Abstract
When funding for Tetrahymena Genome Database (TGD) ended in 2006, no further updates were made to this important community resource and the main database was taken offline in 2008. We have restored and updated this important resource for use by the Tetrahymena research community. We have also retooled the TGD website (now TGD Wiki) to allow members of the community to directly update the information presented for each gene, including gene names, descriptions and Gene Ontology annotations, from a web browser. Maintenance of genome annotations by the authors generating and publishing primary data, rather than dedicated scientific curators, is a viable alternative for the upkeep of genomes, particularly for organisms with smaller research communities. By combining simple, intuitive displays with the powerful search functions made possible by its underlying relational database, TGD Wiki has been designed to maximize participation by bench scientists in the development of their community bioinformatics resource.
Database URL: http://ciliate.org
Introduction
The genome of the ciliated protozoan Tetrahymena thermophila was sequenced at The Institute for Genomic Research (TIGR) in 2003 (1). Over the years Tetrahymena has been a leading model organism for the study of telomeres, ribozymes, heterochromatin formation, transcription regulation, cilia and cellular motility, and a host of other basic biological processes (2). From 2004 to 2009, Tetrahymena Genome Database (TGD; http://www.ciliate.org) was housed at Stanford University, where it served as the primary portal to genomic information about this organism. TGD was modeled after Saccharomyces Genome Database (SGD), employing a nearly identical schema and interface to this successful model organism database (MOD) (3). TGD was actively curated until 2006, when funding for the project ended and the curation staff disbanded. The TGD website remained operational for three more years thanks to the efforts of the SGD staff, but despite 478 papers being published on Tetrahymena from 2007 to 2010, none of the gene function data contained in these papers were added to the database. Furthermore, sequence annotation continued at TIGR (now the J. Craig Venter Institute, JCVI) until 2008, producing a series of improved gene models that were never shown at TGD (4).
Because of the importance of TGD to the future of Tetrahymena research, and with renewed support through the NIH-funded Tetrahymena Stock Center (http://tetrahymena.vet.cornell.edu/), we have revived this resource in a user-editable format—a ‘wiki’. The advantages to this format as a means to maintain genomic data are evident and have been touted in a number of review articles (5, 6). Primarily, by empowering the authors of the research to annotate genes they study directly, it is possible for a wiki-based MOD to operate without a dedicated staff that interprets published findings and inputs these data into the database. This model of gene annotation has the potential to cut the operating costs of these expensive resources and place annotation in the hands of the parties most familiar with the genes in question.
In practice, however, community-maintained genome databases have not become the standard. For many organisms, the annotations are displayed in read-only format and sit unchanged following their initial assignment. In the case of model systems such as fly and yeast, the main databases are edited solely by the curation specialists employed by large MODs. The many nuances of gene annotation have led MODs to develop complex annotation strategies and interfaces, many of which are not intuitive and require a fair amount of training for new curators. Recent increases in the amount of data from large-scale experiments have complicated curation efforts as well, often requiring computer skills beyond those of many bench researchers in order to upload the data properly. Finally, the sheer number of annotations that must be made based on publications about yeast, fly, mouse and other well-studied model organisms warrant employing skilled curators to maintain these genomes. These challenges make it difficult to imagine larger MODs switching over to wiki-based annotations.
The Tetrahymena research community is of sufficient size and productivity that a means to update genome annotations is necessary. Though the exact number of people studying a particular organism is difficult to determine, the Tetrahymena community was estimated at 300 researchers in the 2002 genome sequencing white paper proposal to NHGRI (www.lifesci.ucsb.edu/~orias/ftp/Tetrahymena_White_Paper.doc). This number is supported by more recent data based on publication rates in Pubmed. The Saccharomyces research community was estimated in 2007 at 9447 (7), and a Pubmed search returns 16 493 papers published from 2007 to 2010 using the keyword ‘Saccharomyces’. Using the rate of 0.57 publications per community member over this span, the Tetrahymena community (478 publications) can be estimated at 272. By comparison, the Dictyostelium community has been estimated at ‘500 researchers in 90 laboratories worldwide’ (http://dictybase.org/StockCenter/AboutStockCenter.htm); using this metric yields 514 members (902 publications).
