Abstract
Monoacylglycerol lipase (MGL) is primarily responsible for the hydrolysis of 2-arachidonoylglycerol (2-AG), an endocannabinoid with full agonist activity at both cannabinoid receptors. Increased tissue 2-AG levels consequent to MGL inhibition are considered therapeutic against pain, inflammation, and neurodegenerative disorders. However, the lack of MGL structural information has hindered the development of MGL-selective inhibitors. Here, we detail a fully refined homology model of MGL which preferentially identifies MGL inhibitors over druglike noninhibitors. We include for the first time insight into the active-site geometry and potential hydrogen-bonding interactions along with molecular dynamics simulations describing the opening and closing of the MGL helical-domain lid. Docked poses of both the natural substrate and known inhibitors are detailed. A comparison of the MGL active site to that of the other principal endocannabinoid metabolizing enzyme, fatty acid amide hydrolase (FAAH), demonstrates key differences which provide crucial insight toward the design of selective MGL inhibitors as potential drugs.
Keywords: MGL, comparative model, molecular dynamics, principal component analysis, docking
Introduction
The endocannabinoid signaling system is comprised of two main G-protein coupled cannabinoid receptors (CB1 and CB2), their principal endogenous ligands [the endocannabinoids N-arachidonoylethanolamine (anandamide) and 2-arachidonoylglycerol (2-AG)], and the proteins involved in endocannabinoid synthesis, transformation and transit. Two key enzymes that inactivate endocannabinoids and terminate their signaling are fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MGL). Although FAAH can inactivate 2-AG, it is mainly responsible for degrading anandamide. MGL, along with other MGL-like esterases such as ABHD6 and ABHD12 [1], predominately catalyzes 2-AG hydrolysis in vivo [2-4]. FAAH has been molecularly characterized [5], and a 3D crystal structure has been determined [6]. FAAH knockout and transgenic mouse models have been developed [7, 8], and potent, selective FAAH inhibitors have been reported [9-11]. Far less is known about MGL. Although an experimentally derived structure of MGL is currently unavailable, sequencing, mutational and inhibition data have afforded initial insight into the structural features of MGL. The core tertiary structure of lipases is an α/β hydrolase fold [12], which consists of a core of β sheets surrounded by α helices, with a highly conserved active-site GXSXG motif. MGL also possesses the common lipase HG-dipeptide motif [13]. Like most lipases, MGL is thought to have a helical domain, or lid, covering the active site [13]. Site-directed mutagenesis experiments have confirmed the catalytic triad as S122, D239 and H269 [14]. Previously, a comparative model of MGL based on chloroperoxidase L from Streptomyces lividans was reported [15]. However, the authors describe the model as a crude estimate, and following a virtual screening procedure they themselves were unable to identify any MGL inhibitors [15].
Anandamide is a partial agonist primarily selective for CB1, whereas 2-AG is a potent agonist at both CB1 and CB2 [16, 17]. By inhibiting catalytic 2-AG inactivation by MGL and thereby potentiating tissue 2-AG tone to therapeutic levels, MGL inhibitors have the potential to treat pain, stress-related disorders, and neurodegenerative diseases [18-21]. The selectivity of such an agent for MGL versus FAAH and the cannabinoid receptors is critical, since truly selective MGL inhibition would potentiate endocannabinoid-system activity only at sites where 2-AG is being produced, including those sites where 2-AG production is stimulated to protect against a pathological insult. Furthermore, a selective MGL inhibitor may avoid any adverse motor and psychotropic side-effects such as those associated with nonselective cannabinoid-receptor agonists. Only recently has the high-yield bacterial expression and single-step purification of human MGL been reported [22], along with the proteomic characterization of MGL’s active site [23]. An accurate 3D MGL structure is not available, and as yet just one potent and selective MGL inhibitor, JZL184, has been described [24].
Four general classes of small molecules inhibit MGL [13]: nonspecific serine hydrolase inhibitors; 2-AG and 1-AG substrate analogs; de novo inhibitors such as AM6701 [23, 25] (an isomer of LY2183240 [26]), URB602 [20], and JZL184 [24]; and agents such as maleimides that target essential sulfydryl groups. Of these, members of the de novo inhibitor class have generated particular interest. LY2183240, initially characterized as an anandamide transport inhibitor with analgesic activity in rodents [26], was later shown to target FAAH and other brain serine hydrolases including MGL [27]. LY2183240 consists of two isomers [25] the less polar of which, 5-((biphenyl-4-yl)methyl)-N,N-dimethyl-2H-tetrazole-2-carboxamide (AM6701; MGL inhibition IC50 = 0.9 nM), is the more potent MGL inhibitor [23, 25]. Proteomic analysis of AM6701-treated MGL has shown that inhibition involves carbamylation of the active site serine (S122) [23]. URB602 has been reported as a moderately active MGL inhibitor (IC50 = 75 μM [28] or 28 μM [20], depending on the MGL source), although its selectivity for MGL versus FAAH is a matter of debate [29, 30]. It has been shown that MGL inhibition by URB602 proceeds via a rapid, noncompetitive, and partially reversible mechanism, suggesting that URB602, unlike AM6701, does not covalently modify MGL [31]. JZL184 is a recently reported, potent MGL inhibitor (IC50 = 6nM) identified through competitive activity-based protein profiling methods. JZL184 exhibits selectivity for MGL over FAAH, ABHD6, CB1, CB2, diacylglycerol lipase-α and diacylglycerol lipase-β and is believed to inhibit MGL via a covalent mechanism of inactivation [24].
