Abstract
Natural killer T (NKT) cells recognize glycolipids produced by Sphingomonas bacteria, and these glycolipids contain C6-oxidized sugars, either glucuronic acid or galacturonic acid, linked to ceramides. Glycolipids with gluco stereochemistry are the most prevalent. Multiple studies have demonstrated that galactosylceramides are more potent stimulators of NKT cells than their glucose isomers. To determine if this stereoselectivity is retained in the context of the C6-oxidized sugars found in bacterial glycolipids, we prepared two sets of gluco and galacto-glycolipids oxidized at their C6 positions and compared their NKT stimulatory properties. In the context of carboxylic acid groups at C6, gluco stereochemistry gave the more potent responses. We also prepared bacterial glycolipids containing more complex ceramide groups to determine if these chains impact NKT cell responses.
Natural killer T (NKT) cells are a subset of T cells and play a key role in regulating immune responses.1-3 This regulation impacts responses to infection, tumor growth, and autoimmune diseases. In addition, NKT cell stimulation has been shown to improve the efficacy of vaccines.4 Consequently, there has been considerable interest in the means by which these cells can be stimulated to produce the cytokines that in turn control responses of other aspects of the immune systems in higher organisms. NKT cells are stimulated via presentation of glycolipids by CD1d, and a limited number of glycolipids and lipid polyols have been identified as antigens for NKT cells.5 These include alpha-glycosylceramides,6 alpha-glycuronosylceramides,7,8 alphagalactosyldiacylglycerols, 9 isogloboside iGb3,10 and threitolceramides.11
The first identified NKT cell antigens were isolated from a marine sponge,12 and while potent stimulators of NKT cells, these glycolipid are not considered “natural” antigens. Structure-activity studies of the sponge-derived glycolipids, in the context of anti-tumor properties, led to the generation of alpha-galactosylceramide KRN700013 (Figure 1). This glycolipid has been the primary antigen used in elucidating antigen presentation by CD1d and NKT cell responses.
Figure 1.
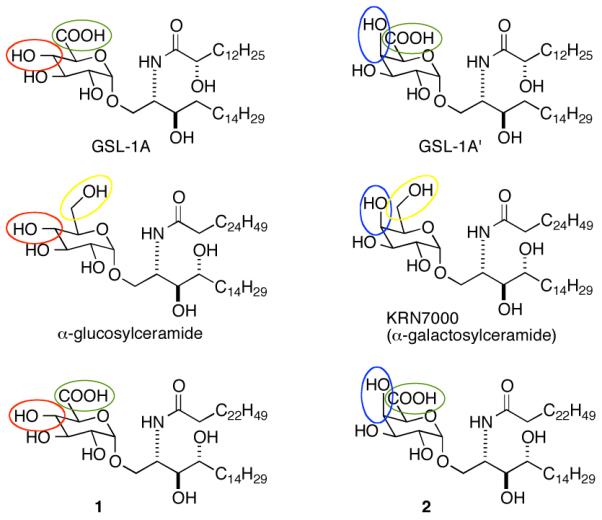
Structures of glycolipids from Sphingomonas (GSL-1A and GSL-1A’), NKT cell agonists α-glucosylceramide, KRN7000 and glycolipids 1 and 2.
The first natural, exogenous antigens for NKT cells were identified among the membrane components of the Sphingomonadaceae family of bacteria (including Sphingomonas spp.).7,8 The bacteria in this family are Gram-negative (possess two lipid bilayers); however, their outer membrane is not made up of the lipopolysaccharide found in many Gram-negative bacteria (e.g., Escherichia coli). In its place, bacteria in this family produce a group of glycuronosylceramides including GSL-1A and GSL-1A’ (Figure 1).14
Sphingomonas bacteria are ubiquitous. They are found in water sources,15 on household items,16 and are one of the most prevalent airborne forms of life.17 Consequently, humans are routinely exposed to these bacteria. There is evidence that these bacteria modulate immune responses18 and that glycolipids from Sphingomonas can trigger allergic asthma.19
To better understand the NKT cell stimulatory properties of the GSLs produced by Sphingomonas, we20 and Kronenberg et al.21 reported the synthesis and NKT cell stimulatory activity of some of the glycolipids produced by this type of bacteria. Both studies showed that sugars appended on GSL-1A decrease activity significantly, and that GSL-1A and GSL-1A’ were less stimulatory than KRN7000. Kronenberg et al. also showed that GSL-1A is somewhat less stimulatory than GSL-1A’.
