Abstract
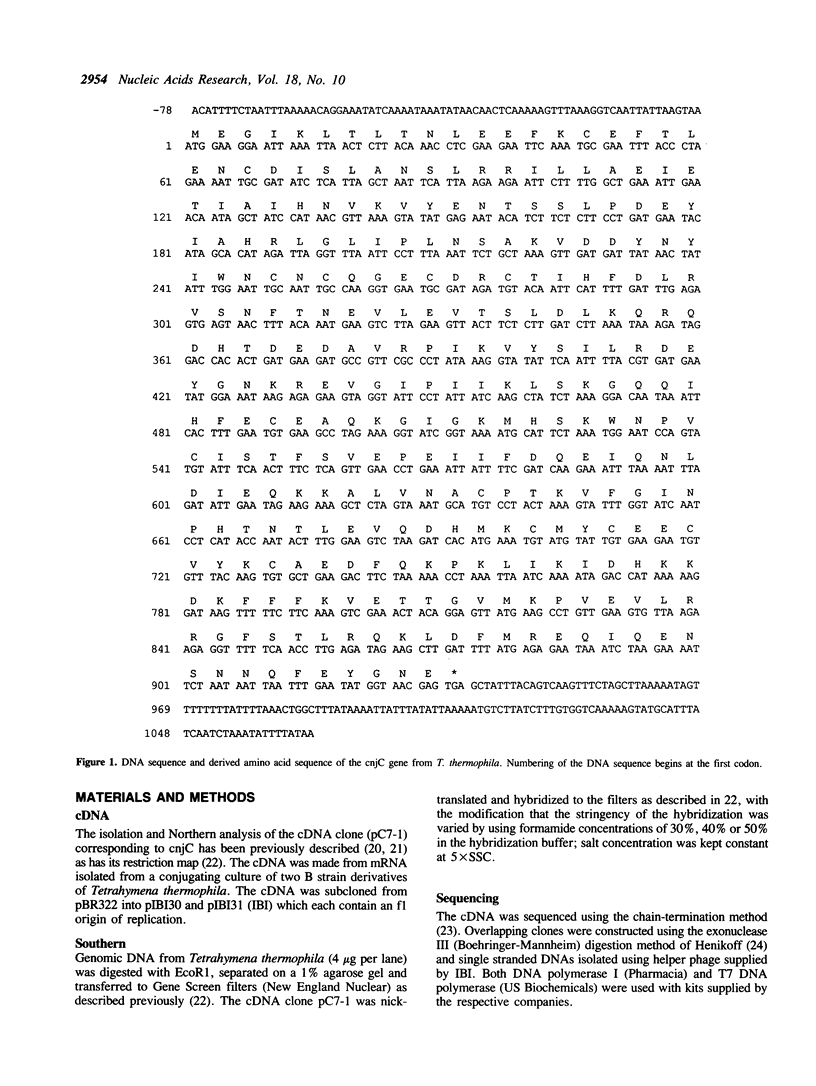
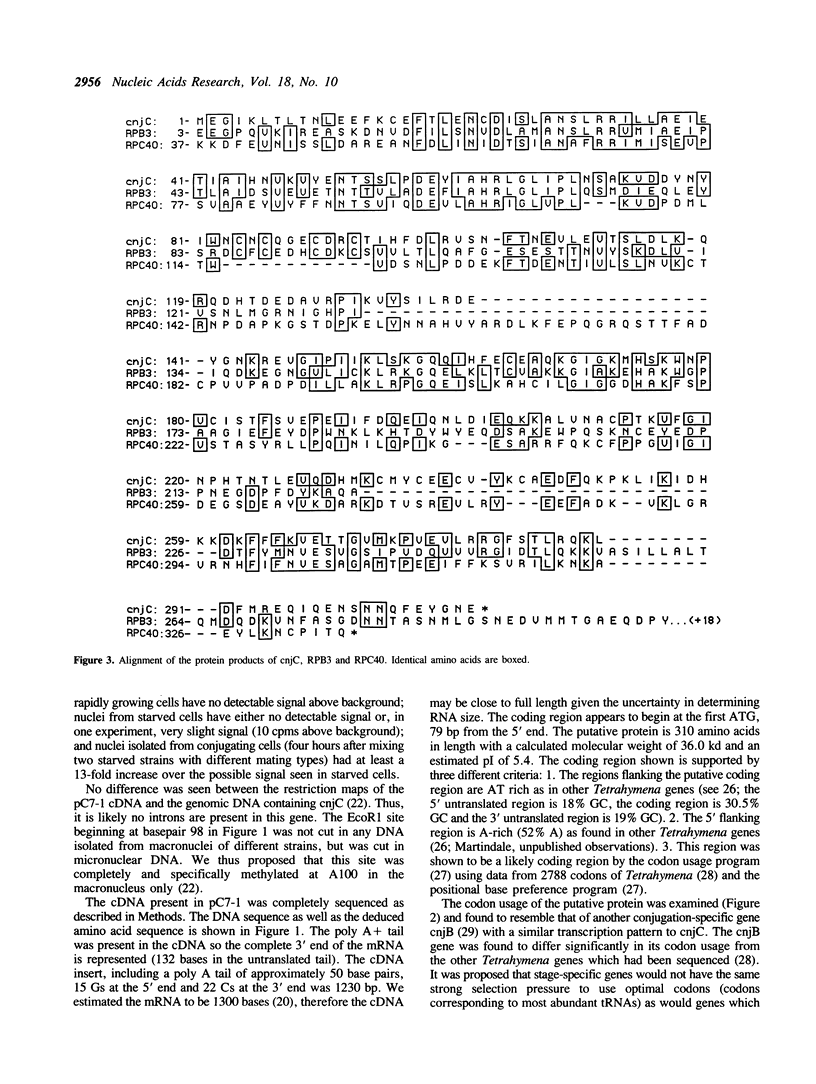
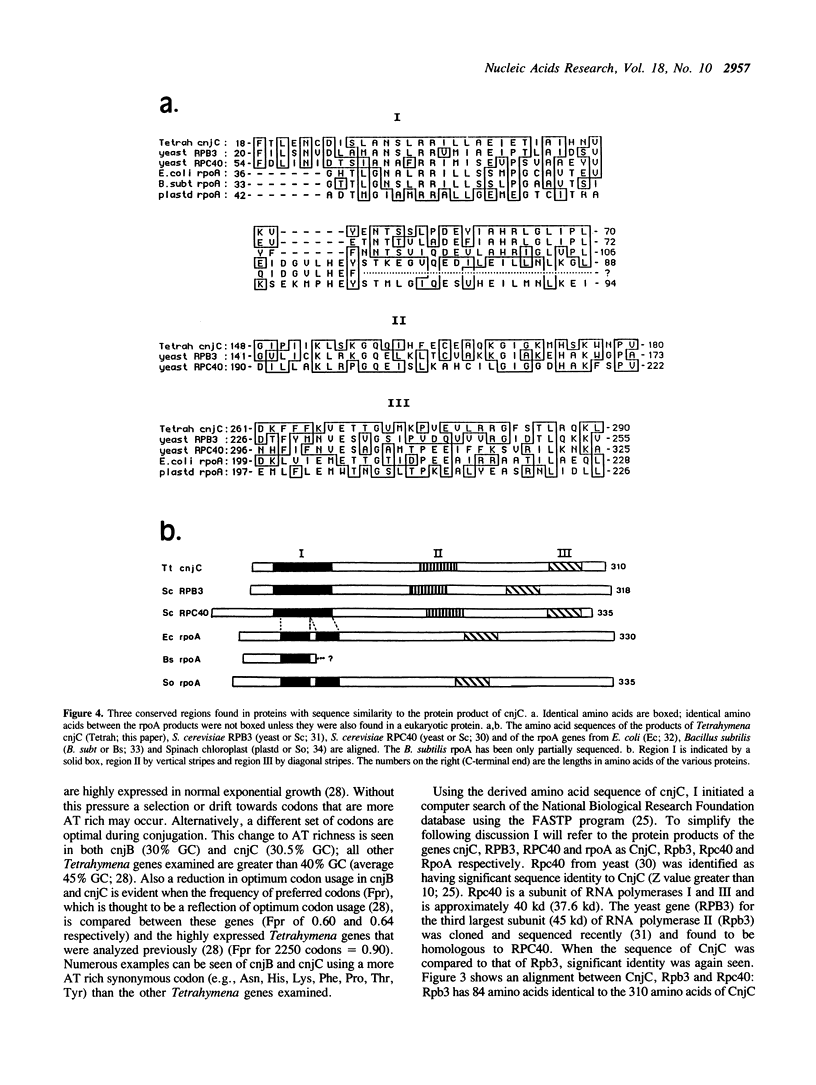
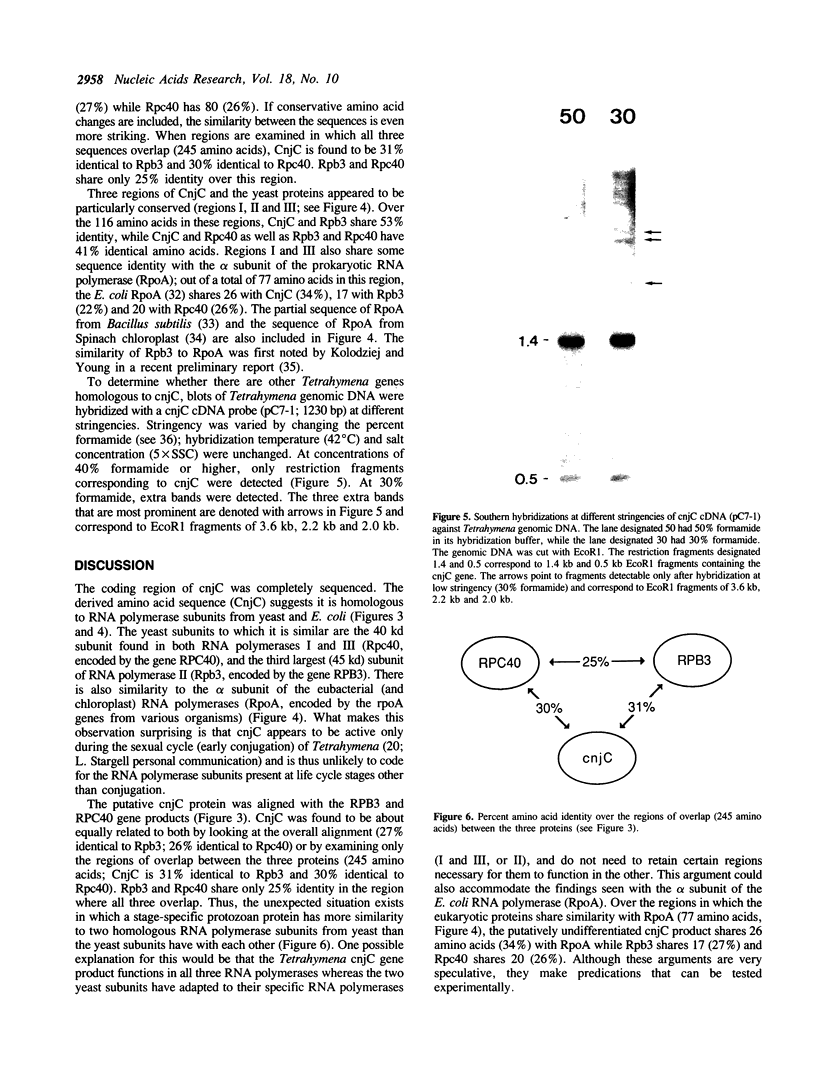
The cnjC gene from the protozoan Tetrahymena thermophila was completely sequenced. The deduced gene product was found to have significant sequence similarity to the yeast and prokaryotic RNA polymerase subunits involved with subunit assembly. Since cnjC is active only during the sexual stage (conjugation) of Tetrahymena's life cycle, these results indicate it may be part of a novel type of transcriptional control. The yeast proteins to which the Tetrahymena cnjC is homologous are the 40 kd protein of RNA polymerases I and III (coded for by gene RPC40) and the third-largest subunit of RNA polymerase II (coded for by gene RPB3). The degree of similarity of the cnjC protein to the two yeast subunits was found to be greater than the similarity of the two yeast subunits to each other. The alpha subunit of the core RNA polymerase from prokaryotes (coded for by gene rpoA) was found to have regions of similarity to the cnjC protein as well as to the subunits encoded by RPC40 and RPB3. Regions of high conservation among the four proteins are noted. The significance of these results is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Buhler J. M., Huet J., Davies K. E., Sentenac A., Fromageot P. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J Biol Chem. 1980 Oct 25;255(20):9949–9954. [PubMed] [Google Scholar]
