Abstract
Mitochondrial DNA (mtDNA) defects are an important cause of disease and may underlie aging and aging-related alterations 1,2. The mitochondrial theory of aging suggests a role for mtDNA mutations, which can alter bioenergetics homeostasis and cellular function, in the aging process 3. A wealth of evidence has been compiled in support of this theory 1,4, an example being the mtDNA mutator mouse 5; however, the precise role of mtDNA damage in aging is not entirely understood 6,7.
Observing the activity of respiratory enzymes is a straightforward approach for investigating mitochondrial dysfunction. Complex IV, or cytochrome c oxidase (COX), is essential for mitochondrial function. The catalytic subunits of COX are encoded by mtDNA and are essential for assembly of the complex (Figure 1). Thus, proper synthesis and function are largely based on mtDNA integrity 2. Although other respiratory complexes could be investigated, Complexes IV and II are the most amenable to histochemical examination 8,9. Complex II, or succinate dehydrogenase (SDH), is entirely encoded by nuclear DNA (Figure 1), and its activity is typically not affected by impaired mtDNA, although an increase might indicate mitochondrial biogenesis 10-12. The impaired mtDNA observed in mitochondrial diseases, aging, and age-related diseases often leads to the presence of cells with low or absent COX activity 2,12-14. Although COX and SDH activities can be investigated individually, the sequential double-labeling method 15,16 has proved to be advantageous in locating cells with mitochondrial dysfunction 12,17-21.
Many of the optimal constitutions of the assay have been determined, such as substrate concentration, electron acceptors/donors, intermediate electron carriers, influence of pH, and reaction time 9,22,23. 3,3'-diaminobenzidine (DAB) is an effective and reliable electron donor 22. In cells with functioning COX, the brown indamine polymer product will localize in mitochondrial cristae and saturate cells 22. Those cells with dysfunctional COX will therefore not be saturated by the DAB product, allowing for the visualization of SDH activity by reduction of nitroblue tetrazolium (NBT), an electron acceptor, to a blue formazan end product 9,24. Cytochrome c and sodium succinate substrates are added to normalize endogenous levels between control and diseased/mutant tissues 9. Catalase is added as a precaution to avoid possible contaminating reactions from peroxidase activity 9,22. Phenazine methosulfate (PMS), an intermediate electron carrier, is used in conjunction with sodium azide, a respiratory chain inhibitor, to increase the formation of the final reaction products 9,25. Despite this information, some critical details affecting the result of this seemly straightforward assay, in addition to specificity controls and advances in the technique, have not yet been presented.
Protocol
1. Tissue preparation for cryosectioning
Sacrifice the animal by either cervical dislocation or decapitation, in accordance with available ethical permit.
Quickly collect tissues of interest (e.g. brain), and rapidly freeze on dry ice (tissues may require freezing in isopentane or propane chilled with liquid nitrogen to obtain optimal morphology). Store tissues in aluminum foil at -80 °C until ready to section.
Embed frozen tissue in preparation for cryosectioning.
Collect 14-μm cryostat sections at -21 °C (may need to adjust temperature ± 1-2 °C). Thaw sections onto slides using heating block, and store slides without coverslipping at -20 °C until ready to use.
2. COX histochemistry
Allow slides to dry at room temperature for 1 hour. Put slides in a slide-staining chamber with wet filter paper, cut into strips. To obtain consistent results in each experiment, it is recommended to process a maximum of ten slides per experiment to minimize time delays.
Prepare under a chemical hood 1X DAB, 100 μM cytochrome c in 0.1 M PBS pH=7.0. Vortex quickly.
Add 2 μg bovine catalase (2 μg ml-1 or approximately 4 IU ml-1). Mix well by vortexing to break up all the grains of catalase.
Apply 150- 200 μL of incubation medium to each slide, use pipette tip to spread evenly onto all sections.
Incubate the slides for 40 minutes at 37 °C.
Remove excess solution from the slides. Wash slides 4 times, 10 minutes each time, in 0.1 M PBS pH=7.0.
Return slides to slide-staining chamber with wet paper strips.
3. SDH histochemistry
Prepare under a chemical hood 1.5 mM NBT, 130 mM sodium succinate, 0.2 mM PMS, and 1.0 mM sodium azide in 0.1 M PBS pH=7.0. Take caution to shield the PMS from light. Vortex quickly.
