Abstract
Throughout mammalian evolution, recombination between the two sex chromosomes was suppressed in a stepwise manner. It is thought that the suppression of recombination led to an accumulation of deleterious mutations and frequent genomic rearrangements on the Y chromosome. In this article, we review three evolutionary aspects related to genomic rearrangements and structures, such as inverted repeats (IRs) and palindromes (PDs), on the mammalian sex chromosomes. First, we describe the stepwise manner in which recombination between the X and Y chromosomes was suppressed in placental mammals and discuss a genomic rearrangement that might have led to the formation of present pseudoautosomal boundaries (PAB). Second, we describe ectopic gene conversion between the X and Y chromosomes, and propose possible molecular causes. Third, we focus on the evolutionary mode and timing of PD formation on the X and Y chromosomes. The sequence of the chimpanzee Y chromosome was recently published by two groups. Both groups suggest that rapid evolution of genomic structure occurred on the Y chromosome. Our re-analysis of the sequences confirmed the species-specific mode of human and chimpanzee Y chromosomal evolution. Finally, we present a general outlook regarding the rapid evolution of mammalian sex chromosomes.
Keywords: Ectopic gene conversion, evolutionary strata, inverted repeats, palindrames, recombination suppression.
RECOMBINATION SUPPRESSION AND PAB FORMATION
Suppression of recombination is crucial for sex chromosomal differentiation and for the proper transmission of sex chromosomes to the next generation. In eutheria (placental mammals), suppression of recombination between the sex chromosomes occurs in a stepwise manner. This was first observed by Lahn and Page in 1999 [1], who showed that 19 pairs of X-Y human gametologs (homologs originating from a pair of autosomal ‘proto-sex chromosomes’) can be categorized into four groups according to the extent of synonymous nucleotide divergence between each gametologous pair and the location of corresponding genes on the X chromosome. The average nucleotide divergence in each group differs significantly from other groups and decreases in a stepwise manner according to the group position along the X chromosome. Each group is called a “stratum”. The number of synonymous divergence per site (ds) of gametologs in each stratum is as follows: ds of stratum 1, stratum 2, stratum 3 and stratum 4 are ~ 1.0, ~0.5, ~0.3 and ~0.1, respectively. Based on their ds values between gametologs, stratum 3 likely emerged after the divergence between eutherian mammals and marsupials, and stratum 4 likely emerged between prosimian and simian primates [2, 3]. Later, Ross et al. [4] found that stratum 5, in which d is ~0.08, might have been generated after the divergence of New World monkeys and Catarrhini (Old World monkeys and hominoids). In fact, the nucleotide sequences of the marsupial genome [5] confirmed the absence of strata 3, 4, and 5 on the marsupial X chromosome. The sex chromosomal region containing these strata originated from an autosome, and indeed, homologs of genes in strata 3, 4, and 5 are present on autosomes in the marsupial genome [5,6]. The region corresponding to strata 3, 4 and 5 is added to an ancient sex chromosome (the present marsupial type X chromosome) prior to the eutherian radiation, about 100 million years ago (mya) [7].
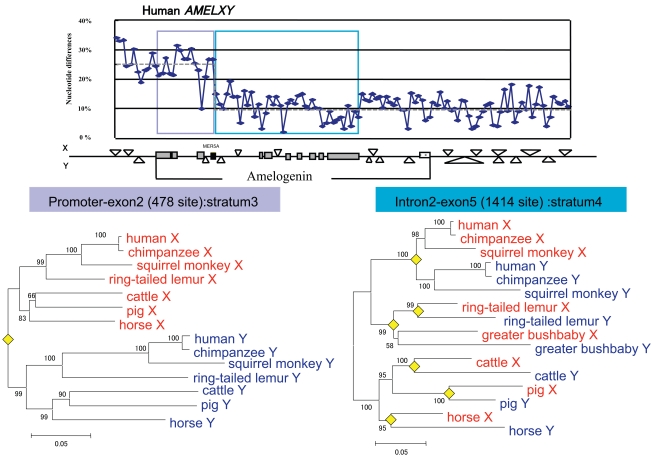
Previous analyses that aimed at identifying strata on sex chromosomes were performed using only the coding sequences (CDS) of genes. However, if each stratum is generated by stepwise recombination suppression, non-coding sequences in each stratum should show the pattern of nucleotide divergence variation similar to that observed for CDS. To clarify this, we compared the available non-coding nucleotide sequences of strata 3, and 4 (including stratum 5) on the short arm of the human X chromosome with that of the corresponding region on the Y chromosome [8]. We confirmed that stepwise differentiation occurred and the presence of clear boundaries among strata 3, 4, and the pseudoautosomal region (PAR) in non-coding regions. One remarkable finding reported in this study is the identification of a boundary between strata 3 and 4 in intron 2 of AMELX and AMELY (Fig. 1) in humans. We also confirmed the presence of the same boundary in other mammals [3].
