology for biomedical resource annotation and retrieval: challenges and lessons learned. [Google Scholar]
- 12.Gaudet P, Bairoch A, Field D, et al. Towards BioDBcore: a community-defined information specification for biological databases. Database. 2011;2011 doi: 10.1093/database/baq027. baq027; doi:10.1093/database/baq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galperin MY, Fernández-Suárez XM. The 2012 Nucleic Acids Research Database Issue and the online Molecular Biology Database Collection. Nucleic Acids Res. 2012;40:D1–D8. doi: 10.1093/nar/gkr1196. [DOI] [PMC free article] [PubMed] [Google Scholar]