Here, we present a fully refined homology model of human MGL together with docking poses of 2-AG, AM6701 and JZL184. The model is robust under a 5-ns molecular dynamics (MD) simulation in explicit water, and normal-mode analysis of these simulations provides insight into the conformational transitions necessary for MGL function. A virtual screen of a modest database verifies the binding site structure, with known MGL inhibitors being preferentially identified over druglike noninhibitors. Knowledge of the specific make-up of the MGL binding site and the overall fold of the enzyme will greatly assist the development of novel, selective MGL inhibitors with pharmacotherapeutic potential.
Methods
Homology modeling
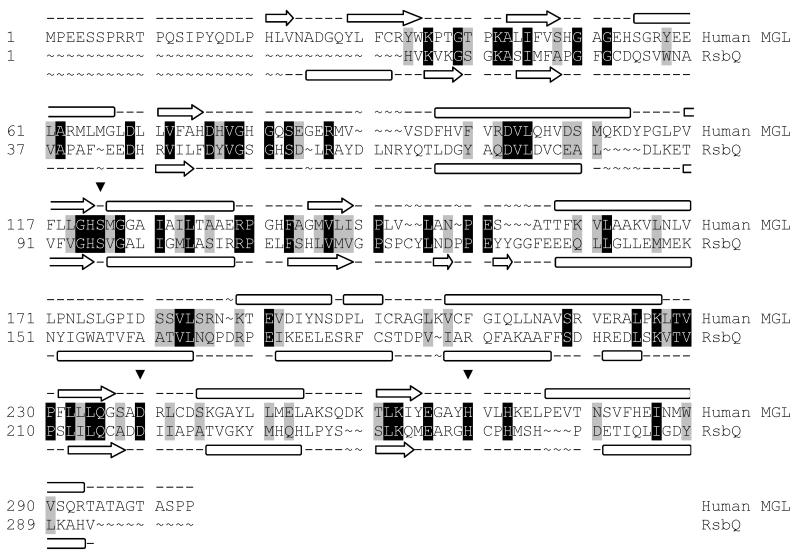
The sequence for human MGL was taken from the SWISS-PROT protein sequence database (primary accession number Q99685). The refined model of MGL was constructed using the native crystal structure of RsbQ from Bacillus subtilis (PDB ID: 1WOM) [32] as a template in prime [33]. The template used in this work affords higher sequence similarity and fewer gaps when compared to the MGL sequence than the template used in the previously reported model [15]. An initial BLAST alignment between the two sequences was adjusted by taking secondary structure into account using SSpro and PSIPRED [34]. This alignment was further refined manually to account for tertiary structure (19% identity, 35% homology, 10% gaps; Fig 1). After construction of the initial model, the loops between helix A and sheet 4, sheet 4 and helix B, helix B and sheet 5, sheet 6 and helix D’1, helix D’1 and helix D’2, helix D’2 and helix D’3, helix D’3 and helix D’4, helix E and sheet 8, and sheet and helix F were refined using an ab initio loop prediction algorithm (see Fig 2 for naming). The loop refinement step deletes the loop and reconstructs it from a backbone dihedral library; the loop is then exhaustively sampled to identify the lowest energy conformation. All other loops featured mainly homologous residues and contained no gaps or insertions. The protein underwent a truncated-Newton energy minimization, using the OPLS_2000 all-atom force field and a Generalized Born continuum solvation model.
Fig. 1.