Fig. (1). Nucleotide differences between mammalian AMEL gene regions.
Nucleotide differences between AMELX and AMELY of humans (top) and the AMEL phylogeny in strata 3 (bottom left) and 4 (bottom right) in mammals are shown. The graph shows the extent of nucleotide difference per site (ordinate) in non-overlapping windows of 100 bp. The abscissa shows the position in AMEL genes. A map of AMELX and AMELY with surrounding regions is shown at the bottom of the graph. Light and dark blue rectangles indicate the regions used for phylogenetic analysis. In the map, the boxes with shading indicate exons of AMELs. The filled box shows a transposable element, MER5, which is located at the strata boundary. Two phylogenies are shown for strata 3 and 4. The filled diamond in both trees reveals the divergence between the X (red) and the Y (blue) gametologs.
This finding is significant in the context of understanding the mechanism by which recombination suppression occurred. One previously proposed, and likely, mechanism is that of an “inversion” on the Y chromosome: the inversion is a possible direct cause to suppress recombination. However, the observation of a boundary in the intron of intact (i.e., functional) AMELX and AMELY suggests that an inversion cannot explain how the strata 3 and 4 boundary was formed. Instead, an alternative mechanism is likely required. Although we currently do not have a clear idea about this alternative mechanism, some clues might be revealed by investigating the current boundary of recombination suppression, or pseudoautosomal boundary (PAB), which is located between stratum 4 (or 5) and the PAR.
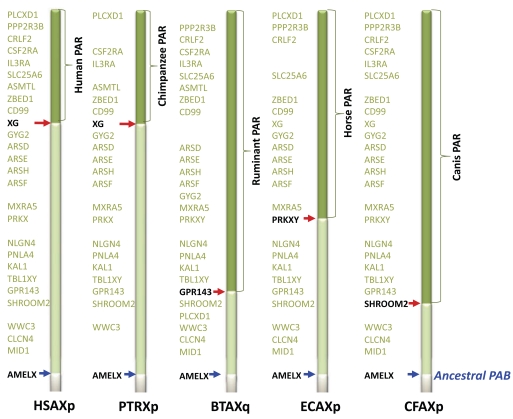
As shown in Fig. (2), the present PABs vary among different species, in that the position of the PAB on the X chromosome appears to be order- or species-specific. Currently, the full nucleotide Y chromosome sequences of many species have not yet been published. Therefore, fluorescent in situ hybridization (FISH) has been the primary method used to identify the PAB locations and genes nearby the PABs [9]. Interestingly, the SRY (Sex determining Region Y) gene is located near the PAB in humans and cats [10]. SRY is a gene encoding a transcription factor that is a primary male determining factor in eutherians. The linkage between male determination-related genes, such as SRY and RNA binding motif protein (RBMY) [3,11], is important in sex determination. Therefore, recombination with X chromosome was likely suppressed by natural selection [12]. When SRY or other male determination-related genes are placed into the PAR of the Y chromosome, the position in the PAR would become a PAB. In this regard, it will be interesting to learn more about the function of genes near PABs on the Y chromosomes of different mammals.
Fig. (2). Comparative gene map of pseudoautosomal boundaries among selected mammals.
Gene maps of the human (HSA), chimpanzee (PTR), cow (BTA), horse (ECA) and dog (CFA) pseudoautosomal boundaries (PABs) and adjacent regions are shown. The positions of PABs and AMELX (ancient PABs) are indicated by red and blue arrows, respectively. The gene order and contents of the stratum (shown in light green) adjacent to PAB and PAR (dark green) have been well conserved in mammals.
For humans and chimpanzees, the location and nucleotide sequences of PABs on the X and Y chromosomes are available, making it possible to search for clues about how recombination was suppressed. In both humans and chimpanzees, PAB is located in the XG gene in both the X and Y chromosomes, and SRY is inserted into intron 3 of the XG gametolog on the Y chromosome, in an orientation opposite to that of XG [13]. Due to this insertion, the human XG gametolog (XGPY2) on the Y chromosome likely lost the downstream region, from intron 3 to exon 10, and became truncated. Interestingly, the results of previous FISH analyses indicated that the XG on the Y chromosome has been duplicated near the centromere (XGPY) [13]. This duplication has been confirmed by BLAST analysis of the human Y chromosome sequences. In this article, we will refer to the sequence in the original PAR, including XGPY2, as PARY1, and the duplicated sequence as PARY1L. In the human Y chromosome, PARY1L is about 40 kb long. XGPY in PARY1L ranges from exon 2 to exon 6 and exhibits 92% similarity to XG. On the other hand, XGPY2 in PARY1 bears an identical sequence from exon 1 to exon 3 to XG. The presence of intron 3 to exon 6 in XGPY suggests that the duplication might have occurred prior to the loss of exons 3 to 10 in XGPY2. The similar duplication of an approximately 40-kb region was also observed in the chimpanzee Y chromosome [14, 15]. The nucleotide divergence between the PARY1 and PARY1L sequences in humans and chimpanzees is 0.087 ± 0.002 and 0.078 ± 0.001, respectively, suggesting that the duplication occurred concurrently with the formation of stratum 5 (~0.08 of the nucleotide divergence between X and Y gametologs) that is located adjacent to PARY. The nucleotide divergence between the X and Y chromosomal regions also indicates that the duplication occurred before the divergence of Old World monkeys and hominoids. If this duplication triggered the formation of the present PAR, then it would be predicted that the PAB in macaques is also located in XG, whereas the PAB might be located in different regions in New World monkeys. A detailed examination of PARY1 and their flanking regions in Old World and New World monkeys will be necessary to fully understand the relationship between the duplication and suppression.
