Abstract
A simple method is described for generating nested deletions from any fixed point in a cloned inset. Starting with a single-stranded phagemid template, T4 DNA polymerase is used to extend an annealed primer. This leads to a fully double-stranded circular molecule with a nick or small gap just 5' to the primer. Exonuclease III initiates progressive digestion from the resulting 3' end. Removal of timed aliquots and digestion with a single-strand specific endonuclease leads to a series of linear nested fragments having a common end corresponding to the 5' end of the primer. These molecules are circularized and used to transform cells, providing large numbers of deletion clones with targeted breakpoints. The 6-step procedure involves successive additions to tubes, beginning with a single-stranded template and ending with transformation; no extractions, precipitations or centrifugations are needed. Results are comparable to those obtained with standard Exonuclease III-generated deletion protocols, but there is no requirement for restriction endonuclease digestion or for highly purified double-stranded DNA starting material. This procedure provides a strategy for obtaining nested deletions in either direction both for DNA sequencing and for functional analysis.
Full text
PDF
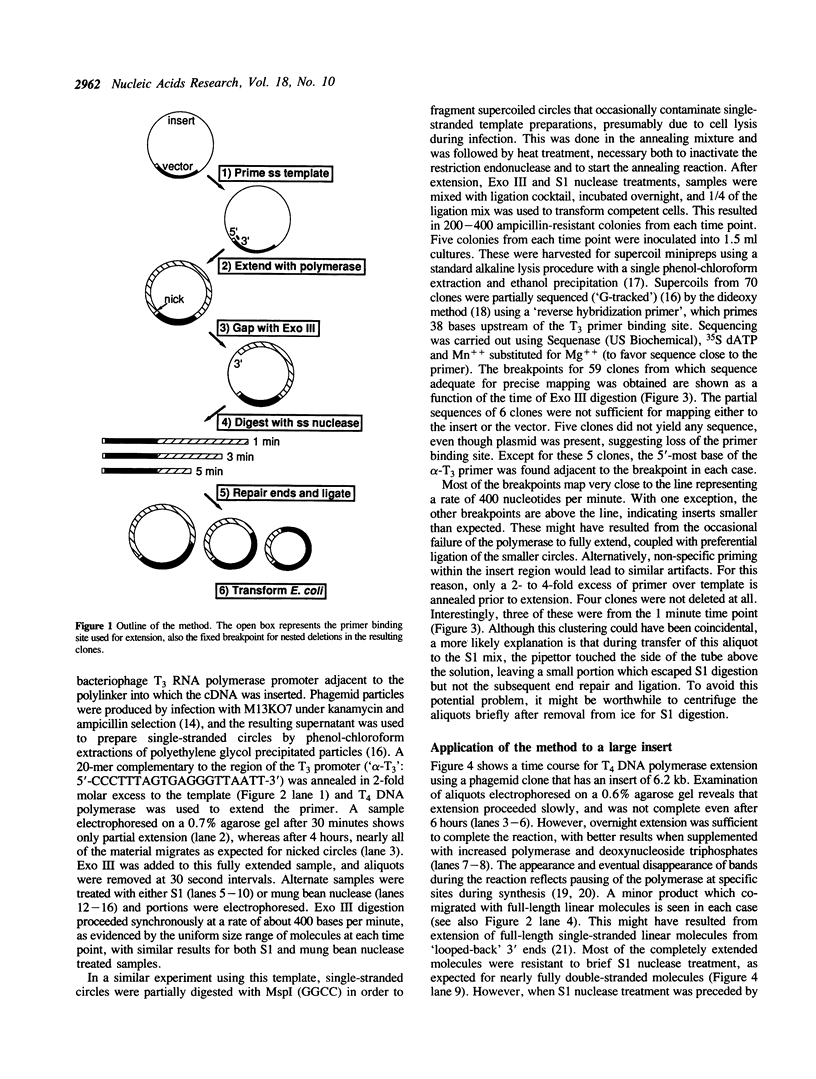

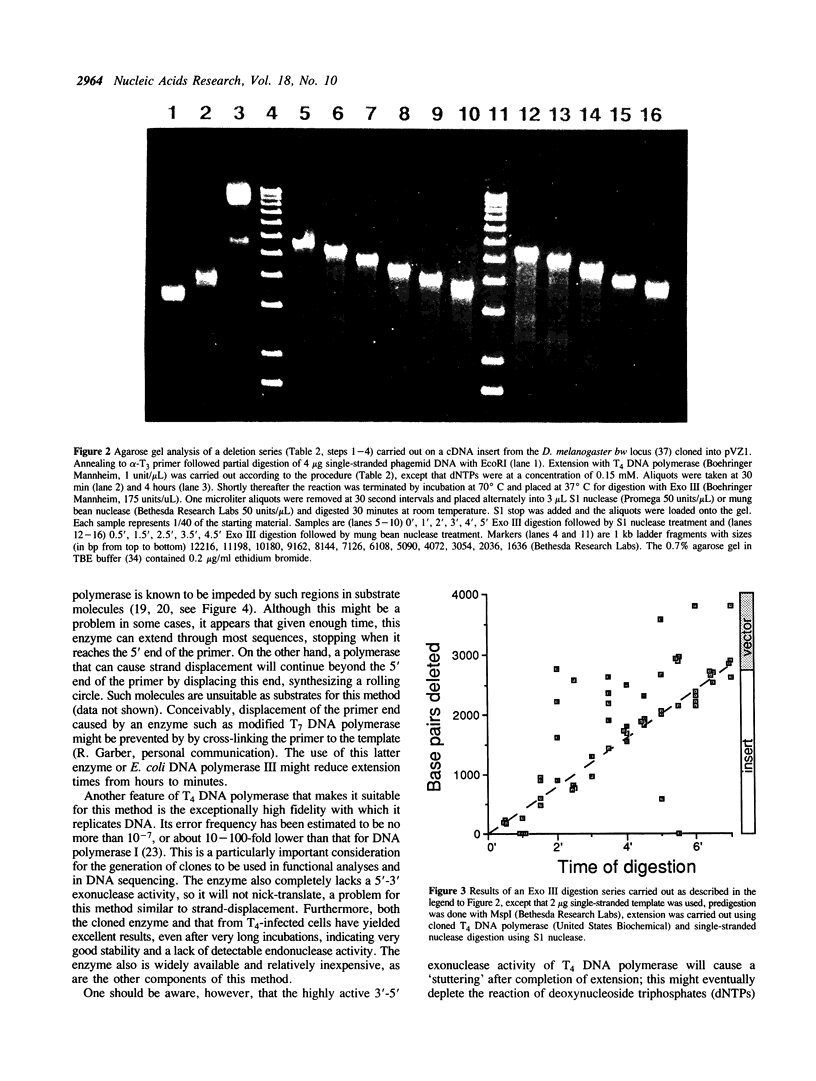
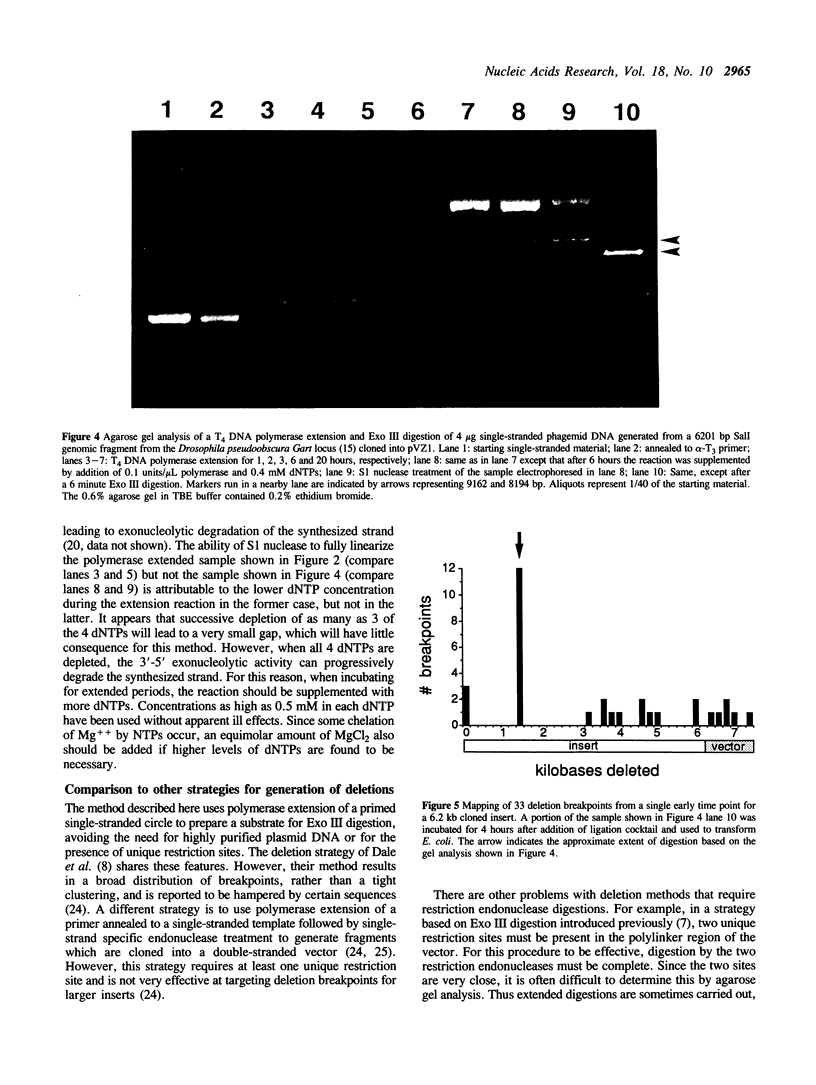

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed A. A rapid procedure for DNA sequencing using transposon-promoted deletions in Escherichia coli. Gene. 1985;39(2-3):305–310. doi: 10.1016/0378-1119(85)90328-2. [DOI] [PubMed] [Google Scholar]
- Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981 Jul 10;9(13):3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcak G. J., Wolf R. E., Jr A method for unidirectional deletion mutagenesis with application to nucleotide sequencing and preparation of gene fusions. Gene. 1986;49(1):119–128. doi: 10.1016/0378-1119(86)90391-4. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Bevan M. Kilo-sequencing: an ordered strategy for rapid DNA sequence data acquisition. Nucleic Acids Res. 1983 Jan 25;11(2):349–368. doi: 10.1093/nar/11.