Abstract
B cells start their life with low affinity antibodies generated by V(D)J recombination. However, upon detecting a pathogen, the variable (V) region of an immunoglobulin (Ig) gene is mutated approximately 100,000-fold more than the rest of the genome through somatic hypermutation (SHM), resulting in high affinity antibodies1,2. In addition, class switch recombination (CSR) produces antibodies with different effector functions depending on the kind of immune response that is needed for a particular pathogen. Both CSR and SHM are initiated by activation-induced cytidine deaminase (AID), which deaminates cytosine residues in DNA to produce uracils. These uracils are processed by error-prone forms of repair pathways, eventually leading to mutations and recombination1-3.
Our current understanding of the molecular details of SHM and CSR come from a combination of studies in mice, primary cells, cell lines, and cell-free experiments. Mouse models remain the gold standard with genetic knockouts showing critical roles for many repair factors (e.g. Ung, Msh2, Msh6, Exo1, and polymerase η)4-10. However, not all genes are amenable for knockout studies. For example, knockouts of several double-strand break repair proteins are embryonically lethal or impair B-cell development11-14. Moreover, sometimes the specific function of a protein in SHM or CSR may be masked by more global defects caused by the knockout. In addition, since experiments in mice can be lengthy, altering expression of individual genes in cell lines has become an increasingly popular first step to identifying and characterizing candidate genes15-18.
Ramos – a Burkitt lymphoma cell line that constitutively undergoes SHM – has been a popular cell-line model to study SHM18-24. One advantage of Ramos cells is that they have a built-in convenient semi-quantitative measure of SHM. Wild type cells express IgM and, as they pick up mutations, some of the mutations knock out IgM expression. Therefore, assaying IgM loss by fluorescence-activated cell scanning (FACS) provides a quick read-out for the level of SHM. A more quantitative measurement of SHM can be obtained by directly sequencing the antibody genes.
Since Ramos cells are difficult to transfect, we produce stable derivatives that have increased or lowered expression of an individual gene by infecting cells with retroviral or lentiviral constructs that contain either an overexpression cassette or a short hairpin RNA (shRNA), respectively. Here, we describe how we infect Ramos cells and then use these cells to investigate the role of specific genes on SHM (Figure 1).
Protocol
1. Preparing samples and cells
Perform a medium scale preparation of DNA (midiprep) of the overexpression or shRNA-containing constructs and the retroviral packaging vector pKat2 or the lentiviral packaging vectors pVSV-G, pMDLg/pRRE, and pRSV-Rev25. Use 6 μg DNA for each construct and either 4 μg of pKat2 (for retroviral infections) or 1.5 μg DNA for each of the pVSV-g, pMDLg/pRRE, and pRSV-Rev packaging vectors (for lentiviral infections).
Prepare BOSC 23 media. For transfection, use both unamended Dulbecco′s modified Eagle′s medium (DMEM) as well as complete DMEM. To make complete DMEM, add 1% HEPES buffer, 1% penicillin-streptomycin-glutamine (PGS), and 10% newborn calf serum (NCS) to a bottle of DMEM.
Prepare Ramos media. For the infection step, make complete RPMI-1640 medium (RPMI) by adding 1% HEPES, 1% PGS, and 10% fetal bovine serum (FBS) to a bottle of RPMI. For single cell seeding step, make "conditioned" media by growing Ramos cells in complete RPMI for 2 days, spinning down the cells down at 400 X g for 4 minutes and filtering the supernatant with a 0.2 μm filter into a sterile bottle. Like other complete media, conditioned media is stored at 4°C until needed.
Split 3 x 106 BOSC 23 cells into a 100-mm plate in 10 mL complete DMEM the day before transfection so that the cells will be at 50-60% confluency the day of transfection. Make one plate of cells for each vector you plan on transfecting. It is important that the cells have not been kept in culture for too long; use low passage number cells or recently thawed cells for both the transfection and infection steps to insure a good infection.
Include controls for your experiments. For transfections and infections, create a negative control (untransfected or uninfected cells) and a positive control (virus that expresses GFP and puromycin-resistance gene). For shRNA knock-down experiments, infect a well with a scrambled or irrelevant shRNA control.
2. Transfecting BOSC 23 cells
Warm the bottles of unamended DMEM and complete DMEM at 37°C.