Apply 150-200 μL of incubation medium to each slide, use pipette tip to spread evenly onto all sections.
Incubate the slides for 40 minutes at 37 °C.
Remove excess solution from the slides. Wash slides 4 times, 10 minutes each time, in 0.1 M PBS pH=7.0.
Dehydrate the slides for 2 minutes in the following concentrations of ethanol: 70%, 70%, 95%, 95%, 99.5%. Then allow 10 minutes in an additional 99.5% step.
Place slides in xylene for 10 minutes. Mount with Entellan and coverslip. Allow the slides to dry overnight, or at least 1-2 hours in a ventilated area.
4. Determination of mitochondrial dysfunction
The amount of mitochondrial dysfunction is indicated by the amount of cellular blue staining. To semi-quantify these amounts, slides should be coded and visualized under bright-field microscopy. Semi-quantification should be performed on a blind basis using a scale, for example from 0-4 (0, no blue staining; 4, only blue staining). It is best to perform this kind of semi-quantification on several sections from a given subject/animal to calculate a mean value for each subject/animal.
Statistics should be performed using a non-parametric test, such as Mann-Whitney or Kruskal-Wallis.
5. Appropriate specificity controls
For specificity controls for COX activity, repeat "COX histochemistry" steps, and add 2.5 mM sodium azide, a terminal respiratory chain inhibitor.
For specificity controls for SDH activity, repeat "SDH histochemistry" steps with the removal of sodium succinate and the addition of 50 mM malonate, a competitive inhibitor of SDH.
Wash and dehydrate sections in an ethanol series, and then mount and coverslip the slides as described in steps 3.4 - 3.6.
6. Representative Results:
The overall scheme of the COX/SDH double-labeling histochemical assay is illustrated in Figure 2. Representative examples of appropriate COX/SDH double-labeling histochemistry in brain sections from wild-type and prematurely aging mtDNA mutator mice are shown in Figure 3. The dark brown staining in wild-type mice (Figure 3, left panel) showed normal COX activity. Cells with respiratory chain deficiencies, indicated by the blue staining, were revealed in 12 week-old mtDNA mutator mice, and these deficiencies became more widespread as mtDNA mutator mice aged to 46 weeks (Figure 3, center and right panel).
Examples of inappropriate COX/SDH double-labeling in brain sections from wild-type mice due to insufficient COX labeling are shown in Figure 4. Inadequate incubation time for the demonstration of COX activity, or reducing the availability of molecular oxygen by coverslipping the slide during incubation, resulted in a reduced deposition of the DAB reaction product, and thus allowed for formation of the blue formazan end product during the SDH incubation.
COX and SDH activities can also be investigated separately (Figure 5, left and center); however, the sequential labeling is helpful in identifying cells with COX deficiencies, due to the formation of the blue precipitate during the SDH incubation (Figure 3, center and right). Specificity controls for COX and SDH activities can also be done (Figure 5, right).
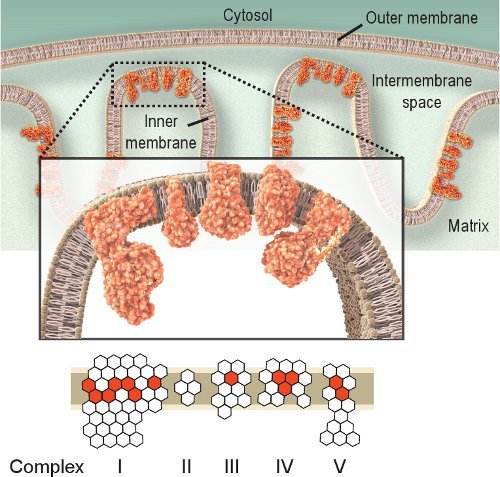 Figure 1.
Mitochondrial respiratory Complexes I-V. The mitochondrial respiratory chain is located within the inner membrane and includes five complexes. The purpose of the respiratory chain is to transport electrons from Complex I to IV and in doing so it creates a proton gradient across the inner membrane used by Complex V (ATPase) to produce ATP. Red hexagons represent subunits encoded by mtDNA. White hexagons represent subunits encoded by nuclear DNA (note that Complex II is completely encoded from the nuclear genome). Thus, mutations in the mitochondrial genome could cause dysfunction of the respiratory chain due to mutations in the subunits of the respiratory chain complexes.