The work of the Tetrahymena research community produces a sizable amount of data that can be used to improve the annotation of the genome. Furthermore, unlike the highly studied genomes of Saccharomyces and Drosophila, relatively few Tetrahymena genes have been studied individually, and large-scale analyses are not yet common. Functional annotations of most Tetrahymena genes have therefore remained untouched by curators since their initial creation. At the moment about 17 000 of the 24 725 gene models are still labeled as ‘hypothetical proteins’. These genes would benefit from even the most rudimentary input by the community.
With these challenges and opportunities in mind, we have developed a simplified website and an easy curation interface that allows users to update the most vital information about each gene: gene names, descriptions, Gene Ontology annotations and Pubmed references. We have also included a field to display longer free-text annotations for each gene or family of genes, to capture information that does not fit easily into the other categories. By creating a user-friendly website and simplifying the annotation process, we hope to encourage contributions from bench researchers to keep the database updated and relevant. We believe this experiment in community annotation will be a model for newly established databases handling the genomes of other species.
Database construction
Unlike many of the existing biological wikis that are based on the popular Mediawiki platform, we have chosen to develop custom software for displaying, editing, and searching the data in TGD Wiki. Though Mediawiki offers contributors the ability to easily add annotations to genes or other items detailed by its web pages, the format of these annotations is often free-form. A lack of structure can complicate searching and standardization of the database contents. The use of a relational database to store specific types of annotations avoids this problem.
TGD began as a clone of the services provided by the SGD, which is designed around an Oracle relational database. TGD initially copied the database schema (http://www.yeastgenome.org/cgi-bin/viewSchema.pl) and website programming used by SGD in 2004 (the ‘bud’ schema), with minor customization to accommodate the Tetrahymena data. A similar strategy was used in the development of other successful MODs for organisms such as Dictyostelium (8), Candida (9) and Aspergillus (10). When hosting of the TGD database transitioned to its current home at Bradley University, the Oracle database management system (DBMS) was replaced with MySQL, an open-source DBMS. Prior to copying the data from the Oracle database to MySQL, we made several additional schema changes and called the new, migrated database ‘tgd’. With the new focus on community annotation, an additional database, ‘tgdAdmin’, was designed to hold user account information. The tgd and tgdAdmin databases together contain the annotation information and attributions shown on the TGD Wiki website.
The tgd schema is designed with a focus on the Feature table, which holds identifying information for the 24 725 protein-coding genes identified by JCVI in the most recent round of gene model annotation. Each of these features (genes) is linked to other tables containing reference information, GO terms, domain names, homologs, alternative gene names (aliases), DNA and protein sequences, and free-text comments. These tables can be searched separately for particular terms, as shown in Figure 1. Storage of data in specific tables also allows us to implement data constraints to prevent loading of improperly formatted data, either by individuals in the community or by web vandalism. For example, the constraints include limiting the primary gene descriptions to 240 characters, limiting gene names to three capital letters followed by a number (the standardized naming conventions for Tetrahymena genes) (11), and requiring properly formatted GO annotations. Guidelines for formatting annotations in TGD Wiki are available on the web at: http://ciliate.org/index.php/show/editguide.
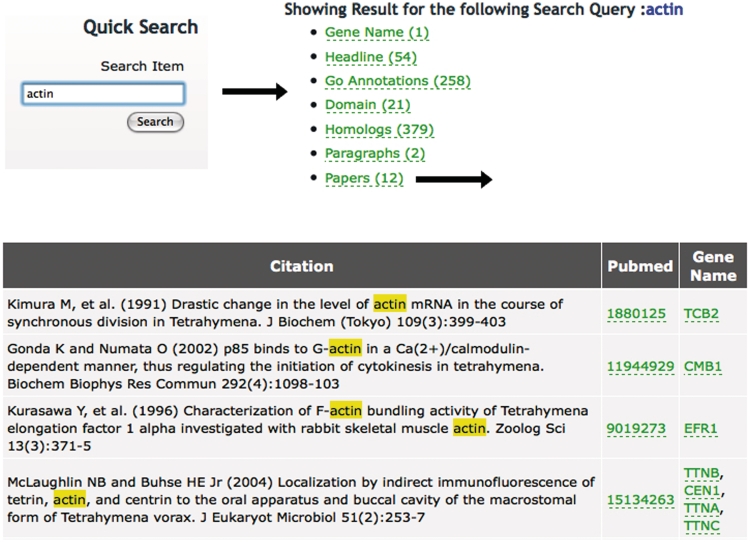
Figure 1.