Remove media from the BOSC 23 cells and replace with 5 mL of complete DMEM. Return the cells to a 37°C incubator until use in step 2.6.
Warm the bottle of FuGENE 6 at room temperature for 5 minutes then briefly vortex (<1 second).
Aliquot 600 μL unamended DMEM into a 1.5 mL microcentrifuge tube.
Dilute 30 μL FuGENE 6 into the 600 μL unamended DMEM, vortex briefly (<1 second), and then incubate at room temperature for five minutes. Be careful to pipette directly into the media. Do not let the FuGENE 6 touch the sides of the tube.
Add 6 μg construct and either 4 μg pKat2 (retroviral infections) or 1.5 μg each of pVSV-G, pMDLg/pRRE, and pRSV-Rev (lentiviral infections) to the DMEM/FuGENE 6 mix, vortex briefly (<1 second), and then incubate at room temperature for 25 minutes.
Add the DNA/DMEM/FuGENE 6 mix to the BOSC 23 cells and return to a 37°C incubator for 24 hours.
After 24 hours, add 5 mL complete DMEM to the transfected cells and return to a 37°C incubator for another 24 hours.
3. Harvesting viral particles
Harvest the 10 mL of media from the transfected BOSC 23 cells with a 10 mL syringe.
Attach a 0.45 μm filter to the end of the syringe and filter the media into a clean 15 mL polypropylene tube.
Use the viral media immediately or freeze as 1 mL aliquots in 2 mL cryogenic vials at -80°C.
After harvesting the viral media, confirm that efficient transfection has occurred in the positive control cells by FACS. This can also be done for all samples if the construct contains either a fluorescent (e.g. GFP) or a cell-surface marker (e.g. Thy1).
4. Infecting B cells
Seed 2 mL of 2 x 105 cells/mL Ramos cells in complete RPMI into a 6-well plate 24 hours before infection. Again, be sure to use low passage number cells or recently thawed cells to insure good infection.
If using frozen aliquots of virus, thaw the viral media quickly at 37°C.
Mix 1 mL viral media with 3 μL of 10 mg/mL polybrene. The final concentration of polybrene should be 10 μg/mL in a total volume of 3 mL.
Add the virus/polybrene mix to the cells and mix well by pipetting. As mentioned earlier, one well should be used as an uninfected control; add 1 mL complete RPMI and 3 μL polybrene to this well instead of 1 mL virus. Be sure to infect one well with a matched negative control. We also infect one well with a GFP-containing vector, to determine infection efficiency by FACS.
Centrifuge the 6-well plate at 3000 X g for 90 minutes at room temperature.
Return the plate to a 37°C incubator for 48 hours.
After 48 hours, spin down the infected cells in a 15 mL polypropylene tube at 400 X g for 4 minutes at room temperature. Remove the media by aspiration, being sure to not disturb the cell pellet. Resuspend the cells in fresh RPMI.
Confirm the efficiency of infection in the positive control sample by FACS. As before, you can do this for all your samples if the construct contains either a fluorescent (e.g. GFP) or a cell-surface marker (e.g. Thy1).
Start selection of cells (if appropriate). Our shRNA-containing constructs all contain a puromycin resistance cassette. We select shRNA-infected Ramos cells with 0.25 μg/mL puromycin. We find that derivatives of Ramos cells sometimes show altered susceptibility to puromycin and, therefore, we perform kill curves to optimize the concentration of puromycin for each derivative. Also treat uninfected control cells with puromycin to confirm that selection is effective.
5. Single cell seeding
Single seeding can be performed in one of two ways: by FACS sorting or manually.
FACS: Use a FACS sorter with a plate adapter to seed 1 cell/well in a 96-well plate pre-filled with 100 μL of 40% RPMI/40% conditioned media/20% FBS. This method gives a lot more single cell clones per plate.
Manually: Count the cells. Dilute an aliquot of cells to 10 cells/mL in 40% RPMI/40% conditioned media/20% FBS. Use 10 mL of diluted cells for each seeded plate. Different clones will show variable recovery at this stage therefore it is useful to set up several plates at different concentrations (ranging from 3 to 20 cells/mL). Use the one that produces fewer outgrowths to avoid clones arising from multiple cells). Use a multichannel pipette to seed 96-well plates with 100 μL of diluted cells.