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., McGraw R. A., Edgell M. H., Hutchison C. A., 3rd A directed nucleotide-sequencing approach for single-stranded vectors based on recloning intermediates of a progressive DNA synthesis reaction. Gene. 1988 Jul 30;67(2):159–168. doi: 10.1016/0378-1119(88)90393-9. [DOI] [PubMed] [Google Scholar]
- Chang G. J., Johnson B. J., Trent D. W. Site-specific oligonucleotide-directed mutagenesis using T4 DNA polymerase. DNA. 1988 Apr;7(3):211–217. doi: 10.1089/dna.1988.7.211. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Dreesen T. D., Johnson D. H., Henikoff S. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol Cell Biol. 1988 Dec;8(12):5206–5215. doi: 10.1128/mcb.8.12.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eghtedarzadeh M. K., Henikoff S. Use of oligonucleotides to generate large deletions. Nucleic Acids Res. 1986 Jun 25;14(12):5115–5115. doi: 10.1093/nar/14.12.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M., Lucas Z. J., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXV. Purification and properties of deoxyribonucleic acid polymerase induced by infection with phage T4. J Biol Chem. 1968 Feb 10;243(3):627–638. [PubMed] [Google Scholar]
- Henikoff S., Eghtedarzadeh M. K. Conserved arrangement of nested genes at the Drosophila Gart locus. Genetics. 1987 Dec;117(4):711–725. doi: 10.1093/genetics/117.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hong G. F. A systemic DNA sequencing strategy. J Mol Biol. 1982 Jul 5;158(3):539–549. doi: 10.1016/0022-2836(82)90213-3. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E., Alberts B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981 Apr 25;256(8):4087–4094. [PubMed] [Google Scholar]
- Konings R. N., Verhoeven E. J., Peeters B. P. pKUN, vectors for the separate production of both DNA strands of recombinant plasmids. Methods Enzymol. 1987;153:12–34. doi: 10.1016/0076-6879(87)53045-2. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Loeb L. A., Goodman M. F. On the fidelity of DNA replication. The accuracy of T4 DNA polymerases in copying phi X174 DNA in vitro. J Biol Chem. 1984 Feb 10;259(3):1539–1545. [PubMed] [Google Scholar]