Using the Quick Search tool at TGD Wiki. Entering a search term (‘actin’ in this example) returns the list of annotation types shown at TGD Wiki and the number of times the term appears in each category. Following the links to these annotation types shows the entry where this term is found and lists the genes it is used to annotate. In this case, 12 articles mention the word ‘actin’ in the title. Following the ‘Papers’ link shows their citation information, links to Pubmed and links to the Gene Pages of the specific genes mentioned in each paper.
Website displays
The TGD Wiki display and update interfaces were programmed in PHP with an eye toward future enhancements. Keeping the ability to set layout and formatting preferences simple has been a priority from the beginning of the project, and the style of the website can be changed easily by editing a template file. Not only does this allow the look of the TGD Wiki website to be updated very easily, it also simplifies the process of creating new genome wiki websites using the TGD Wiki software.
The overall format of TGD Wiki is similar to that of most other MODs. The front page, http://ciliate.org, is the main portal to the website and offers links to a database query tool (Quick Search), the genome browser and the BLAST server (Figure 2). The front page also shows messages to the community that can be added and updated by TGD Wiki staff using a web-based interface, and provides links to useful Tetrahymena-related resources. The left hand sidebar, which remains part of most web pages on the site, lists the latest Tetrahymena papers and most recent gene updates entered into the database.
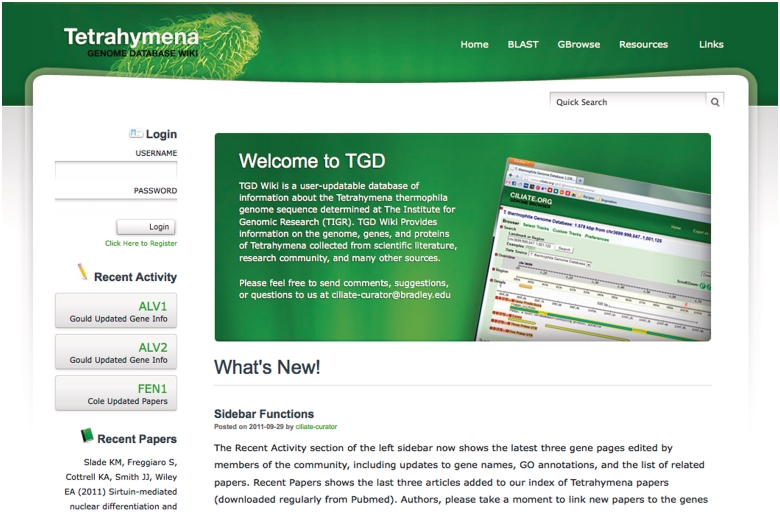
Figure 2.
TGD Wiki home page. The latest community annotations are highlighted in the Recent Activity section, with the three most recent Tetrahymena papers available in Pubmed shown below. The Quick Search tool, administrator posts, and editor login are also found on this page, as are links to BLAST, GBrowse and external web sites.
Navigating the website using Quick Search, the genome browser, or the BLAST server leads ultimately to the individual pages that show annotations for each gene. The ‘Gene Page’ presents annotations of various types in a standardized format (Figures 3 and 4); our software does not allow users to manipulate the outline of the web pages. Though this limits the extent to which community members can add new types of annotations to the page, it guarantees that each gene will be displayed in a uniform manner and ensures that all of the annotations shown are properly categorized for searching purposes. At this time, all information about a particular gene is shown on a single page that is divided into several sections. These sections are described below.