Figure 1.
Mitochondrial respiratory Complexes I-V. The mitochondrial respiratory chain is located within the inner membrane and includes five complexes. The purpose of the respiratory chain is to transport electrons from Complex I to IV and in doing so it creates a proton gradient across the inner membrane used by Complex V (ATPase) to produce ATP. Red hexagons represent subunits encoded by mtDNA. White hexagons represent subunits encoded by nuclear DNA (note that Complex II is completely encoded from the nuclear genome). Thus, mutations in the mitochondrial genome could cause dysfunction of the respiratory chain due to mutations in the subunits of the respiratory chain complexes.
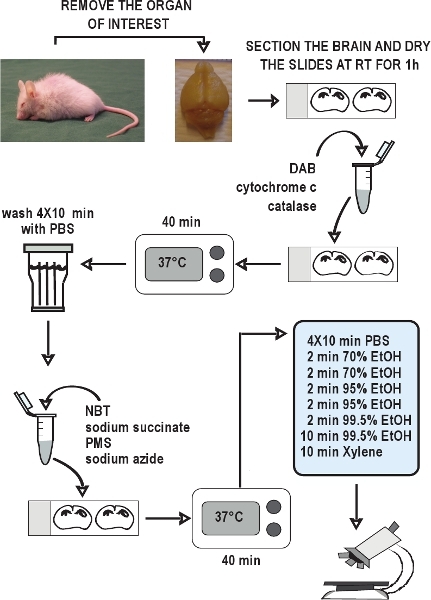 Figure 2.
Flow chart of the COX/SDH double-labeling histochemical assay. Dissect the organs of interest, rapidly freeze the tissues on dry ice, and store them at - 80 °C. Collect cryostat sections and keep at - 20 °C until use. Allow sections to air-dry at room temperature for 1 hour. Prepare the incubation medium for COX histochemistry, apply it to the slides, and incubate for 40 minutes at 37 °C. Wash the sections in PBS 4 times for 10 minutes each wash. Prepare the incubation medium for SDH histochemistry, apply it to the slides, and again incubate for 40 minutes at 37 °C. Wash the sections again in PBS, dehydrate in an ethanol series, and then mount and coverslip the slides. The COX/SDH double-labeled sections are ready to view under bright-field microscopy within 1-2 hours.
Figure 2.
Flow chart of the COX/SDH double-labeling histochemical assay. Dissect the organs of interest, rapidly freeze the tissues on dry ice, and store them at - 80 °C. Collect cryostat sections and keep at - 20 °C until use. Allow sections to air-dry at room temperature for 1 hour. Prepare the incubation medium for COX histochemistry, apply it to the slides, and incubate for 40 minutes at 37 °C. Wash the sections in PBS 4 times for 10 minutes each wash. Prepare the incubation medium for SDH histochemistry, apply it to the slides, and again incubate for 40 minutes at 37 °C. Wash the sections again in PBS, dehydrate in an ethanol series, and then mount and coverslip the slides. The COX/SDH double-labeled sections are ready to view under bright-field microscopy within 1-2 hours.
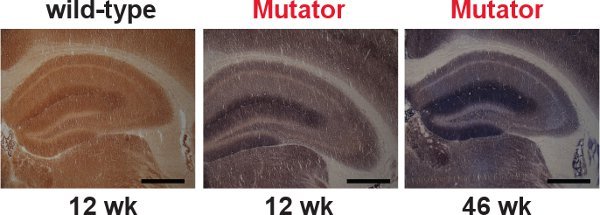 Figure 3.
Representative examples of COX/SDH double-labeling. Brain sections from wild-type and prematurely aging mtDNA mutator mice were sequentially labeled for COX and SDH activities. (Scale bar: 200 μm.) Normal COX activity (indicated by dark brown color) was shown in hippocampus from wild-type mice (left). COX deficiencies (indicated by blue color) were revealed in hippocampus from mtDNA mutator mice (center and right). There was a further decrease in COX activity by 46 weeks of age in mtDNA mutator mice, suggesting widespread exacerbation of respiratory chain dysfunction. The observed mitochondrial dysfunction in mtDNA mutator mice 12 is caused by high levels of mtDNA point mutations as well as increased levels of linear deletions 5.