Modeling alignment for the sequences of human MGL (Q99685) and RsbQ from Bacillus subtilis (PDB ID: 1WOM). Aligned query and template residues that are identical are highlighted in black; conserved residues (according to the BLOSUM62 scoring matrix), in grey. The predicted secondary structure for MGL and the experimentally determined secondary structure for RsbQ are shown above and below the sequence, respectively. β sheets are denoted by an arrow, and α helices by a block. The residues of the catalytic triad are marked with an inverted triangle (▼)
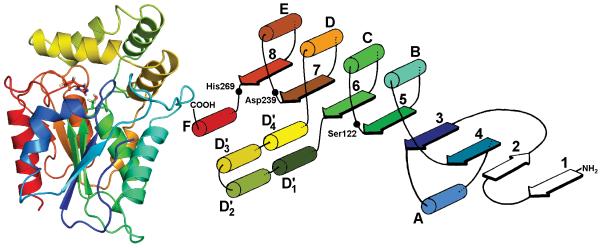
Fig. 2.
Tertiary structure and schematic diagram of MGL. The catalytic triad S122, D239 and H269 is shown in stick representation. The enzyme takes theα/β hydrolase fold, with four helices (D’1-D’4) forming a lid covering the active site. Due to lack of sequence homology, a structure prediction prior to Y34 is not possible. The coloring of the structure is equivalent for both representations
Molecular dynamics
To establish the stability of the model, a 5 ns MD simulation was performed on the MGL structure in an explicit water environment. For comparison, a similar simulation was carried out on the RsbQ crystal structure. The enzyme was solvated in a box of water extending at least 5 Å from the protein. The system was made electrically neutral by replacing two water molecules at the most positive electrostatic potential with two chloride ions (13 sodium ions required for RsbQ). Each system was minimized with steepest descents to relax unfavorable contacts between molecules. To achieve stability during production dynamics, equilibration MD was performed for 1 ns, with all heavy atoms in the enzyme positionally restrained with a force constant of 1000 kJ mol-1 nm-2. Unconstrained production MD was performed on each system for 5 ns.
All MD simulations were performed in the NPT ensemble and periodic boundary conditions. A temperature of 300 K was maintained by a Berenden-thermostat [35] with time constant 0.1 ps. Isotropic pressure coupling was used, with the reference pressure set at 1.0 bar and time constant 5 ps. Coulomb and short-range neighbor list cut-offs were both set to 0.9 nm, and Lennard-Jones cut-offs were set to 1.2 nm. The electrostatic interactions were computed with the Particle-Mesh Ewald (PME) method [36, 37], with an interpolation order of 4 and a maximum grid spacing of 0.12 nm. A time-step of 2 fs was used, and pair lists were updated every 10 steps. The LINCS algorithm [38] was used to preserve bond lengths. The GROMOS96 united-atom force field [39] and the simple point charge (SPC) water model [40] were used throughout. All simulations were carried out with the GROMACS program version 3.2.1 [41, 42].
Snapshots were taken every 100 ps during production MD (51 in total) and clustered into six bins using the atomic RMS between heavy atoms of residues within 6 Å of S122 with Xcluster [43]. Xcluster is an agglomerative hierarchical clustering method which uses the single-linkage method of calculating distances between clusters [44]. The bin leaders were snapshots from 0, 100, 500, 1200, 4500 and 4900 ps. Each snapshot represented a similar arrangement of amino acids and the key difference between bin leaders was the volume of the binding pocket. Movement of the lid region during MD affected the accessibility and magnitude of the binding site. Each bin leader was prepared as a docking receptor. 2-AG was docked to each receptor to elucidate the optimal conformation for structure-based drug design. The snapshot from 0 ps, which represented a more open binding site conformation, was the most appropriate for further docking studies. Principal component analysis was performed by Dynamite [42, 45, 46].
Docking
Ligands were prepared for docking using the LipPrep protocol [47] and the OPLS_2005 force field. For comparison, the ligands were also docked to FAAH (PDB ID:1MT5) [6]. Chain A from the crystal structure was used and the covalent ligand, methyl arachidonyl fluorophosphonate, was removed. Hydrogens were then added and minimized to yield the catalytic triad (S241, S217, K142) in the pre-inhibition orientation. Ligands were docked using the extra precision (XP) procedure in Glide [48].
Virtual screen
A database of 1000 decoy ligands was taken from the Glide enrichment studies [49, 50] and was seeded with 2-AG and 12 known MGL inhibitors from the literature [2, 20, 23-25, 28, 51-56]. The full set of structures and affinity data are provided in the Supporting Information. The full database of 1013 compounds was docked using the XP procedure in Glide [48]. The successfully docked compounds were sorted based upon the Glide scores. This ranked list was then used to generate enrichment and receiver operating characteristic (ROC) curves. An enrichment curve represents the proportion of active molecules retrieved from a certain fraction of the screened compounds, whereas, a ROC curve illustrates both sensitivity (ability to identify true positives) and selectivity (ability to avoid false positives). The area under the ROC curve (AUC) quantifies the enrichment value of the model, an AUC value greater than 0.9 is considered excellent whereas a value below 0.5 would be no better than random.