Place the plate in a 37°C incubator for 2 weeks.
Check individual wells under the microscope to confirm the presence of cells one day after seeding. Single cells can be tricky to find due to the need to focus at the correct depth. Focusing on the well numbers imprinted at the bottom of the 96-well plate should help. Cells are easier to find when seeded by the FACS sorter since most wells have a cell.
After 2 weeks, growing colonies can be barely seen by naked eye by looking at the plates from below. They are much easier to see under the microscope. Colonies tend to grow along the edges of the well and typically most colonies in a plate grow along the same edge in different wells. Mark wells with robust cell growth and transfer the cells to 24-well plates in 1 mL RPMI and return to a 37°C incubator.
Maintain cells at 1 x 105 to 1 x 106 cells/mL. Change the media by spinning the cells at 400 X g for 4 minutes at room temperature and removing the media as needed. Replace with 1 mL fresh RPMI.
After several days of growth, transfer the cells to a 6-well plate and continue growing in a 37°C incubator.
After 3 weeks, analyze the clones for IgM loss (as described below).
Continue to grow the cells for 2 more weeks and then analyze the clones again at the 5 week time point.
6. Analysis: IgM loss (3 weeks and 5 weeks)
Aliquot 5 x 105 cells from each well into a 1.5 mL microcentrifuge tube and spin the cells down at 400 X g for 4 minutes at room temperature. Use cells from each of the control and shRNA-knock-down wells, as well as one sample for an unstained control.
Wash the cells in 1 mL of PBS and then spin again at 400 X g for 4 minutes at room temperature.
Resuspend in 50 μL of PBS (unstained control) or PBS + 1:20 dilution of PE-labeled α-human IgM antibody. Incubate on ice for 1 hour.
Spin the cells down at 400 X g for 4 minutes at room temperature.
Wash the cells twice with 1 mL of PBS and spin for 400 X g for 4 minutes each time at room temperature.
Remove the supernatant and resuspend in 100 μL of PBS. Transfer the resuspended stained cells to a FACS tube.
Analyze the cells using a FACS machine.
Compare the percentage of IgM- cells from the controls vs. the shRNA knocked-down cells. As noted before, IgM loss as measured by FACS is a reliable semi-quantitative measure of SHM in Ramos cells.
7. Analysis: Confirm knock-down by quantitative RT-PCR (5 weeks)
Aliquot 3 x 106 cells from each well and spin down at 400 X g for 4 minutes at room temperature.
Prepare RNA from the aliquot of cells. We use TRIzol according to manufacturer's recommendations.
Prepare cDNA from the RNA prep. We use 2 μg RNA and Superscript II according to manufacturer's recommendations.
Perform a quantitative PCR using the cDNA to confirm knock-down. We use GAPDH as a normalization control, and the SYBR FAST qPCR kit in a 10 μL total reaction volume.
8. Analysis: Genomic sequencing (5 weeks)
Aliquot 5 x 106 cells from each well and spin down at 400 X g for 4 minutes at room temperature.
Isolate genomic DNA from these cells. We use the Wizard SV genomic DNA purification kit according to manufacturer's recommendations.
PCR amplify the genomic DNA for the Ig V genes19 (Table 1) and any other genes of interest using the KAPA HiFi DNA polymerase. We use this polymerase because of its low error-rates, but any high-fidelity PCR polymerase should be acceptable.
Gel purify the PCR products on a 1% agarose gel using the QIAquick gel extraction kit.
Some polymerases add a single deoxyadenosine (A) residue to the 3′ ends of the PCR products, while others do not. These 3′ A tails are necessary for the TOPO TA cloning step. The KAPA HiFi polymerase does not leave 3′ A tails. A-tail the PCR products by incubating 25 μL gel-purified PCR product with 3 μL 10x Taq reaction buffer, 1 μL 10 mM dNTPs, and 1 unit Taq polymerase at 72°C for 30 minutes.
Clone the gel-purified PCR products using the TOPO TA Cloning kit. Transform the products from the TOPO reaction into the provided E. coli DH5α-T1R competent cells and plate on LB/agar plates containing 100 μg/mL ampicillin and supplemented with 1.6 mg of X-gal according to manufacturer's recommendations.
Send individual colonies for sequencing.