GENE CONVERSION BETWEEN THE X AND Y CHROMOSOMES
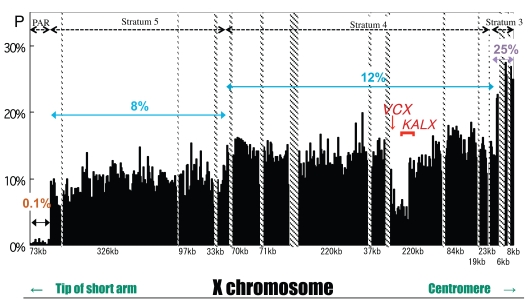
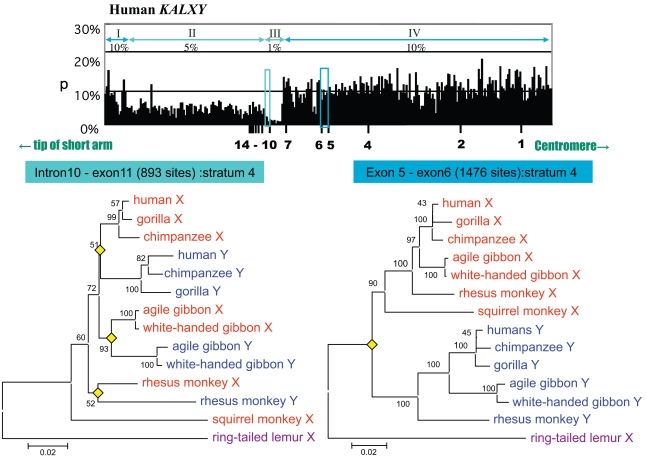
Except for PAR, genetic exchange between X and Y gametologous sequences, including non-coding regions, should only be observed rarely due to the suppression of recombination. However, a ~100-kb region is found to have significantly low nucleotide divergence compared with neighboring regions in the stratum 4 [3, 4,16]. Fig. (3) shows the levels of nucleotide divergence between the human sequence along the short arm of the X chromosome and its gametologous sequence on the Y chromosome. Stepwise changes in nucleotide divergence along the chromosomal arm are observed. The region with a divergence of 0.1% corresponds to PAR, the 8% divergent region corresponds to stratum 5, the 12% divergent region corresponds to stratum 4, and the 25% divergent region corresponds to stratum 3. Notably, there is a sub-segment with a significantly low extent of nucleotide divergence (1–5%) in the 12% region compared with the divergence at neighboring sequences. This sub-segment contains the KALX/Y and VCX/Y genes (see enlargement of KAL-coding and its flanking region shown in Fig. 4). This sub-segment can be further divided into four smaller regions (regions I to IV) corresponding to differences in nucleotide divergence. Regions I and IV have approximately 10% nucleotide divergence, whereas regions II and III have 5% and 1% divergence, respectively. Considering that both the telomeric and centromeric parts of regions II and III show 10% divergence, a decrease in the mutation rate is unlikely, rather, frequent gene conversions likely occurred in these regions. To test this idea, phylogenies for regions III and IV were constructed using other primate sequences, including lemurs and New World monkeys (Fig. 4). As expected, the tree corresponding to region IV shows a divergence of X and Y sequences prior to the divergence of simian primates, whereas the topology of the tree for region III revealed at least three independent gene conversions between X and Y sequences in the lineage leading to Old World monkeys, gibbons, and other hominoids. Further analysis revealed evidence for seven additional but independent X-Y conversion events during the evolution of Catarrhini [16]. The estimated rate of conversion is approximately 0.1/gene /million years (myr), assuming that the divergence of Old World monkeys, gibbons, orangutans, gorillas and chimpanzees from humans occurred 23, 20, 13, 7 and 6 myr, respectively [17,18].
Fig. (3). Summary map of nucleotide differences between the human X and Y chromosomes.