Results and discussion
MGL structure
MGL takes the α/β hydrolase fold commonly found in lipases (Fig. 2). Due to a lack of sequence similarity prior to Y34 of MGL, the two N-terminus β-sheets (sheets 1 and 2) are not present in the model. The catalytic triad residues (S122, D239, H269) are ideally positioned to increase S122 nucleophility. The GHSMG pentapeptide motif (residues 120-124) is highly conserved in the lipase family (GXSXG), and this sharp γ turn positions the catalytic S122 optimally between H269 and the binding pocket. The HG dipeptide (H49-G50) is located in the loop between sheet 3 and helix A, which flanks the binding pocket. In this position the amide backbones of the HG dipeptide can stabilize the oxyanion intermediate formed during the hydrolysis reaction. The four α-helices, D’1-D’4 (T158-V217), form a lid which covers the catalytic triad.
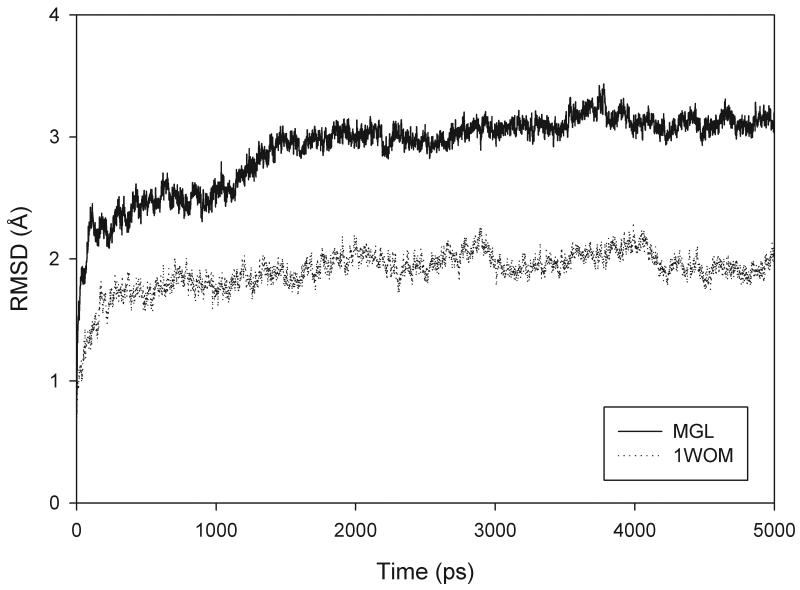
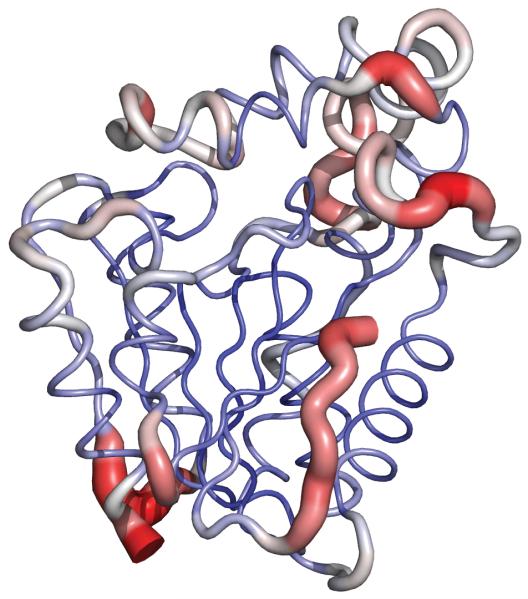
The MD simulation of the MGL model was stable, with both the energy of the system and the temperature remaining constant throughout the simulation. The root-mean-squared deviation (RMSD) of the Cα atoms to the model was calculated for MGL and similarly to the crystal structure for the simulation of RsbQ (Fig. 3). During the 5-ns of production MD, MGL remains well folded and stable. Although the RMSD for the MGL model is higher than that of RsbQ, inspection of the trajectory showed that the largest fluctuations were at the N- and C-termini, the hinge region of the lid, and the loop between helix E and sheet 8. The core, including the loops containing the catalytic triad and the residues comprising the binding pocket, exhibited very little deviance during the simulations. The relative rigidity of the MGL core during MD justifies use of the MGL model for docking experiments (Fig. 4).
Fig. 3.
Plot reflecting the CαRMSD fluctuations between the equilibrium structure of MGL (solid line) and template structure of RsbQ (PDB ID 1WOM; dotted line) and respective trajectory snapshots versus time
Fig. 4.