α-Galactosylceramide is a much more potent stimulator of NKT cells than α-glucosylceramide (Figure 1).6 Among Sphingomonas bacteria, GSL-1A, with gluco stereochemistry, is generally more predominant than GSL-1A’, which has galacto stereochemistry (Figure 1).14,22. If one of the major roles of NKT cells is surveillance for Sphingomonas and related bacteria, it would be expected that the more predominant glycolipid (GSL-1A (gluco stereochemistry)) would be a more potent stimulator than the less common isomer (GSL-1A’ (galacto stereochemistry)). Among the Sphingomonas glycolipids, C6 in the sugars is oxidized, and we questioned if oxidation at this position might alter recognition of gluco vs. galacto selectivity in NKT cell stimulation. To answer this question, we prepared GSL-1A, GSL-1A’ and glycolipids 1 and 2 (Figure 1). KRN-7000 and the Sphingomonas glycolipids differ in the ceramide portions of the molecules, and ceramide structure has been shown to play role in NKT cell responses to glycolipids. Compounds 1 and 2 contain the glucoronic and galacturonic acids found in GSL-1A and GSL-1A’ and the ceramide found in KRN7000. These compounds proved useful in dissecting the impact of the acid groups in the sugars on NKT cell responses.
To determine the full impact of differences in the ceramide portions of glycolipids from Sphingomonas on NKT cell stimulation, we also prepared GSL-1B and GSL-1C (Figure 2), in which the sphinganine chain is elongated and modified as compared to GSL-1A.14 These two glycolipids are produced by bacteria but in lower amounts as compared to GSL-1A. The syntheses of GSL-1B and GSL-1C and NKT cell stimulatory activities of these two compounds have not been reported.
Figure 2.
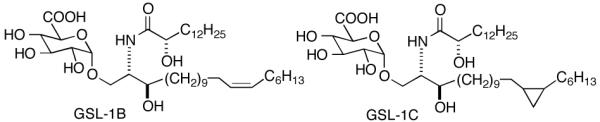
Structures of GSL-1B and GSL-1C .
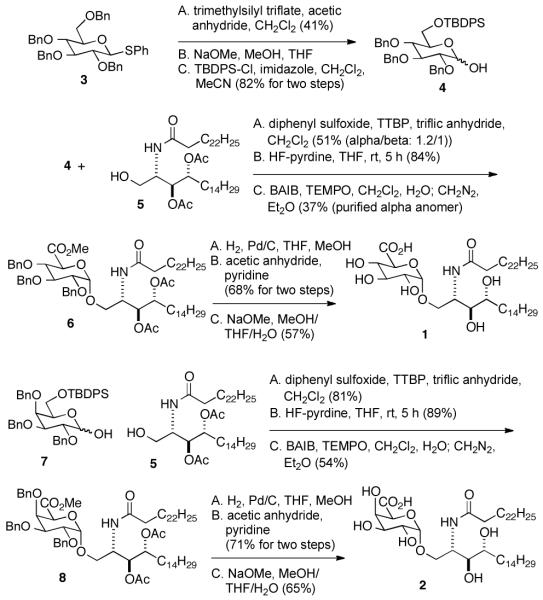
Multiple syntheses of GSL-1A and GSL-1A’ have been reported (for examples see refs. 19, 20, 22 and 23). Alpha-glycoside bond formation with glucuronic and galacturonic acid derivatives is typically complicated by low anomeric selectivities. Pilgrim and Murphy23 reported an ingenious means of generating the alpha-anomer in high yields in the synthesis of GSL-1A and GSL-1A’. This group formed the corresponding beta anomer in good yield, then used chelation-induced anomerization to give the alpha anomer in very high yield. Alternatively, GSL-1 can be formed in reasonable yield by oxidizing glucose to glucuronic acid after glycoside bond formation.24 We used the latter approach for the glycolipids 1 and 2 (Scheme 1) and the former approach for synthesis of GSL-1B and GSL-1C (Scheme 2).