Figure 3.
Representative examples of COX/SDH double-labeling. Brain sections from wild-type and prematurely aging mtDNA mutator mice were sequentially labeled for COX and SDH activities. (Scale bar: 200 μm.) Normal COX activity (indicated by dark brown color) was shown in hippocampus from wild-type mice (left). COX deficiencies (indicated by blue color) were revealed in hippocampus from mtDNA mutator mice (center and right). There was a further decrease in COX activity by 46 weeks of age in mtDNA mutator mice, suggesting widespread exacerbation of respiratory chain dysfunction. The observed mitochondrial dysfunction in mtDNA mutator mice 12 is caused by high levels of mtDNA point mutations as well as increased levels of linear deletions 5.
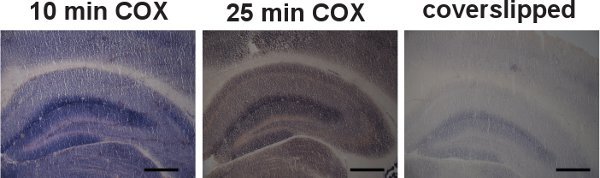 Figure 4.
Examples of inappropriate COX/SDH double-labeling. Brain sections from wild-type mice were sequentially labeled for COX and SDH activities. (Scale bar: 200 μm.) Inadequate incubation times (10 and 25 minutes) for the demonstration of COX activity resulted in a reduced deposition of the brown DAB reaction product, compared to the 40-minute incubation time (left and center). The shortened incubation times allowed for the formation of the blue formazan end product during the SDH incubation, misleadingly suggesting the presence of cells with COX deficiencies. Coverslipping the slides during the COX incubation also resulted in inaccurate formation and deposition of the DAB reaction product (right).
Figure 4.
Examples of inappropriate COX/SDH double-labeling. Brain sections from wild-type mice were sequentially labeled for COX and SDH activities. (Scale bar: 200 μm.) Inadequate incubation times (10 and 25 minutes) for the demonstration of COX activity resulted in a reduced deposition of the brown DAB reaction product, compared to the 40-minute incubation time (left and center). The shortened incubation times allowed for the formation of the blue formazan end product during the SDH incubation, misleadingly suggesting the presence of cells with COX deficiencies. Coverslipping the slides during the COX incubation also resulted in inaccurate formation and deposition of the DAB reaction product (right).
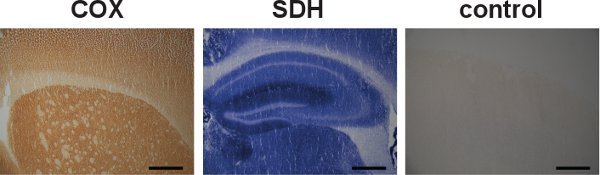 Figure 5. Individual COX and SDH labeling and specificity control. Brain sections from wild-type mice were separately labeled for COX and SDH activities, indicated by the dark brown color and the blue color, respectively (left and center). Although COX and SDH activities can be individually labeled, the sequential labeling has proved to be advantageous in locating cells with mitochondrial dysfunction. An example of a specificity control for COX and SDH activities in brain from a wild-type mouse showed absence of labeling (right). (Scale bar: 200 μm.)
Figure 5. Individual COX and SDH labeling and specificity control. Brain sections from wild-type mice were separately labeled for COX and SDH activities, indicated by the dark brown color and the blue color, respectively (left and center). Although COX and SDH activities can be individually labeled, the sequential labeling has proved to be advantageous in locating cells with mitochondrial dysfunction. An example of a specificity control for COX and SDH activities in brain from a wild-type mouse showed absence of labeling (right). (Scale bar: 200 μm.)
Discussion
The combined COX/SDH histochemical method enables the visualization of cells with mitochondrial dysfunction. This technique, with early studies dating back to 1968, remains popular, with many considering it the "gold standard" for identifying mitochondrial diseases in patients14,19,26,27. It is now frequently used to investigate mtDNA mutation-driven aging and aging-related disorders 12,13,18,20,21,24. The COX/SDH double-labeling method is often used in parallel with other techniques to identify specific mtDNA mutations and to further investigate the mitochondrial respiratory enzymes, such as oximetric measurements and spectrophotometric enzyme analysis 28,29.