Average backbone structure for MGL snapshots during molecular dynamics simulations. A red, wider tube indicates greater RMSD across the trajectory, whereas a narrow blue tube shows greater stability
Close inspection of the binding site during MD showed that without a ligand present, the lid (T158-V217) closes down upon the active site, narrowing the binding cavity and eventually preventing access to the catalytic serine. It is possible that interfacial activation could cause the lid to open, allowing the ligand access to the active site [13, 29]. The ‘closed’ state does not present a viable conformation for structure-based drug design.
Principal component analysis
By reducing the dimensionality of the MD trajectory, it is possible to describe the dynamic motions of the MGL structure. The first three PCA modes accounted for 65% of the total dynamics, with the motion dominated by the first eigenvector (50%). This describes the movement of the lid and hinge region, as MGL undergoes a breathing-type motion. Although the amplitude of motion is relatively small (in the 1 Å range), a clear lid opening and closing is apparent. Accompanying this domain motion is a swinging action of K255-T260 (the loop between helix E and sheet 8). There are close steric interactions between this loop and W289 of helix F, and these appear to drive a pumping motion of this helix, which is partially responsible for the placement of catalytic H269.
MGL binding site
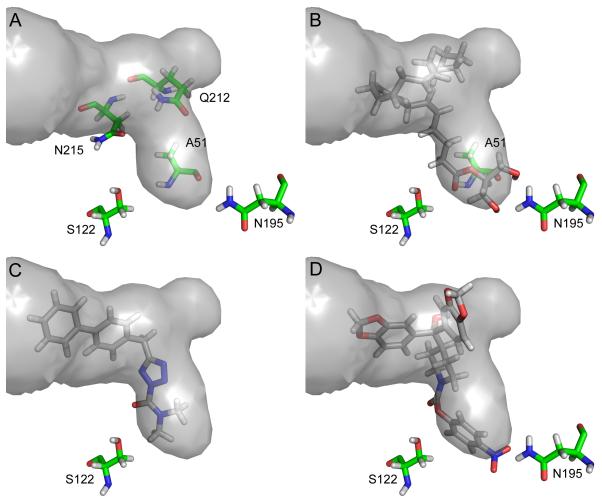
The MGL binding site is a single hydrophobic channel, consisting of an L-shaped main channel with a sub-pocket at the turn (Fig. 5a). The main channel measures ~ 11 Å to the bend, where it widens from a radius of ~ 2.5 Å to ~ 3.5 Å. The catalytic S122 is positioned ~ 3.6 Å from the hydrophilic closed end. In addition to S122, the backbone of A51 and the amine side chain group of N195 provide hydrogen-bonding opportunities at the closed end of the binding site. N215 and Q212 positioned near the bend in the binding channel also present opportunities for hydrogen bonding. N215 accepts hydrogen bonds through its side-chain carbonyl, and Q212 both donates hydrogen bonds through its amide side chain and accepts them through its backbone carbonyl. Several aromatic residues are within 6 Å of the binding pocket. H121, H272, and catalytic H269 are near the closed end of the binding pocket, and F209 is in the lid region.
Fig. 5.
The MGL binding pocket is shown in gray, and the predicted binding modes of 2-AG (b), AM6701 (c) and JZL184 (d) are shown in stick representation. Residues that may be important for hydrogen bonding are shown in stick representation (green carbons). Design of more potent and selective MGL inhibitors could be facilitated by taking advantage of hydrogen-bonding opportunities or by occupying the sub-pocket
Docking of 2-AG and known MGL inhibitors
The head group of 2-AG occupies the hydrophilic region of the binding pocket described by H121, N195 and D180. From above, on the lid side of the pocket, two hydrogen bonds are formed from Hδ1 and Hδ2 of N195 to the oxygens of the hydroxyl groups. From below, the carbonyl backbone oxygen of A51 forms bifurcated hydrogen bonds to the hydroxyl groups of 2-AG. A hydrogen bond formed between the amide backbone of A51 and the carbonyl of 2-AG helps to position the substrate for nucelophilic attack from S122. The aliphatic tail of 2-AG lies in the hydrophobic region of the binding pocket described by in part by A51, L148, A151, V161, I179, V183, L199, A216 and L241. Although the arachidonoyl chain tail is highly flexible with many rotatable bonds the binding channel confines the magnitude of possible conformations. In the top pose 2-AG assumes the binding pocket shape, with the tail curling back on itself to occupy the sub-pocket characterized by A164 and K165 (Fig. 5b). 2-AG did not result in a docking pose to FAAH at the XP level (2-AG weakly inhibits FAAH by 37% at 100mM [2]).
AM6701 conforms to the L shape of the binding site, and contacts of AM6701 with the binding site are similar to those seen with the 2-AG docking pose (Fig. 5c). However, the close contacts with A164 and K165 are lost as the sub-pocket is not occupied. Instead the biphenyl moiety, which extends further up the binding pocket, makes additional contacts with A156, T157 and K160. As expected, AM6701 also docked successfully to FAAH (FAAH IC50 = 33 nM [25]).