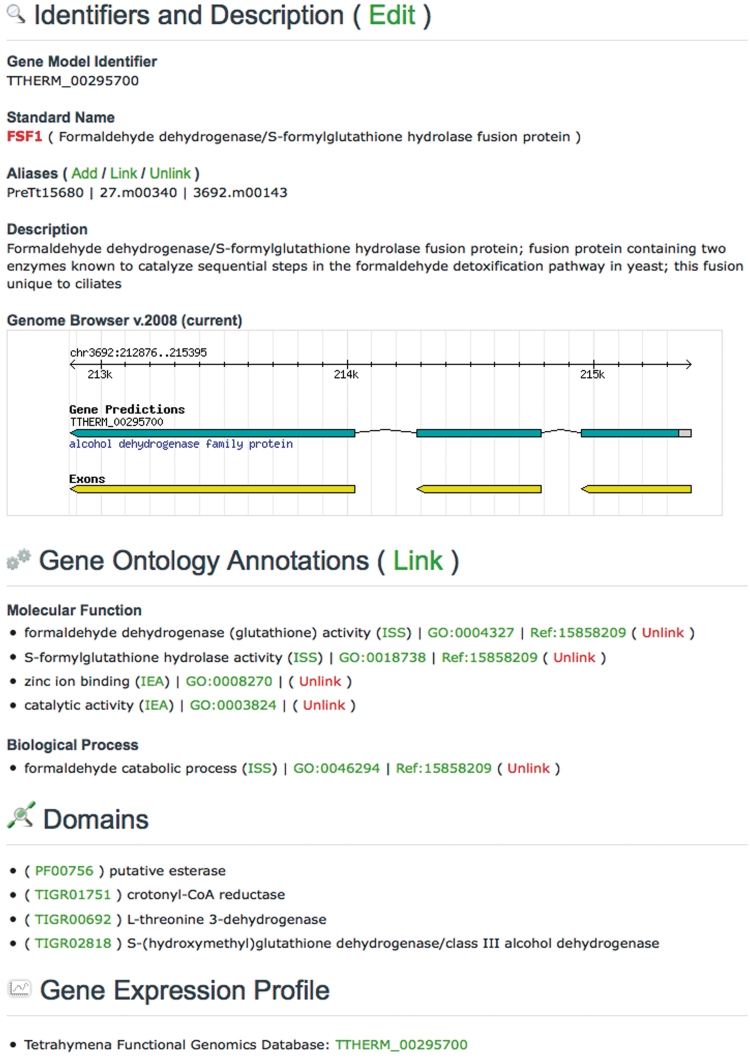
Figure 3.
An abbreviated TGD Wiki Gene Page showing the Edit options.
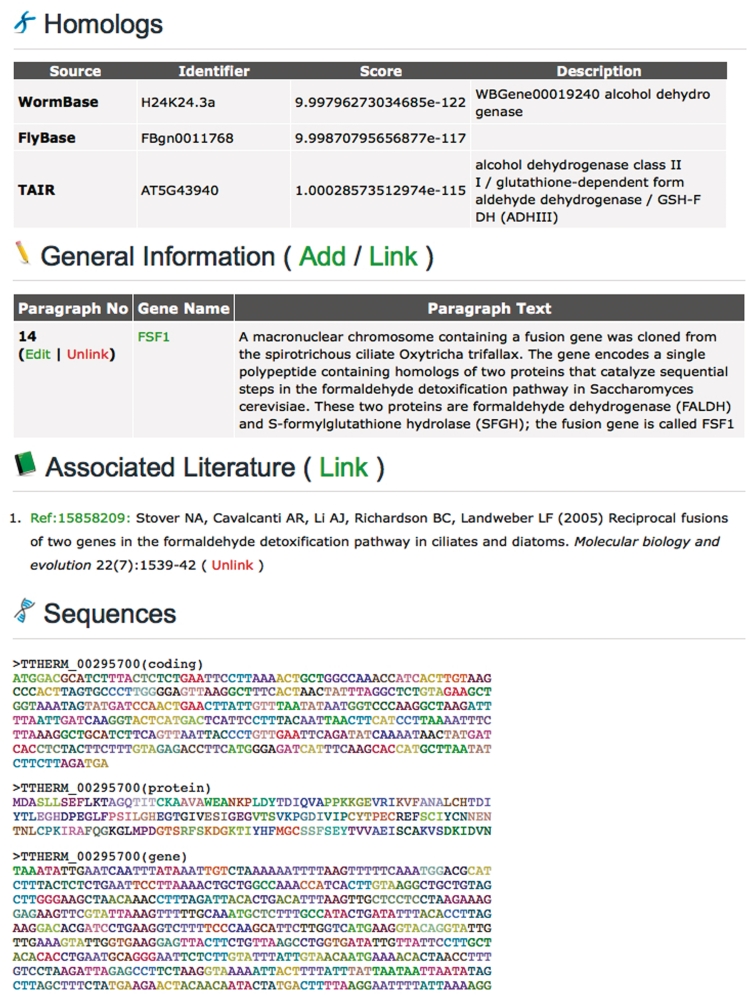
Figure 4.