In spite of its long-standing use, important details, simple specificity controls, and advances to improve the method have not yet been presented. In regards to handling of the tissue, it is important to use fresh-frozen tissue with this technique, because although COX will survive the formalin fixation, SDH will not. Although this is a limitation of the technique, no post-mortem delay has been found to interfere with this method 24, but it is suggested to quickly remove the organs of interest to preserve morphology. However, continuous freeze-thaw cycles of the tissues and sections should be avoided. Tissue samples may be frozen on dry ice; however, some tissues may require freezing in isopentane or propane cooled with liquid nitrogen to obtain optimal morphology and to avoid freezing artifacts. The appropriate thickness of the cryostat sections, depending on tissue-type, should also be determined. It is recommended to collect between 8 - 14-μm sections, thin enough for penetration of the solutions but thick enough to preserve anatomical features after dehydration. The suggested cryostat temperature of -21 °C may need to be adjusted ± 1-2 °C. If the specimen is too cold, the section may curl, if it is too warm, the section may stick to the knife. With regard to handling of the slides, the use of a grease pen, which is often beneficial in keeping the incubation medium on the slide, should be avoided. Instead, use 150- 200 μl of incubation medium per slide, depending on the number of sections. The use of a coverslip during incubation should also be avoided until the slide is ready to be mounted, because molecular oxygen must be present during the COX labeling step (Figure 4). Lastly, it is suggested to use the commercially available DAB solution (e.g. Sigma), a safer alternative to preparing DAB solution from the crystalline solid. DAB solution can also be made from available tablets, but inconsistent COX labeling results were found when this tablet-based DAB solution was used.
To obtain consistent and reliable results from this biochemical assay, the following points are recommended. Use freshly prepared PBS and ethanol in each experiment. Prepare stock solutions of cytochrome c, NBT, PMS (shield from light), sodium succinate, sodium azide, and malonate in 0.1 M PBS pH=7.0, adjusting the pH to 7.0 with 0.1 M HCl or 0.1 M NaOH. Aliquot and store the stock solutions at -20 °C, and rapidly thaw the solutions just prior to use. To obtain consistent results in each experiment, it is recommended to process a maximum of ten slides per experiment to minimize time delays. There is some variability between experiments; therefore, sections from each subject/animal should be repeated at least three-to-four times, and appropriate controls should be used in each experiment. Lastly, as the incubation times do influence the amount of COX-labeling (Figure 4, left and middle panels) and SDH-labeling, it is not recommended to vary the incubation times by more than 5% (2 mins).
The combined COX/SDH protocol presented here is based on principles previously described 9,22,23,25. As with many technqiues, variations of this protocol, such as excluding the addition of PMS and sodium azide, using an aqueous mountant, and varying incubation and rinse times, do exist and may work equally as well.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by the National Institute of Aging (AG04418), National Institute on Drug Abuse, National Institute of Health-Karolinska Institutet Graduate Partnerships Program, Karolinska Institutet, Swedish Research Council, Swedish Brain Power, and Swedish Brain Foundation. Many thanks to Mattias Karlen and Dr. Giuseppe Coppotelli for creative support with Figure 1 and 2, respectively; Karin Pernold for technical assistance; and Drs. Barry J. Hoffer, Lars Olson, and Nils-Göran Larsson for much helpful advice and discussion.