Perform sequence analysis to determine types and locations of mutations.
9. Representative Results:
As noted before, we use a positive control viral vector that expresses both GFP as well as a puromycin-resistance gene. We typically see 50-75% GFP+ cells two days after transfection. In our infections, we typically see 50-70% cells are GFP+ two days after infection but before selection. After selection is complete, >95% of cells are GFP+.
As representative experiments, we show the effects of overexpressing AID or knocking down a repair factor in Ramos cells. Specifically, we transfected a retroviral overexpression vector for AID linked via an IRES site to Thy1.1 along with the retroviral packaging vector pKat2 into BOSC 23 cells. Virus-containing media was filtered and then used to infect Ramos cells. Both wild type (WT) and AID overexpressing (AIDhi) cells were single cell seeded and grown for 3 weeks, at which point they were analyzed for gene expression by qRT-PCR (Figure 2) and for IgM loss by FACS (Figure 3). For the FACS staining, the cells were incubated with both 1:20 diluted PE-labeled α-human IgM antibody as well as 1:400 diluted FITC-labeled α-rat CD90/mouse CD90.1 (Thy1.1) antibody. In a separate experiment, lentiviruses containing either an irrelevant shRNA (control) or an shRNA against a high-fidelity DNA repair factor expected to protect against SHM were made in BOSC 23 cells, and then infected into an AIDhi clone of Ramos cells. Again, gene expression (Figure 4) and IgM loss (Figure 5) were analyzed in single cell clones after 3 weeks. Since the repair factor is thought to provide protection against mutations during SHM, we see an increase in IgM loss and SHM in the absence of the factor. If we had knocked-down a factor involved in generating SHM-associated mutations, we would have seen a decrease in IgM loss instead.
As expected, our qRT-PCR results show a marked increase in AID expression following infection of cells with an AID overexpression vector (Figure 2), while levels of the DNA repair factor drop in the presence of a targeted shRNA against that factor (Figure 4). Because individual clones may vary, you might see some variation in gene expression levels. Because gene expression can vary, it is necessary to confirm the expression in each clone you plan on analyzing further. Different shRNAs against the same gene might have different effects on gene expression as well, so you should screen several shRNAs to determine which has the greatest impact. Knockdown may also be confirmed at the protein level by immunoblotting. Likewise, individual clones might vary greatly in levels of IgM loss. Some clones might demonstrate a "jackpot" effect, where an IgM mutation occurred in one of the earliest cell divisions - for example, you will see >50% IgM loss simply because one of the cells became IgM- at the two-cell stage. The influence of these clones is minimized by calculating the median for several single cell clones.
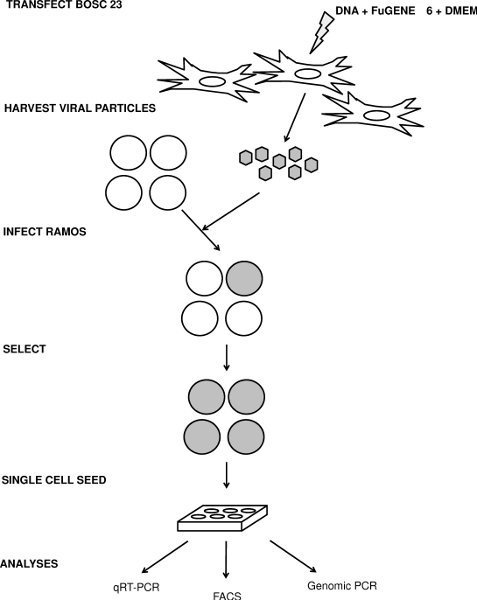 Figure 1. Schematic for the transfection, infection and analysis of IgM loss. See text for details.
Figure 1. Schematic for the transfection, infection and analysis of IgM loss. See text for details.
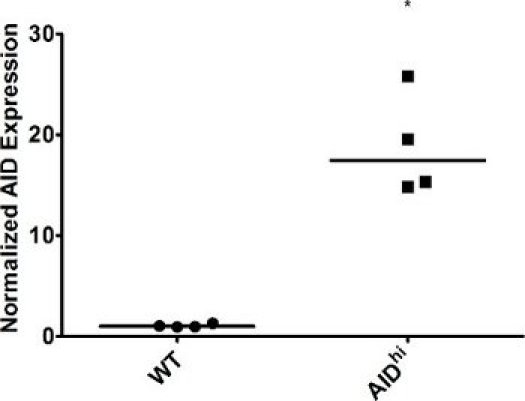 Figure 2. AID overexpression in Ramos cells. WT and AIDhi Ramos cells were single cell cloned and grown for 3 weeks, at which point AID expression in individual clones was measured by qRT-PCR. GAPDH expression was used for normalization. WT was set to 1. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student′s t-test.