JZL184 represents a ligand with increased steric bulk over AM6701 and 2-AG. The extra bulk fills the binding site more completely and one 1,3-benzodioxole moiety makes additional contacts with the lid region of MGL. There is also a weak hydrogen bond from N195 to the p-nitro substituent (Fig. 5d). The relatively more narrow binding channels of FAAH create steric clash with JZL184, which precludes the ligand from achieving a docking pose to FAAH at the XP level (FAAH IC50 = 4 μM [24]).
Virtual screening results
The MGL homology model is able to preferentially identify MGL inhibitors over druglike noninhibitors. The screen showed that four of the known inhibitors (31%) were recovered with 6% of the false positives being identified. All the known inhibitors were identified in the top 49% of the database. Structures and affinity data for the known inhibitors are provided in the Supporting Information. The AUC was 0.73, which is fair but we believe that difficulties in database construction due to the paucity of potent, selective MGL inhibitors have negatively impacted the virtual screening evaluation. Many of the potent MGL inhibitors also inhibit other serine esterases with similar effect, indicating that the electrophilic character of the reactive moiety in the inhibitor is also significant. Additionally, of those inhibitors which showed more selective inhibition many were of low affinity (1 – 100 μM). Although many docking methods perform well for inhibitors in the low nanomolar range, it is common for performance to drop away as the activity decreases [57]. However, it is encouraging that the recently reported MGL selective inhibitor JZL184 (IC50 = 6 nM [24]) was ranked in the top 2% of screened compounds.
Toward MGL inhibitor selectivity
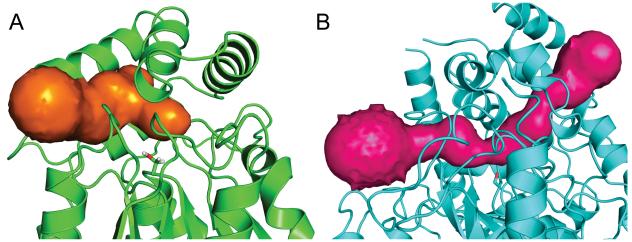
FAAH has both an acyl chain binding (ACB) and cytoplasmic access (CA) channel which narrow and meet at the catalytic S241, whereas MGL has a single binding channel (Fig. 6). The ability to increase steric bulk on either side of the substrate carbonyl allows for generation of selective FAAH inhibitors. Whereas the binding site of MGL cannot accommodate large moieties on both sides of the site for nucleophilic attack, an MGL inhibitor can be placed into one channel of FAAH. This structural difference may account for the lack of MGL-selective inhibitors relative to the potent, FAAH-selective inhibitors reported in the literature [58].
Fig. 6.
The binding channel of MGL (a) and binding channels of FAAH (b) calculated by CAVER [59]
Steric considerations could increase inhibitor selectivity for MGL. In FAAH, the ACB and CA channels narrow dramatically at the catalytic serine (~ 3 Å across); in MGL, the binding site at S122 is more open (~5 Å across). Hence, an increase of steric bulk around the site for nucleophilic attack may preclude FAAH interaction with the inhibitor. Occupation of the sub-pocket of MGL (as, for example, in the case of the 2-AG tail) may increase the hydrophobic contacts and thus the binding affinity of an inhibitor, restricting binding to FAAH. It is also important to maximize hydrogen-bonding interactions between the enzyme and substrate. The hydrogen-bonding motif created by A51 and N195 at the end of the MGL binding pocket cradles the 2-AG head group. If it were possible to exploit these interactions and those available from Q212 and N215, whether through rational modification of existing inhibitors or design of novel scaffolds, appreciable MGL selectivity should be attainable.
Conclusion
A refined homology model of MGL has been produced to help inform the discovery of potent and selective MGL inhibitors. Following its initial modeling, the enzyme underwent 5-ns of molecular dynamics in an explicit water environment. Analysis of the resulting structure showed MGL to take the α/β hydrolase fold, with four helices forming a lid covering the active site. The docking poses of the natural substrate 2-AG and de novo inhibitors AM6701 and JZL184 highlight opportunities for increased MGL affinity and selectivity. Catalytic S122 of MGL is positioned at the bottom of a single channel, whereas the catalytic triad of FAAH lies between two channels. Potent, MGL inhibitors with selectivity over FAAH may be realized by exploiting this structural difference as well as the binding sub-pocket, increased volume around the catalytic site, and potential hydrogen-bonding opportunities.