- Burton Z. F., Ortolan L. G., Greenblatt J. Proteins that bind to RNA polymerase II are required for accurate initiation of transcription at the adenovirus 2 major late promoter. EMBO J. 1986 Nov;5(11):2923–2930. doi: 10.1002/j.1460-2075.1986.tb04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisek L. J., Corden J. L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989 Jun 29;339(6227):679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Faust D. M., Renkawitz-Pohl R., Falkenburg D., Gasch A., Bialojan S., Young R. A., Bautz E. K. Cloning and identification of the gene coding for the 140-kd subunit of Drosophila RNA polymerase II. EMBO J. 1986 Apr;5(4):741–746. doi: 10.1002/j.1460-2075.1986.tb04276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Bowen J. K., Bannon G. A., Gorovsky M. A. Unusual features of transcribed and translated regions of the histone H4 gene family of Tetrahymena thermophila. Nucleic Acids Res. 1987 Jan 12;15(1):141–160. doi: 10.1093/nar/15.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Riva M., Sentenac A., Fromageot P. Yeast RNA polymerase C and its subunits. Specific antibodies as structural and functional probes. J Biol Chem. 1985 Dec 5;260(28):15304–15310. [PubMed] [Google Scholar]
- Ishihama A. Promoter selectivity of prokaryotic RNA polymerases. Trends Genet. 1988 Oct;4(10):282–286. doi: 10.1016/0168-9525(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Kolodziej P., Young R. A. RNA polymerase II subunit RPB3 is an essential component of the mRNA transcription apparatus. Mol Cell Biol. 1989 Dec;9(12):5387–5394. doi: 10.1128/mcb.9.12.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. RNA and protein synthesis during meiotic prophase in Tetrahymena thermophila. J Protozool. 1985 Nov;32(4):644–649. doi: 10.1111/j.1550-7408.1985.tb03094.x. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Bruns P. J. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: identification of mRNA species present exclusively during meiosis. Mol Cell Biol. 1983 Oct;3(10):1857–1865. doi: 10.1128/mcb.3.10.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W. Codon usage in Tetrahymena and other ciliates. J Protozool. 1989 Jan-Feb;36(1):29–34. doi: 10.1111/j.1550-7408.1989.tb02679.x. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Martindale H. M., Bruns P. J. Tetrahymena conjugation-induced genes: structure and organization in macro- and micronuclei. Nucleic Acids Res. 1986 Feb 11;14(3):1341–1354. doi: 10.1093/nar/14.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D. W., Taylor F. M. Multiple introns in a conjugation-specific gene from Tetrahymena thermophila. Nucleic Acids Res. 1988 Mar 25;16(5):2189–2201. doi: 10.1093/nar/16.5.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D. W., Hayward R. S. Nucleotide sequence of the rpoA-rplQ DNA of Escherichia coli: a second regulatory binding site for protein S4? Nucleic Acids Res. 1984 Jul 25;12(14):5813–5821. doi: 10.1093/nar/12.14.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Sopta M., Burton Z. F., Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989 Oct 5;341(6241):410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Sugai T., Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in Tetrahymena pyriformis. J Protozool. 1974 Oct;21(4):542–548. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Suh J. W., Boylan S. A., Price C. W. Gene for the alpha subunit of Bacillus subtilis RNA polymerase maps in the ribosomal protein gene cluster. J Bacteriol. 1986 Oct;168(1):65–71. doi: 10.1128/jb.168.1.65-71.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilamitjana J., Baltz T., Baltz D., Barreau C. Probing eukaryotic RNA polymerases B with monoclonal antibodies. FEBS Lett. 1983 Jul 25;158(2):343–348. doi: 10.1016/0014-5793(83)80610-3. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Zheng X. M., Moncollin V., Egly J. M., Chambon P. A general transcription factor forms a stable complex with RNA polymerase B (II). Cell. 1987 Jul 31;50(3):361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]