Nucleotide differences per site (ordinate) between the short arm of the human X chromosome and the corresponding region in the Y chromosome. The region is divided into four sub-regions according to the number of nucleotide differences (p-distance). From the tip of the short arm (abscissa), the four regions have p-distances of 0.1%, 8%, 12%, and 25%, respectively. The p-distances were calculated for nonoverlapping windows of 1 kb. In the middle of the region with a p-distance of 12%, there is a smaller region with significantly lower pdistance than in neighboring regions. A red arrow and a red square bracket show the position of VCX and KALX genes, respectively.
Fig. (4). Nucleotide differences in the human KAL gene region and corresponding regions in primates.
Nucleotide differences per site in a low p-distance region (top) in the human KAL gene and its flanking region, as well as the phylogeny of regions III and IV of primates (bottom). Bars at the bottom of the top graph represent exons in KALX. The tree was constructed using Neighbor joining based on the p-distances of KAL genes from several primates. The ring-tailed lemur KALX sequence was used to root each tree. The yellow diamond represents the divergence between KALX and KALY.
The ongoing gene conversion between the X and Y can be observed between KALX and KALY in extant human populations. Since KALY is a pseudogene, whereas KALX is functional, conversion of KALY to KALX can potentially destroy the functional KALX, a phenomenon that can result in Kallmann syndrome. Kallmann syndrome is a genetic disease characterized by a deficiency of gonadotropin-releasing hormone (GnRH), resulting in a decreased function of the glands producing sex hormones. In one patient with Kallmann syndrome, exon 14 in KALX was replaced by the homologous segment in KALY [19].
A second example of ongoing gene conversion events in humans are those that occurred between VCX and VCY, which was inferred from shared SNPs between the two genes [20]. VCX and VCY are located near the end of the low nucleotide divergence region. Phylogenetic analysis of the genes also reveals that gene conversion has occurred in both chimpanzees and humans, as well as in an ancestral population of both species [16, 21]. The overall rate of X to Y conversion was estimated to be 1.8 ( 10-7 /bp/year for VCX and VCY [20]. Because the size of these genes is approximately 1.5 kb, the conversion rate is likely 3 ( 10-4 /gene/year, which is much higher than the rate observed for KALX/Y.
The region of frequent gene conversion is confined to the ~100-kb region. Frequent conversion in this region might be due to the presence of sequence motifs or “hot spots” for non-allelic homologous recombination (NAHR) [22]. In fact, NAHR motifs have been conserved in regions flanking human VCX and VCY [23], as well as in VCX/Y of chimpanzee and macaque VCX. The presence of NAHR in primate VCX/Y might promote more frequent gene conversion than what is observed in KALX/Y. On the other hand, the presence of a LINE element in KALX/Y sequences might also promote gene conversion between the two genes [16].
EVOLUTION OF PALINDROMES ON SEX CHROMOSOMES
Palindromes (PDs) are composed of repeated sequences located in opposite directions, i.e. inverted repeats (IRs) with an interval of unrelated sequences [24]. Each repeated sequence is called an “arm” and a pair of arms forms a “stem” in a PD. A sequence sandwiched by two IRs forms is referred to as a “loop”. Warburton et al. [24] found many IRs on the X and Y chromosomes, which are likely to form PDs. The high level of nucleotide similarity between arms of the PDs appears to be due to gene conversion between them rather than recent divergence [24]. Interestingly, genes in IRs on the X or Y chromosome tend to be members of gene families that are mainly expressed in the testis and cancer cells [24]. However, we note that the overall gene content in PDs on the X and Y chromosomes is quite different. On the X chromosome, each PD is usually composed of members from a single gene family, whereas on the Y chromosome, each PD often contains members from several different gene families.
One example of genes, found on the human X chromosome PD, is the melanoma antigen A (MAGE-A) gene subfamily, which is located at around 150 Mb, near the tip of the long arm of the chromosome. The PD is about ~100 kb in size and seven genes and pseudogenes are located on this PD: MAGE-A2, A2B, A3, A6, A12, psA (a MAGE-A pseudogene) and psAL (‘MAGE-A pseudogene-like’) [25]. The MAGE-A subfamily is specific to eutherian mammals, and phylogenetic analysis shows that the MAGE-A members in a given eutherian species form a species-specific cluster. However, PDs that include a MAGE-A gene cluster have not been detected in eutherian mammals other than primates, especially other than Catarrhini. Among the seven members of the human MAGE-A family, A12 is located in a loop region, and the A2 and A2B, A3 and A6, and psA and psAL pairs are located on symmetrical positions on the arm of the PD. Moreover, nucleotide sequences of the A2 and A2B pair, and those of the psA and psAL pair are almost identical, whereas those of A3 and A6 are not. The underlying biology that explains the divergence between A3 and A6 has been discussed in another paper [25]. In brief, MAGE-A encodes epitopes that work in cancer immunity and negative selection against homogenization appears to operate to maintain a variety of epitopes among cancer cells. Negative selection against homogenization of sequences is also observed in immunoglobulin genes [26].