Supplementary Material
Acknowledgements
This work has been supported by grants from the National Institutes of Health, National Institutes on Drug Abuse: DA3801, DA07312 and DA000493. We thank Ioannis Karageorgos and Drs. Nikolai Zvonok, Lakshmipathi Pandarinathan, and David Janero for useful discussions.
References
- 1.Blankman JL, Simon GM, Cravatt BF. Chem Biol. 2007;14:1347. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghafouri N, Tiger G, Razdan RK, Mahadevan A, Pertwee RG, Martin BR, Fowler CJ. Br J Pharmacol. 2004;143:774. doi: 10.1038/sj.bjp.0705948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Proc Natl Acad Sci USA. 2002;99:10819. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinh TP, Kathuria S, Piomelli D. Mol Pharmacol. 2004;66:1260. doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- 5.Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Nature. 1996;384:83. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- 6.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Science. 2002;298:1793. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 7.Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Proc Natl Acad Sci USA. 2001;98:9371. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cravatt BF, Saghatelian A, Hawkins EG, Clement AB, Bracey MH, Lichtman AH. Proc Natl Acad Sci USA. 2004;101:10821. doi: 10.1073/pnas.0401292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero FA, Du W, Hwang I, Rayl TJ, Kimball FS, Leung D, Hoover HS, Apodaca RL, Breitenbucher JG, Cravatt BF, Boger DL. J Med Chem. 2007;50:1058. doi: 10.1021/jm0611509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mor M, Rivara S, Lodola A, Plazzi PV, Tarzia G, Duranti A, Tontini A, Piersanti G, Kathuria S, Piomelli D. J Med Chem. 2004;47:4998. doi: 10.1021/jm031140x. [DOI] [PubMed] [Google Scholar]
- 11.Ahn K, Johnson DS, Fitzgerald LR, Liimatta M, Arendse A, Stevenson T, Lund ET, Nugent RA, Nomanbhoy TK, Alexander JP, Cravatt BF. Biochemistry. 2007;46:13019. doi: 10.1021/bi701378g. [DOI] [PubMed] [Google Scholar]
- 12.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. Protein Eng. 1992;5:197. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 13.Viso A, Cisneros JA, Ortega-Gutierrez S. Curr Top Med Chem. 2008;8:231. doi: 10.2174/156802608783498032. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C. J Biol Chem. 1997;272:27218. doi: 10.1074/jbc.272.43.27218. [DOI] [PubMed] [Google Scholar]
- 15.Saario SM, Poso A, Juvonen RO, Jarvinen T, Salo-Ahen OMH. J Med Chem. 2006;49:4650. doi: 10.1021/jm060394q. [DOI] [PubMed] [Google Scholar]
- 16.Gonsiorek W, Lunn C, Fan XD, Narula S, Lundell D, Hipkin RW. Mol Pharmacol. 2000;57:1045. [PubMed] [Google Scholar]
- 17.Hillard CJ. Prostaglandins Other Lipid Mediat. 2000;61:3. doi: 10.1016/s0090-6980(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 18.Comelli F, Giagnoni G, Bettoni I, Colleoni M, Costa B. Br J Pharmacol. 2007;152:787. doi: 10.1038/sj.bjp.0707425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohmann AG. Br J Pharmacol. 2007;150:673. doi: 10.1038/sj.bjp.0707153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, Krey JF, Walker JM, Holmes PV, Crystal JD, Duranti A, Tontini A, Mor M, Tarzia G, Piomelli D. Nature. 2005;435:1108. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- 21.Saario SM, Laitinen JT. Basic Clin Pharmacol Toxicol. 2007;101:287. doi: 10.1111/j.1742-7843.2007.00130.x. [DOI] [PubMed] [Google Scholar]
- 22.Zvonok N, Williams J, Johnston M, Panarinathan L, Janero DR, Li J, Krishnan SC, Makriyannis A. J Proteome Res. 2008;7:2158. doi: 10.1021/pr700839z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zvonok N, Williams J, Johnston M, Panarinathan L, Karageorgos I, Janero DR, Krishnan SC, Makriyannis A. Chem Biol. 2008;15:854. doi: 10.1016/j.chembiol.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavón FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Nat Chem Biol. 2009;5:37. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortar G, Cascio MG, Moriello AS, Camalli M, Morera E, Nalli M, Di Marzo V. Eur J Med Chem. 2008;43:62. doi: 10.1016/j.ejmech.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Moore SA, Nomikos GG, Dickason-Chesterfield AK, Schober DA, Schaus JM, Ying BP, Xu YC, Phebus L, Simmons RMA, Li D, Iyengar S, Felder CC. Proc Natl Acad Sci USA. 2005;102:17852. doi: 10.1073/pnas.0507470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexander JP, Cravatt BF. J Am Chem Soc. 2006;128:9699. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]