Scheme 1.
Synthesis of glycolipids 1 and 2.
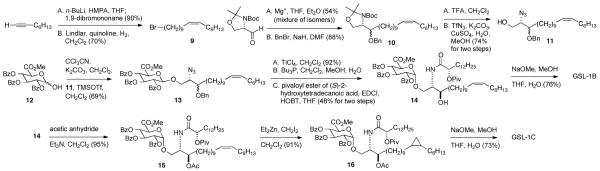
Scheme 2.
Synthesis of GSL-1B and GSL-1C.
Oxidation at C6′′ after glycoside bond formation required preparation of C6-orthogonally protected glycoside donor 4 (Scheme 1). To access this compound, we used thioglycoside 3 and treated it with TMS-triflate in acetic anhydride to give the corresponding 1,6-diacetate. Though the yield of this reaction is moderate, the starting material is simple to prepare in large scale, and purification of the product is straightforward. Removal of the acetates and selective protection at C6′′ gave 4 in good overall yield. Surprisingly, coupling of 4 with 5 gave the corresponding glycoside with little anomeric selectivity. The anomers were not separated but were carried forward through deprotection at C6′′, oxidation and ester formation giving 6. At this point, separation of the alpha and beta anomers was easily accomplished. Reductive deprotection was followed by peracylation to provide a compound that was easily chromatographable. The final deprotection gave 1. For the synthesis of 2 (Scheme 1), compound 7 was prepared as described for 4. The remaining steps paralleled those used in the preparaton of 2.
Synthesis of the sphinganine for GSL-1B and GSL-1C began with 1-octyne (Scheme 2). Deprotonation and reaction with 1,9-dibromononane followed by reduction with Lindlar’s catalyst gave 9. Grignard formation and reaction with Garner’s aldehyde gave the corresponding alcohol as a mixture of diastereomers that were difficult to separate. Consequently, the mixture was carried on through benzylation (giving 10), liberation of the amine and alcohol at C1 and azide formation yielding 11. Glycoside formation with 12 gave 13 in reasonable yield with good anomeric selectivity. Isomerization, giving the alpha anomer, azide reduction, and amide formation gave 14, which was readily separated from the minor diastereomer. Treatment with sodium methoxide in the presence of a small amount of water gave GSL-1B. Preparation of GSL-1C required protection of the alcohol at C3 in 14 followed by cyclopropane formation using Simmons-Smith chemistry. In the identification of GSL-1C, the relative stereochemistry of the cyclopropyl group was not determined.14 The diastereomers formed from cyclopropanation of 16 proved to be inseparable, and in the NMR spectra of 16, individual diastereomers were not observed. Deprotection of 16 gave GSL-1C.
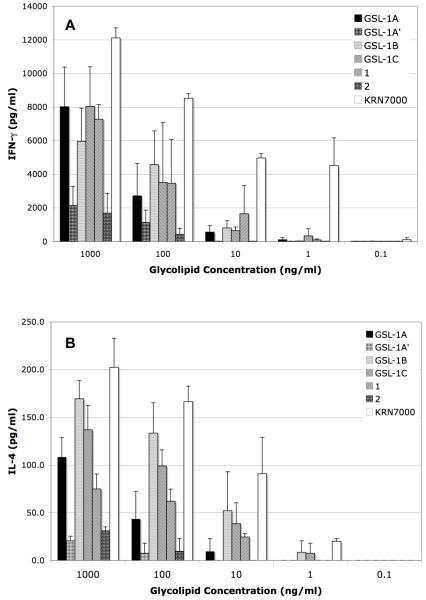
Stimulation of NKT cells by glycolipids depends upon presentation by the protein CD1d on antigen presenting cells. Dendritic cells are the primary presenters of glycolipids to NKT cells,1 and these and NKT cells are found in relatively high abundance in the spleen. Murine (B6 mice) splenocytes were used to measure NKT cell stimulation by the synthesized glycolipids. Stimulation is quantified in terms of cytokine release, and proinflammatory and immunomodulatory cytokines are typified by IFN-γ and IL-4, respectively.20 The relative amounts of these cytokines released by NKT cells determine the types of immune responses that follow stimulation. Cytokine release was measured using quantitative ELISA.