References
- Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- Cottrell DA. Role of mitochondrial DNA mutations in disease and aging. Ann. NY Acad. Sci. 2000;908:199–207. doi: 10.1111/j.1749-6632.2000.tb06647.x. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria. J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial genetics - a paradigm for aging and degenerative diseases. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- Trifunovic A. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Ameur A. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011;7:e1002028–e1002028. doi: 10.1371/journal.pgen.1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- DiMauro S, Bonilla E, Zeviani M, Nakagawa M, DeVivo DC. Mitochondrial myopathies. Ann. Neurol. 1985;17:521–538. doi: 10.1002/ana.410170602. [DOI] [PubMed] [Google Scholar]
- Old SL, Johnson MA. Methods of microphotometric assay of succinate dehydrogenase and cytochrome c oxidase activities for use on human skeletal muscle. Histochem. J. 1989;21:545–555. doi: 10.1007/BF01753355. [DOI] [PubMed] [Google Scholar]
- Chaturvedi RK. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum. Mol. Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar D. Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell. Metab. 2009;10:131–138. doi: 10.1016/j.cmet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Ross JM. High brain lactate is a hallmark of aging and caused by a shift in the lactate dehydrogenase A/B ratio. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20087–20092. doi: 10.1073/pnas.1008189107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crugnola V. Mitochondrial respiratory chain dysfunction in muscle from patients with amyotrophic lateral sclerosis. Arch. Neurol. 2010;67:849–854. doi: 10.1001/archneurol.2010.128. [DOI] [PubMed] [Google Scholar]
- Nonaka I. Muscle pathology in cytochrome c oxidase deficiency. Acta. Neuropathol. 1988;77:152–160. doi: 10.1007/BF00687425. [DOI] [PubMed] [Google Scholar]
- DiMauro S. Mitochondrial encephalomyopathies. Neurol. Clin. 1990;8:483–506. [PubMed] [Google Scholar]
- Bonilla E. New morphological approaches to the study of mitochondrial encephalomyopathies. Brain. Pathol. 1992;2:113–119. doi: 10.1111/j.1750-3639.1992.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Brierley EJ, Johnson MA, Lightowlers RN, James OF, Turnbull DM. Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann. Neurol. 1998;43:217–223. doi: 10.1002/ana.410430212. [DOI] [PubMed] [Google Scholar]
- Borthwick GM, Johnson MA, Ince PG, Shaw PJ, Turnbull DM. Mitochondrial enzyme activity in amyotrophic lateral sclerosis: implications for the role of mitochondria in neuronal cell death. Ann. Neurol. 1999;46:787–790. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Gellerich FN. Mitochondrial respiratory rates and activities of respiratory chain complexes correlate linearly with heteroplasmy of deleted mtDNA without threshold and independently of deletion size. Biochim. Biophys. Acta. 2002;1556:41–52. doi: 10.1016/s0005-2728(02)00305-5. [DOI] [PubMed] [Google Scholar]
- Larsson NG. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman AM, Karnovsky MJ, Wasserkrug HL, Hanker JS. Nondroplet ultrastructural demonstration of cytochrome oxidase activity with a polymerizing osmiophilic reagent, diaminobenzidine (DAB) J. Cell. Biol. 1968;38:1–14. doi: 10.1083/jcb.38.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowitz V, Brooke M. Saunders; 1973. Muscle Biopsy: A Modern Approach. [Google Scholar]
- Cottrell DA. Cytochrome c oxidase deficient cells accumulate in the hippocampus and choroid plexus with age. Neurobiol. Aging. 2001;22:265–272. doi: 10.1016/s0197-4580(00)00234-7. [DOI] [PubMed] [Google Scholar]
- Blanco CE, Sieck GC, Edgerton VR. Quantitative histochemical determination of succinic dehydrogenase activity in skeletal muscle fibres. Histochem. J. 1988;20:230–243. doi: 10.1007/BF01747468. [DOI] [PubMed] [Google Scholar]
- Moraes CT, Ricci E, Bonilla E, DiMauro S, Schon EA. The mitochondrial tRNA(Leu(UUR)) mutation in mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes (MELAS): genetic, biochemical, and morphological correlations in skeletal muscle. Am. J. Hum. Genet. 1992;50:934–949. [PMC free article] [PubMed] [Google Scholar]
- Petruzzella V. Extremely high levels of mutant mtDNAs co-localize with cytochrome c oxidase-negative ragged-red fibers in patients harboring a point mutation at nt 3243. Hum. Mol. Genet. 1994;3:449–454. doi: 10.1093/hmg/3.3.449. [DOI] [PubMed] [Google Scholar]
- Tulinius MH, Holme E, Kristiansson B, Larsson NG, Oldfors A. Mitochondrial encephalomyopathies in childhood. I. Biochemical and morphologic investigations. J. Pediatr. 1991;119:242–250. doi: 10.1016/s0022-3476(05)80734-6. [DOI] [PubMed] [Google Scholar]
- Haas RH. The in-depth evaluation of suspected mitochondrial disease. Mol. Genet. Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