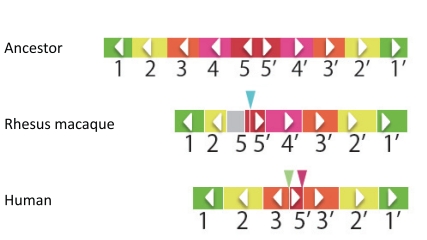
Based on the level of nucleotide divergence among members at asymmetrical positions, the divergence among MAGE-A members in humans is estimated to have occurred in a stem lineage of Catarrhini. The estimated time of human PD formation on the X chromosome predicted the presence of an orthologous PD in the macaque genome. As expected, a region containing several MAGE-A genes was identified in the Rhesus macaque genome. However, orthologous relationship between macaque and human MAGE-A genes is not obvious from the extent of nucleotide divergence between homologous regions and phylogenetic analysis: the divergence between a pair of possible orthologs is unexpectedly large [25]. Furthermore, the form of PD in macaques is expected to differ from that in humans: in macaques, high extents of sequence similarity between pairs among MAGE-A genes are not observed, except for the psA and psAL pair, which are located at the outermost part of the PD. Thus, the predicted PD in macaques has a relatively short stem and large loop compared with that in humans. When we compared the PDs of the two species, we found species-specific deletions (Fig. 5). Interestingly, these species-specific deletions were observed not only in these two species, but also in other primates, such as chimpanzees and orangutans. Although the nucleotide sequences of the corresponding region in chimpanzees and orangutans are not yet complete, the species-specific deletion was inferred from a comparison of duplicated units in each species. The species-specific deletions are indicative of PD instability.
Fig. (5). Duplicated units and specific deletions in humans, macaques, and an inferred common ancestor.
The arrangement of seven duplicated units of palindromes (PDs) in humans and macaques, as well as an inferred arrangement in a common ancestor. Same-color rectangles indicate regions with a high level of similarity in a conspecific genome and orthologs between different species. An arrowhead in each rectangle indicates the direction of the unit. A triangle indicates the position of a deletion. A gray rectangle represents a region in which the nucleotide sequence has not yet been determined. The arrangement present in an ancestor is parsimoniously inferred from comparison of humans and macaques.
On the human Y chromosome, there are eight palindromes on the long arm of the chromosome. We examined the nucleotide divergence between different gene members in different PDs or at asymmetrical positions in a single PD, and concluded that the majority of gene members in PDs were duplicated in the stem lineage of Catarrhini (~30–50 mya) [21]. Notably, the formation of PD on the Y chromosome coincides with that on the X chromosome. Because the formation of PD is dependent on the presence of IRs, this nearly simultaneous formation of PDs on X and Y chromosomes indicates simultaneous segmental duplication events in the ancestor genome, which might be related to a burst of LINE activity in the primate genome [27].
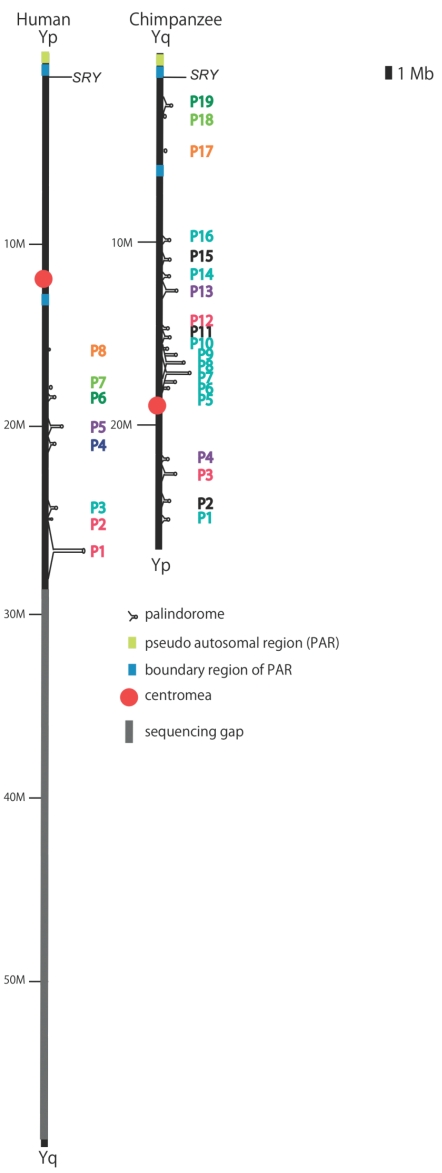
Recently, the nucleotide sequences of the chimpanzee Y chromosome were independently reported by two groups [14, 15]. Both analyses revealed drastic changes in the genomic structures in chimpanzees and/or humans following the divergence of the two species. It is remarkable that a large number of PDs (i.e., 19 PDs) are distributed on the chimpanzee Y chromosome, whereas only eight palindromes have been maintained in humans [2,15]. To investigate the evolutionary mode of structures on the sex chromosome, we examined the origin and relationship between these PDs in chimpanzees and humans. To distinguish human and chimpanzee PDs in the following discussion, we will refer to the PDs in humans as HPD1 to HPD8, and those in chimpanzees as CPD1 to CPD19 (Fig. 6). The eight human PDs and 19 chimpanzee PDs can be classified into three groups by their origin: human-specific, chimpanzee-specific, and shared PDs between the two species. A simple dot-plot analysis between the two chromosomes revealed that HPD 6, 7, and 8 are orthologs of CPD 19, 18, and 17, respectively [14, 15]. Though HPD7 and CPD18 do not contain any genes, these sequences are unique in the genome and are well conserved between the two species. It will be interesting to determine whether this PD structure is conserved in other primates. If the PD sequence is conserved in orangutans or macaques, for example, then the conservation might indicate that these nucleotide sequences have biological significance in these primates.