As expected, stimulation of proinflammatory cytokine IFN-γ by KRN7000 occurred at low concentrations, and as has been observed previously, stimulation by glycolipids from Sphingomonas required higher concentrations (Figure 3A). Nevertheless, substantial responses were observed at 100 ng/mL. Interestingly, the ceramide structure of the glycolipids had only a minor effect on NKT cell stimulation (compare GSL-1A, GSL-1B, GSL-1C, and 1). The greatest impact on IFN-γ release was the sugar stereochemistry; compounds with glucostereochemistry were more stimulatory than those with galacto stereochemistry (compare GSL1A with GSL1A’ and 1 with 2).
Figure 3.
Cytokine production by splenocytes in response to the indicated glycolipids.
The same trend was seen with IL-4 release (Figure 3B); that is, among the Sphingomonas glycolipids, we found glycolipids with gluco stereochemistry to be more stimulatory than galacto. Glycolipids with shorter acyl chains than those found in KRN7000 are associated with higher amounts of IL-4 release, in relation to IFN-γ release.25 Comparision of the impact of the ceramide in 1 and that found in GSL-1B and GSL-1C suggest that there may be a greater propensity for IL-4 production with the latter two glycolipids due to their relatively short acyl chains.
Few “natural” antigens for NKT cells have been identified, and an understanding how these antigens impact human health requires elucidation of relative stimulatory properties of glycolipids found in sources of these antigens. We have demonstrated that the more predominant glycolipid found in Sphingomonas spp. (GSL-1A) stimulates stronger NKT cell responses than its isomer (GSL-1A’) and that the stereoselective recognition of these glycolipids is independent of ceramide structure. GSL-1B and GSL-1C are less predominant than GSL-1A, but both stimulate NKT cell reponses comparable to GSL-1A. These results shed additional light on structural requirements for NKT cell stimulation and may prove useful in development of novel NKT cell antigens and adjuvants.
Supplementary Material
Acknowledgment
Financial support from the National Institutes of Health (NIAID, P01 AI053725) is gratefully acknowledged. We thank Michael E. Fusakio for aiding in measuring cytokine release from splenocytes.
Footnotes
Supporting Information Available Experimental procedures for preparation of GSL-1B, GSL-1C, 1 and 2. 1H and 13C NMR spectra of 1, 2, GSL-1B and GSL-1C. Methods of measuring cytokine production stimulated by CD1d presentation of glycolipids to NKT cells.