An abbreviated TGD Wiki Gene Page showing the Edit options (continued).
Data types and sources
All 24 725 protein-coding genes in Tetrahymena are represented as features in the database, each with its own dedicated Gene Page. The sections of this page have been seeded with gene-specific data collected from a variety of sources. The Identifiers and Description section begins with the Gene Model Identifier (all of which begin with ‘TTHERM’), which was provided by JCVI during the final scheduled gene model annotation in 2008 (v.2008). This is followed by the Standard Name of the gene and a brief description of its three-letter acronym. Except for some legacy gene names, Standard Names follow the naming conventions detailed by the Ciliate Gene Nomenclature Consortium (11). A list of other names used to describe the gene is found in the Aliases section. The current gene models and their historical aliases are listed in a table on the website. Short summaries of gene functions were pulled from the JCVI v.2008 annotation and used to populate the Description section, then augmented with the summaries written by curators in 2006. Gene Names, Aliases and Descriptions are updatable columns in TGD Wiki, and a few of the initial entries have since been edited by the research community.
The Gene Ontology is a controlled vocabulary developed and used by MODs for the purpose of standardizing functional annotations among different organisms (12). Use of these standardized terms at TGD Wiki allows for easy comparisons between Tetrahymena and other GO annotated genomes. Because of their importance to dedicated Tetrahymena researchers and those from other communities, we provide users the ability to update the GO annotations for each gene. The update interface requires editors to enter the GO ID, Pubmed ID of the paper that supports the annotation, and the GO Evidence Code (13). The appropriate terms and links are then generated for display on the web page, eliminating the need for writing and formatting this information. Extensive guidelines on how to properly annotate genes with GO terms are available at the GO website (www.geneontology.org), and the TGD Wiki staff are committed to helping ciliate researchers understand the GO and use the terms for annotation.
The Domains listed for each gene were provided by JCVI and consist of both PFAM (14) and TIGRFAM (15) domains. The identifiers are hyperlinked to the PFAM and TIGRFAM database websites, respectively, where additional information about each domain can be viewed. The Homolog list for each Tetrahymena gene was generated by TGD based on the v.2006 gene model annotation by JCVI. Each Tetrahymena protein sequence was used as a BLASTP query against protein datasets from a variety of organisms of interest to Tetrahymena researchers (Table 1). The top BLAST hit in each of these organisms is shown on the Gene Page. The Gene Expression Profile section shows the conditions in which the gene is found to be expressed in large-scale experiments. This section currently contains links to only one database, Tetrahymena Functional Genomics Database (TetraFGD; formerly Tetrahymena Gene Expression Database) (16), which shows expression profiles of genes during times of growth, starvation and conjugation. The Gene Page will be updated to provide additional links in this section as other resources displaying large-scale expression data emerge.
Table 1.
Species selected for BLAST comparison with T. thermophila protein sequences
| Species | Database | Website |
|---|---|---|
| Arabidopsis thaliana | TAIR (23) | http://www.arabidopsis.org |
| Caenorhabditis elegans | WormBase (24) | http://www.wormbase.org |
| Cryptosporidium parvum | CryptoDB (25) | http://www.cryptodb.org |
| Dictyostelium discoideum | Dictybase (26) | http://www.dictybase.org |
| Drosophila melanogaster | FlyBase (27) | http://www.flybase.org |
| Homo sapiens | IPI (28) | http://www.ebi.ac.uk/IPI |
| Mus musculus | MGI (29) | http://www.informatics.jax.org |
| Paramecium tetraurelia | ParameciumDB (30) | http://paramecium.cgm.cnrs-gif.fr |
| Plasmodium falciparum | PlasmoDB (31) | http://www.plasmodb.org |
| Saccharomyces cerevisiae | SGD (3) | http://www.yeastgenome.org |
| Toxoplasma gondii | ToxoDB (32) | www.toxodb.org |
The Associated Literature section lists published papers that reference the gene. Citation information is downloaded from Pubmed regularly for papers that have the word ‘Tetrahymena’ in the title, abstract, or keywords. Once a paper is loaded into the database, it can then be linked to any gene using a simple interface that requires only the Pubmed ID of the paper to be entered. The appropriate citation, plus a hyperlink to the paper's record in Pubmed, is then displayed on the page. As with the GO Annotations, this simplifies the process of adding new information to the Gene Pages and reduces the chances of adding faulty or non-standard annotations.