Fig. (6). Palindromes on the human and chimpanzee Y chromosomes.
Palindromes (PDs) on the human (left) and chimpanzee (right) Y chromosomes. The blue bars indicate the position of PARY1 and PARY1L. The same-colored number on the right side of each chromosome indicates homologous PDs.
In contrast to the conserved evolutionary mode of HPD 6, 7, 8 and CPD 19, 18, 17, the remaining PDs exhibit rather divergent mode. HPD4 in humans and CPD 2, 11, and 15 in chimpanzees are specific to each species, whereas the remaining PDs share ancestry (Fig. 6). HPD4 contains a set of 16 members from seven gene families (OFD1, USP9Y, XKRY, TCEB1, HSFY, TTY, TRAPPCA2) and among these 16 genes, HSFY1/HSFY2 exist only in HPD4 and thus they have been lost specifically in chimpanzees [15]. Considering that HSFYs are observed widely in mammals, the gene may play an important role in spermatogenesis [28, 29]. It is interesting whether or not there is a gene that can compensate a function of HSFYs in chimpanzees.
On the other hand, CPD 2, 11, 15 are specific to chimpanzees and all contain three pairs of genes: C-terminal-binding protein 2-like (CtBP2L), keratin type I cytoskeletal 18-like (KRT18L), and carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2-like (CTDSP2L). The nucleotide sequences of these PDs in chimpanzees show a quite high level of similarity to one another. It is remarkable that nucleotide sequences not only among CPD 2, 11, and 15, but also those between other PDs on the short and long arm of the Y chromosome show a high level of similarity [15]. Although only partial similarity between different PDs is observed in humans [21,30], the similarity between the entire PDs in chimpanzees was a new finding [15], indicating that entire PDs were recently duplicated in chimpanzees.
Hughes et al. [15] proposed that CPD 2, 11 and 15 arose from an autosome, because homologs of genes in the PDs are located on autosomes. If this is true, we can calculate the synonymous divergence of these genes on CPDs and autosomes, and estimate the emergence time of these PDs. Three genes on CPDs, CtBP2Ls, KRT18L, and CTDSP2L, are nearly identical (~1% nucleotide difference) compared with the corresponding sequences on the chromosome 7. Moreover, all of the homologous genes are located in a region of ~30 kb, near the tip of the short arm of chromosome 7. The extent of nucleotide differences suggests that the formation of these specific CPDs occurred specifically in chimpanzees after the divergence of chimpanzees from humans.
The remaining PDs in humans and chimpanzees share common ancestry. For example, HPD1 and HPD2 show partial similarity to CPD3, HPD5 shows similarity to CPD4 and CPD13, and HPD3 shows similarity to CPD1, CPD5-CPD10, CPD14 and CPD16. HPD3 and nine CPDs contain both RBMY and chromodomain protein Y –linked (CDY) genes and these genes seem to have been multiplicated specifically in chimpanzees through amplification of CPDs.
Instability of Y chromosome appears to be evident from the structural differences found between species as closely related as humans and chimpanzees. By comparing the complete Y chromosome sequence of different primates as well as other mammals, this and other interesting questions about the evolutionary mode of Y chromosome may be addressed in the future.
FUTURE PERSPECTIVES
We would like to specify three main points regarding evolution and the origin of genomic structures on the sex chromosomes. First, we focus on the bias in the number of IRs and PDs on the X and Y chromosomes. Here, we presented a comparison of the numbers and positions of IRs and PDs between the human and chimpanzee genomes. It remains to be determined whether the bias observed in these two species has been evolutionarily conserved. In a preliminary study, IRs or PDs have been found on the X chromosome of other eutherian mammals, but interestingly such genomic structures are rare in marsupial and platypus X chromosome. IRs and PDs have not been detected even on platypus chromosome 6, a proto-sex chromosome in Therians (marsupials and eutherians). If they are truly absent, the presence of PDs on sex chromosomes might be specific to the eutherian lineage. In fact, mouse X chromosome has its own IRs and PDs that are not shared with humans. Furthermore, the genomic structures in the eutherian X chromosome predominantly contain genes in the cancer testis antigen (CTA) gene super-family, exemplified by MAGE gene family members. The origin of CTA family genes is not fully understood. The relationship between genomic structures like PDs and the origin of CTA family genes is an interesting topic to be further explored.