- Lechner R. L., Engler M. J., Richardson C. C. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983 Sep 25;258(18):11174–11184. [PubMed] [Google Scholar]
- Liu Z. J., Hackett P. B. Rapid generation of subclones for DNA sequencing using the reverse cloning procedure. Biotechniques. 1989 Jul-Aug;7(7):722–728. [PubMed] [Google Scholar]
- Misra T. K. A new strategy to create ordered deletions for rapid nucleotide sequencing. Gene. 1985;34(2-3):263–268. doi: 10.1016/0378-1119(85)90135-0. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nakauchi H. An improved method to make sequential deletion mutants for DNA sequencing. Trends Genet. 1989 Oct;5(10):325–325. [PubMed] [Google Scholar]
- Poncz M., Solowiejczyk D., Ballantine M., Schwartz E., Surrey S. "Nonrandom" DNA sequence analysis in bacteriophage M13 by the dideoxy chain-termination method. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4298–4302. doi: 10.1073/pnas.79.14.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. C., Nossal N. G., Englund P. T. Rapid hydrolysis of deoxynucleoside triphosphates accompanies DNA synthesis by T4 DNA polymerase and T4 accessory proteins 44/62 and 45. J Biol Chem. 1982 Feb 10;257(3):1267–1273. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J. A., Blinderman L. A. ExoMeth sequencing of DNA: eliminating the need for subcloning and oligonucleotide primers. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9208–9212. doi: 10.1073/pnas.86.23.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steggles A. W. A rapid procedure for creating nested sets of deletions using mini-prep plasmid DNA samples. Biotechniques. 1989 Mar;7(3):241–242. [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb J. F., Barcak G. J. Regulating the 3'-5' activity of exonuclease III by varying the sodium chloride concentration. Biotechniques. 1989 Oct;7(9):932–933. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wang L. M., Geihl D. K., Choudhury G. G., Minter A., Martinez L., Weber D. K., Sakaguchi A. Y. Site-directed deletion mutagenesis using phagemid vectors and genetic selection. Biotechniques. 1989 Oct;7(9):1000-6, 1008-10. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]