The General Information section is designed for community members to write essays (capped at 5000 characters) about individual genes, gene families, or processes in Tetrahymena. Each paragraph can be linked to all appropriate genes. For example, there are 13 myosin homologs in Tetrahymena (17), and all of them have been linked to a single paragraph describing the entire family. We believe this section will also provide researchers a useful forum for listing any potential problems the gene models shown in the Sequences section may have.
The CDS and protein sequences are displayed at the bottom of each Gene Page and are color-coded by amino acid to help aid in domain identification. In order to avoid confusion, we have not opened the v.2008 CDS, protein and gene sequences shown on the Gene Page to editing by the community. These sequences are also displayed in the TGD Wiki genome browser and BLAST server, as well as at JCVI, GenBank and various other websites. Different research groups perform maintenance of the sequences at all of these resources, and having conflicting versions of gene sequences in the public domain is undesirable. As such we are not currently allowing modification of the actual gene models themselves, but we encourage members of the community to comment on any discrepancies they see, as this information may be useful to other researchers and during future rounds of annotation.
Additional tools: GBrowse and BLAST
The Quick Search tool, which is used for keyword searching through the MySQL database, displays hits in multiple different categories including Gene Name and Domains, and is equipped with an autocomplete feature that allows immediate access to gene pages. Two other portals to the gene annotations are available at TGD Wiki. The updated genome browser now presents the v.2008 Tetrahymena genome sequence and gene models using the GBrowse2 (18) software provided by the Generic MOD project (GMOD). Each graphic is hyperlinked to the web page or chromosomal region of the corresponding gene, and each Gene Page shows a detailed snapshot of the local area where the gene can be found. The BLAST server provides sequence searching of the v.2008 gene models, chromosome sequences, and Trace sequences using NCBI BLAST (19). Of particular importance, the BLAST server offers translated BLAST searching using non-standard genetic codes for both query and subject sequences. Two of the stop codons from the universal code, UAA and UAG, encode glutamine (Q) in Tetrahymena, as they do in many ciliates. Selecting the option ‘Ciliate Nuclear (6)’ allows these codons to be translated properly.
Future directions
The transition of Tetrahymena genome data from their original database and server to the new home of TGD Wiki has been completed. The original information curated by TGD staff has been preserved, and nearly all of the features used by the Tetrahymena research community are still available at the newly designed website. The sequences and large-scale annotations in the database have been updated to the most recent version produced by JCVI. Finally, the ability to update this information has been turned over to the Tetrahymena research community.
With these tasks completed we are now turning our attention to two goals: integrating new sources of data that will make the site more useful and convenient, and promoting use of the community annotation feature among Tetrahymena researchers. Several important large-scale datasets have been produced recently that will be incorporated into TGD Wiki. Foremost among these are the newly available T. thermophila micronuclear genome sequence and the sequences of three closely related Tetrahymena species. Longer-term goals for the project are to create Gene Pages for non-coding RNAs, to display genome comparisons with related species, and to output data to GFF3 and the standardized CHADO schema in order to more easily integrate new tools developed by contributors to GMOD.
TGD Wiki currently receives an average of 1600 visits per month according to Google Analytics, primarily for browsing annotations rather than contributing data. Since October 2010, when the wiki function became operational, we have registered 37 labs and received annotations from 7 labs to 139 genes (74 excluding edits made by TGD Wiki personnel). Though other groups hosting biological wikis have seen sluggish contribution rates as well (20–22), the fact that the sequences, references and tools on the website had not been updated for much of this time may have factored into these numbers. Now that most of the data on the website is current, we intend to step up our campaign to promote contributions. This will include posting a new round of announcements on the community mailing list (maintained by Jacek Gaertig at the University of Georgia), continuing to advertise the site at the biennial FASEB Ciliate Molecular Biology conference and other meetings, reporting new features and milestones on the website, and by contacting individual authors about new publications.