The second point to be determined is the biological importance of IRs or PDs on sex chromosomes. On the Y chromosome, it is thought that IRs or PDs might have developed to compensate for the lack of recombination by providing a mechanism for intra-chromosomal gene exchange. However, this reason does not hold true for the presence of IRs or PDs on the X chromosome. The X chromosome experiences recombination and therefore, compensating for the lack of recombination by gene conversion between PD sequences cannot explain the presence of PDs on X chromosomes. If it is so, the biological significance of having PDs on the X chromosome should be investigated from other viewpoints.
The third point we wish to address is the mechanism of recombination suppression between sex chromosomes. The presence of strata, or stepwise suppression of recombination along the sex chromosomes, is also observed in chickens, dioecious plants and smut fungi [31-33]. In chickens, the sex chromosomes are designated as Z and W, with the ZW genotype corresponding to females and ZZ to males. The chicken sex chromosomes are characterized by nonlinear strata, i.e., the extent of Z-W divergence does not correspond to the location of genes on the Z chromosome [31]. In a dioecious plant, Silene latifolia, the extent of synonymous divergence has increased in proportion to the distance from PAR on the sex chromosome. Although the increase is linear, a distinct stratum has not been observed, suggesting that steps suppressing recombination between the sex chromosomes in this plant are still progressing [32]. In the smut fungus Microbotryum violaceum, the synonymous divergence between sex chromosomes is ~10%, suggesting that sex chromosomes recently differentiated in this species. However, a gradual increase of nucleotide divergence towards a sex-determining (mating type-determining) gene is clearly observed [33].
Suppression of recombination appears to be a crucial step for sex chromosome differentiation in many organisms. It will be interesting to determine whether or not there is a general mechanism of recombination suppression. We know that PAB in the X chromosome is specific for each species. Thus, specific rearrangements at PAB on the Y chromosome should be examined, since they might provide new insights about a common molecular mechanism, other than inversion, that could lead to suppression of recombination between sex chromosomes.
Addressing these questions will shed light on the mechanism of sex chromosomal evolution and will hopefully reveal the broader evolutionary significance of genomic structures like IRs or PDs.
REFERENCES
- 1.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science . 1999;286: 964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 2.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature . 2003;423: 825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 3.Iwase M, Satta Y, Hirai Y, Hirai H, Imai H, Takahata N. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc Natl Acad Sci USA. 2003;100:5258–5263. doi: 10.1073/pnas.0635848100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross MT, Grafham DV, and 284 authors The DNA sequence of the human X chromosome. Nature . 2005;434: 325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delbridge ML, Patel HR, Waters PD, McMillan DA, Graves JAM. Does the human X contain a third evolutionary block? Origin of genes on human Xp11 and Xq28. Genome Res . 2009;19:1350–1360. doi: 10.1101/gr.088625.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veryunes F, Waters PD, Miethke P, Rens , McMillan D, Alsop AE, Grützner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Graves JAM. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res . 2008;18: 965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y, Fujiwara M, Nikaido M, Okada N, Hasegawa M. Interordinal relationships and timescale of eutherian evolution as inferred from mitochondrial genome data. Gene . 2000;259: 149–158. doi: 10.1016/s0378-1119(00)00427-3. [DOI] [PubMed] [Google Scholar]
- 8.Iwase M, Satta Y, Takahata N. Sex-chromosomal differentiation and amelogenin genes in mammals. Mol Biol Evol . 2001;18: 1601–1603. doi: 10.1093/oxfordjournals.molbev.a003948. [DOI] [PubMed] [Google Scholar]
- 9.Das PJ, Chowdhary BP, Ruadsepp T. Characterization of the bovine pseudoautosomal region and comparison with sheep goat and other mammalian pseudoautosomal regions. Cytogenet. Genome Res. 2009;126:139–147. doi: 10.1159/000245913. [DOI] [PubMed] [Google Scholar]
- 10.Murphy WJ, Wilkerson AJP, Raudsepp T, Agarwala R, Schäffer AA, Stanyon R, Chowdhary BP. Novel gene acquisition on carnivore Y chromosomes. PLoS Genetics. 2006;2:353–363. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skrisovska L, Bourgeois CF, Stefl R, Grellscheid S-N, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH-T. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep . 2007;8: 372–379. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M. Linkage modification and sex difference in recombination. Genetics. 1969;63: 681–699. doi: 10.1093/genetics/63.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weller PA, Critcher R, Goodfellow PN, German J, Ellis NA. The human Y chromosome homologue of XG transcription of a naturally truncated gene. Hum Mol Gent . 1995;4: 859–868. doi: 10.1093/hmg/4.5.859. [DOI] [PubMed] [Google Scholar]