- 28.Makara JK, Mor M, Fegley D, Szabo SI, Kathuria S, Astarita G, Duranti A, Tontini A, Tarzia G, Rivara S, Freund TF, Piomelli D. Nat Neurosci. 2005;8:1139. doi: 10.1038/nn1521. [DOI] [PubMed] [Google Scholar]
- 29.Saario SM, Laitinen JT. Chem Biodivers. 2007;4:1903. doi: 10.1002/cbdv.200790158. [DOI] [PubMed] [Google Scholar]
- 30.Vandevoorde S, Jonsson KO, Labar G, Persson E, Lambert DM, Fowler CJ. Br J Pharmacol. 2007;150:186. doi: 10.1038/sj.bjp.0706971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.King AR, Duranti A, Tontini A, Rivara S, Rosengarth A, Clapper JR, Astarita G, Geaga JA, Luecke H, Mor M, Tarzia G, Piomelli D. Chem Biol. 2007;14:1357. doi: 10.1016/j.chembiol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko T, Tanaka N, Kumasaka T. Protein Sci. 2005;14:558. doi: 10.1110/ps.041170005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prime version 1.6. Schrödinger, LLC; New York, NY: 2007. [Google Scholar]
- 34.Jones DT. J Mol Biol. 1999;292:195. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 35.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. J Chem Phys. 1984;81:3684. [Google Scholar]
- 36.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089. [Google Scholar]
- 37.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577. [Google Scholar]
- 38.Hess B, Bekker H, Berendsen HJC, Fraaije J. J Comput Chem. 1997;18:1463. [Google Scholar]
- 39.van Gunsteren WF, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG. Biomolecular Simulation: The GROMOS96 Manual and User Guide. vdf Hochschulverlag AG; Zürich: 1996. [Google Scholar]
- 40.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. In: Intermolecular Forces. Pullman B, editor. Reidel; Dordrecht: 1981. pp. 331–342. [Google Scholar]
- 41.Berendsen HJC, Vanderspoel D, Vandrunen R. Comput Phys Commun. 1995;91:43. [Google Scholar]
- 42.Lindahl E, Hess B, van der Spoel D. Journal of Molecular Modeling. 2001;7:306. [Google Scholar]
- 43.MacroModel XCluster version 9.5. Schrödinger, LLC; New York, NY: 2005. [Google Scholar]
- 44.Shenkin PS, McDonald DQ. J Comput Chem. 1994;15:899. [Google Scholar]
- 45.Barrett CP, Hall BA, Noble MEM. Acta Crystallogr D Biol Crystallogr. 2004;60:2280. doi: 10.1107/S0907444904019171. [DOI] [PubMed] [Google Scholar]
- 46.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 47.LigPrep version 2.2. Schrödinger, LLC; New York, NY: 2005. [Google Scholar]
- 48.Glide version 5.0. Schrödinger, LLC; New York, NY: 2008. [Google Scholar]
- 49.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47:1739. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 50.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. J Med Chem. 2004;47:1750. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 51.Goparaju SK, Ueda N, Yamaguchi H, Yamamoto S. FEBS Lett. 1998;422:69. doi: 10.1016/s0014-5793(97)01603-7. [DOI] [PubMed] [Google Scholar]
- 52.Muccioli GG, Labar G, Lambert DM. Chembiochem. 2008;9:2704. doi: 10.1002/cbic.200800428. [DOI] [PubMed] [Google Scholar]
- 53.Ortar G, Bisogno T, Ligresti A, Morera E, Nalli M, Di Marzo V. J Med Chem. 2008;51:6970. doi: 10.1021/jm800978m. [DOI] [PubMed] [Google Scholar]
- 54.Cisneros JA, Vandevoorde S, Ortega-Gutierrez S, Paris C, Fowler CJ, Lopez-Rodriguez ML. J Med Chem. 2007;50:5012. doi: 10.1021/jm070642y. [DOI] [PubMed] [Google Scholar]
- 55.Kapanda CN, Muccioli GG, Labar G, Draoui N, Lambert DM, Poupaert JH. Med Chem Res. 2009;18:243. doi: 10.1021/jm901323s. [DOI] [PubMed] [Google Scholar]
- 56.Saario SM, Savinainen JR, Laitinen JT, Jarvinen T, Niemi R. Biochem Pharmacol. 2004;67:1381. doi: 10.1016/j.bcp.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Charifson PS, Corkery JJ, Murcko MA, Walters WP. J Med Chem. 1999;42:5100. doi: 10.1021/jm990352k. [DOI] [PubMed] [Google Scholar]
- 58.Vandevoorde S. Curr Top Med Chem. 2008;8:247. doi: 10.2174/156802608783498005. [DOI] [PubMed] [Google Scholar]
- 59.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.