Funding
National Center for Research Resources at the National Institutes for Health (P40 RR019688) and by the Bradley University Special Emphasis Fund (13-31-085). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the NIH. Funding for open access charge: National Center for Research Resources at the National Institutes for Health (P40 RR019688).
Conflict of interest. None declared.
Acknowledgements
The authors would like to thank Mike Cherry, Stuart Miyasato, Gail Binkley and the staff at Saccharomyces Genome Database for maintaining TGD service during the transition to Bradley University. We would also like to thank Anudeep Singh, Alex Uskov, David Scuffham, the students of the 2007-2009 Bradley University Computer Science Capstone Project, the friendly staff at dictyBase, and the members of the Tetrahymena research community.
References
- 1.Eisen JA, Coyne RS, Wu M, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins K, Gorovsky MA. Tetrahymena thermophila. Curr. Biol. 2005;15:R317–R318. doi: 10.1016/j.cub.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 3.Nash R, Weng S, Hitz B, et al. Expanded protein information at SGD: new pages and proteome browser. Nucleic Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coyne RS, Thiagarajan M, Jones KM, et al. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics. 2008;9:562. doi: 10.1186/1471-2164-9-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzberg SL. Genome re-annotation: a wiki solution? Genome Biol. 2007;8:102. doi: 10.1186/gb-2007-8-1-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K. Gene-function wiki would let biologists pool worldwide resources. Nature. 2006;439:534. doi: 10.1038/439534a. [DOI] [PubMed] [Google Scholar]
- 7.Peña-Castillo L, Hughes TR. Why are there still over 1000 uncharacterized yeast genes? Genetics. 2007;176:7–14. doi: 10.1534/genetics.107.074468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kreppel L, Fey P, Gaudet P. dictyBase: a new Dictyostelium discoideum genome database. Nucleic Acids Res. 2004;32:D332–D333. doi: 10.1093/nar/gkh138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnaud MB, Costanzo MC, Skrzypek MS, et al. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 2005;33:D358–D363. doi: 10.1093/nar/gki003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnaud MB, Chibucos MC, Costanzo MC, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Seventh International Meeting on Ciliate Molecular Biology Genetics Nomenclature. Proposed genetic nomenclature rules for Tetrahymena thermophila, Paramecium primaurelia and Paramecium tetraurelia. Genetics. 1998;149:459–462. doi: 10.1093/genetics/149.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 2008;9:509–515. doi: 10.1038/nrg2363. [DOI] [PubMed] [Google Scholar]
- 14.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao W, Xiong J, Bowen J, et al. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams SA, Gavin RH. Myosin genes in Tetrahymena. Cell Motil. Cytoskeleton. 2005;61:237–243. doi: 10.1002/cm.20078. [DOI] [PubMed] [Google Scholar]
- 18.Stein LD, Mungall C, Shu S, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.McIntosh BK, Renfro DP, Knapp GS, et al. EcoliWiki: a wiki-based community resource for Escherichia coli. Nucleic Acids Res. 2011;10 doi: 10.1093/nar/gkr880. doi:10.1093/nar/gkr880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stehr H, Duarte JM, Lappe M, et al. PDBWiki: added value through community annotation of the Protein Data Bank. Database. 2010 doi: 10.1093/database/baq009. 2010, doi:10.1093/database/baq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch R, Welch L. If you build it, they might come. Nat. Rev. Microbiol. 2009;7:90–90. [Google Scholar]
- 23.Swarbreck D, Wilks C, Lamesch P, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris TW, Antoshechkin I, Bieri T, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heiges M, Wang H, Robinson E, et al. CryptoDB: a Cryptosporidium bioinformatics resource update. Nucleic Acids Res. 2006;34:D419–D422. doi: 10.1093/nar/gkj078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fey P, Gaudet P, Curk T, et al. dictyBase–a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tweedie S, Ashburner M, Falls K, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersey PJ, Duarte J, Williams A, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 29.Bult CJ, Kadin JA, Richardson JE, et al. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnaiz O, Cain S, Cohen J, Sperling L. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007;35:D439–D444. doi: 10.1093/nar/gkl777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aurrecoechea C, Brestelli J, Brunk BP, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gajria B, Bahl A, Brestelli J, et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–D556. doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]