- 14.Kuroki Y, Toyoda A, Noguchi H, Taylor TD, Itoh T, Kim D-S: Kim, D.-W. Choi, S.-H. Kim, I.-C. Choi, H.H. Kim, Y.S. Satta Y, Saitou N, Yamada T, Morishita S, Hattori M, Sakaki Y, Park H-S, Fujiyama A. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat Genet. 2006;38: 158–167. doi: 10.1038/ng1729. [DOI] [PubMed] [Google Scholar]
- 15.Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SKM, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, Trask BJ, Mardis ER, Warren WC, Repping S, Rozen S, Wilson RK, Page DC. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463: 536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwase M, Satta Y, Hirai H, Hirai Y, Takahata N. Frequent gene conversion events between the X and Y homologous chromosomal regions in primates. BMC Evol Biol . 2010;10: 225. doi: 10.1186/1471-2148-10-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasegawa M, Thorne JL, Kishino H. Time scale of eutherian evolution estimated without assuming a constant rate of molecular evolution. Genes Genet Syst . 2003;78: 267–283. doi: 10.1266/ggs.78.267. [DOI] [PubMed] [Google Scholar]
- 18.Glazko G V, Nei M. Estimation of divergence times for major lineages of primate species. Mol Biol Evol . 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- 19.Guioli S, Incerti B, Zanaria E, Bardoni B, Frano B, Taylor K, Ballabio A, Camerino G. Kallmann syndrome due to a translocation resulting in an X/Y fusion gene. Nat Genet . 1992;1:337–340. doi: 10.1038/ng0892-337. [DOI] [PubMed] [Google Scholar]
- 20.Trombetta B, Cruciani F, Underhill PA, Sellitto D, Scozzari R. Footprints of X-to-Y gene conversion in recent human evolution. Mol Biol Evol . 2010;27: 714–725. doi: 10.1093/molbev/msp231. [DOI] [PubMed] [Google Scholar]
- 21.Bhowmick BK, Satta Y, Takahata N. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 2007;17: 441–450. doi: 10.1101/gr.5734907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers S, Freeman C, Auton A, Donnelly P, Vean GM. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet . 2008;40: 1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 23.Van Esch H, Hollanders K, Badisco L, Melotte C, Van Hummelen P, Vermeesch JR, Devriendt K, Fryns J-P, Marynen P, Froyen G. Deletion of VCX-A due to NAHR plays a major role in the occurrence of mental retardation in patients with X-linked ichthyosis. Hum Mol Genet. 2005;14:1795–1803. doi: 10.1093/hmg/ddi186. [DOI] [PubMed] [Google Scholar]
- 24.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res . 2004;14: 1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsura Y, Satta Y. Evolutionary history of the cancer immunity antigen MAGE gene family. PLoS ONE . 2011;6: e20365. doi: 10.1371/journal.pone.0020365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nei M, Rooney A P. Concerted and birth-and-death evolution of multigene families. Ann Rev Genet . 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim T-M, Hong S-J, Rhyu M-G. Periodic explosive expansion of human retroelements associated with the evolution of the hominoid primates. J. Korean Med. Sci. 2004;19:177–185. doi: 10.3346/jkms.2004.19.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tessari A, Salata E, Ferlin A, Bartoloni L, Slongo ML, Forest C. Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol. Hum. Reprod. 2004;10:253–258. doi: 10.1093/molehr/gah036. [DOI] [PubMed] [Google Scholar]
- 29.Shinka T, Sato Y, Chen G, Naroda T, Kinoshita K, Unemi U, Tsuji K, Toida K, Iwamoto T, Nakahori Y. Molecular Characterization of heat shock-like factor encoded on the human Y chromosome, and implications for male infertility. Biol. Reprod. 2004;71:297–306. doi: 10.1095/biolreprod.103.023580. [DOI] [PubMed] [Google Scholar]
- 30.Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen R, Page DC. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- 31.Nam K, Ellegren H. The chicken (Gallus gallus) Z chromosome contains at least three nonlinear evolutionary strata. Genetics. 2008;180:1131–1136. doi: 10.1534/genetics.108.090324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergero R, Forrest A, Kamau E, Charlesworth D. Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: Evidence from new sex-linked genes. Genetics. 2007;175:1945–1954. doi: 10.1534/genetics.106.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Votintseva AA, Filatov DA. Evolutionary strata in a small mating-type-specific region of the smut fungus Microbotryum violaceum. Genetics. 2009;182:1391–1396. doi: 10.1534/genetics.109.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]