Literature Cited
- 1.Bendelac A, Savage PB, B P, Teyton L. Annual Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 2.Balato A, Unutmaz D, Gaspari AA. J. Invest. Dermatol. 2009;129:1628–1642. doi: 10.1038/jid.2009.30. [DOI] [PubMed] [Google Scholar]
- 3.Kronenberg M, Kinjo Y. Curr. Opin. Immunol. 2009;21:391–396. doi: 10.1016/j.coi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guillonneau C, Mintern JD, Hubert FX, Hurt AC, Besra GS, Porcelli S, Barr IG, Doherty PC, Godfrey DI, Turner SJ. Proc. Nat’l Acad. Sci. USA. 2009;106:3330–3335. doi: 10.1073/pnas.0813309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage PB, Bendelac A, Teyton L. Chem. Soc. Rev. 2006;35:771–779. doi: 10.1039/b510638a. [DOI] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Tanguchi M. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 7.Kinjo Y, Wu D, Kim G, Xing G-W, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong C-H, Kronenberg M. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 8.Mattner J, DeBord KL, Goff RD, Cantu C, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Ismail N, Walker D, Buetler B, Teyton L, Savage PB, Bendelac A. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 9.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Manachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Buetler B, Wilson IA, Tsuji, Sellati TJ, Wong C-H, Kronenberg M. Nature Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Mattner J, Cantu C, III, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu Y, Teneberg S, Wang, Proia R, Levery SB, Savage PB, Teyton L, Bendelac A. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
- 11.Silk JD, Salio M, Reddy BG, Shepherd D, Gileadi U, Brown J, Masri SH, Pozella P, Ritter G, Besra GS, Jones EY, Schmidt RR, Cerundolo V. J. Immunol. 2008;180:6452–6456. doi: 10.4049/jimmunol.180.10.6452. [DOI] [PubMed] [Google Scholar]
- 12.Natori T, Koezuka Y, Higa T. Tetrahedron Lett. 1993;34:5591–5592. [Google Scholar]
- 13.Morita M, Motoki K, Akimoto K, Natori T, Sakai T, Sawa E, Yamaji K, Koezuka Y, Kobayashi E, Fukushima H. J. Med. Chem. 1995;38:2176–2187. doi: 10.1021/jm00012a018. [DOI] [PubMed] [Google Scholar]
- 14.Kawahara K, Kuraishi H, Zähringer U. J. Industrial Microbiol. & Biotech. 1999;23:408. doi: 10.1038/sj.jim.2900708. [DOI] [PubMed] [Google Scholar]
- 15.Giao MS, Azevedo NF, Wilks SA, Vieira MJ, Keevil CJ. BMC Microbiol. 2011;11:57. doi: 10.1186/1471-2180-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly ST, Theisen U, Angenent LT, Amand AS, Pace NR. Applied Environ. Microbiol. 2004;70:4187–4192. doi: 10.1128/AEM.70.7.4187-4192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlgren C, Bratbak G, Sandaa RA, Thyrhaug R, Zweifel UL. Aerobiologia. 2011;27:107–120. [Google Scholar]
- 18.Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, Scanlon ST, Teyton L, Hart J, Ridgway WM, Wicker LS, Gershwin EM, Bendelac A. Cell Host Microbe. 2008;3:304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer EH, Goya S, Akbari O, Berry GJ, Savage PB, Kronenberg M, Nakayama T, DeKruyff RH, Umetsu DT. Natl Acad. Sci. USA. 2006;103:2782–2787. doi: 10.1073/pnas.0510282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long X, Deng S, Zang Z, Mattner J, Zhou D, McNary N, Goff RD, Teyton L, Bendelac A, Savage PB. Nature Chem. Biol. 2007;3:559–564. doi: 10.1038/nchembio.2007.19. [DOI] [PubMed] [Google Scholar]
- 21.Kinjo Y, Pei B, Sufali S, Raju R, Richardson SK, Imamura M, Fujio M, Wu D, Khurana A, Kawahara K, Wong C-H, Howell AR, Seeberger PH, Kronenberg M. Chem. Biol. 2008;15:654–664. doi: 10.1016/j.chembiol.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naka T, Fujiwara N, Yabuuchi E, Doe M, Kobayashi K, Kato Y, Yano I. J. Bacteriol. 2000;182:2660–2663. doi: 10.1128/jb.182.9.2660-2663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilgrim W, Murphy PV. Org. Lett. 2009;11:939–942. doi: 10.1021/ol802915h. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto N, Kanie O, Huang Y-Y, Fujii E, Watanabe H, Shimamura M. Chem. Biol. 2005;12:677–683. doi: 10.1016/j.chembiol.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Goff RD, Gao Y, Mattner J, Zhou D, Yin N, Cantu C, III, Teyton L, Bendelac A, Savage PB. J. Am. Chem. Soc. 2004;126:13602–13603. doi: 10.1021/ja045385q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.