Figure 2. AID overexpression in Ramos cells. WT and AIDhi Ramos cells were single cell cloned and grown for 3 weeks, at which point AID expression in individual clones was measured by qRT-PCR. GAPDH expression was used for normalization. WT was set to 1. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student′s t-test.
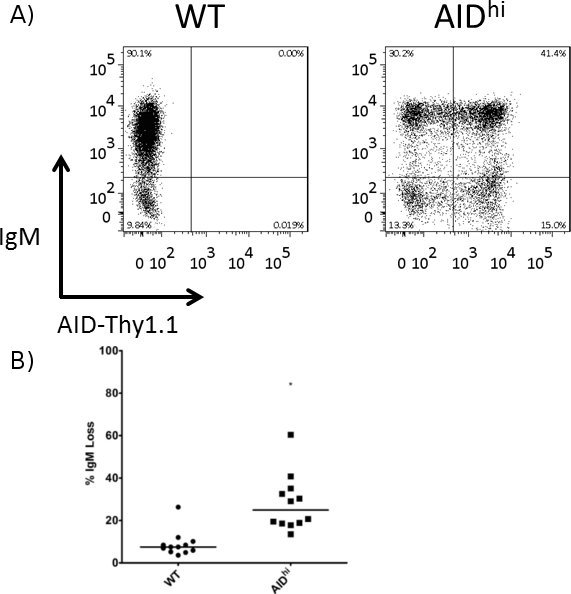 Figure 3. Increased IgM loss in AIDhi cells as compared to WT cells. Surface IgM was measured by FACS at the 3 week time point, and the percentage of IgM loss was calculated for WT and AIDhi cells. (A) Representative FACS plot of a WT clone and an AIDhi clone. The AID overexpression vector has AID linked to Thy1.1 via an IRES site, so Thy1.1 expression is used as a surrogate for AID expression. (B) Quantitation of IgM loss in several WT and AIDhi clones. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student's t-test.
Figure 3. Increased IgM loss in AIDhi cells as compared to WT cells. Surface IgM was measured by FACS at the 3 week time point, and the percentage of IgM loss was calculated for WT and AIDhi cells. (A) Representative FACS plot of a WT clone and an AIDhi clone. The AID overexpression vector has AID linked to Thy1.1 via an IRES site, so Thy1.1 expression is used as a surrogate for AID expression. (B) Quantitation of IgM loss in several WT and AIDhi clones. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student's t-test.
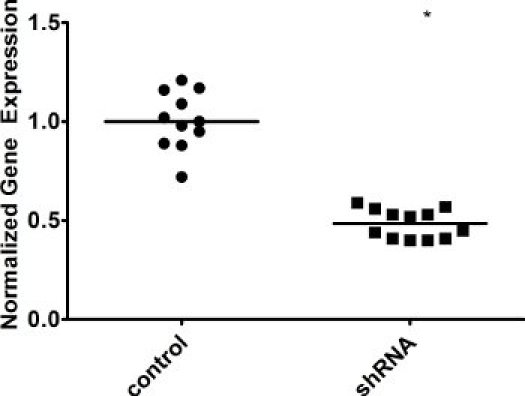 Figure 4. Knockdown of a repair factor in AIDhi cells. Cells were infected with either an irrelevant shRNA control (control) or our shRNA of interest (shRNA), and single cell seeded. After 3 weeks, gene expression in individual clones was measured by qRT-PCR. GAPDH expression was used for normalization. WT was set to 1. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student's t-test.
Figure 4. Knockdown of a repair factor in AIDhi cells. Cells were infected with either an irrelevant shRNA control (control) or our shRNA of interest (shRNA), and single cell seeded. After 3 weeks, gene expression in individual clones was measured by qRT-PCR. GAPDH expression was used for normalization. WT was set to 1. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student's t-test.
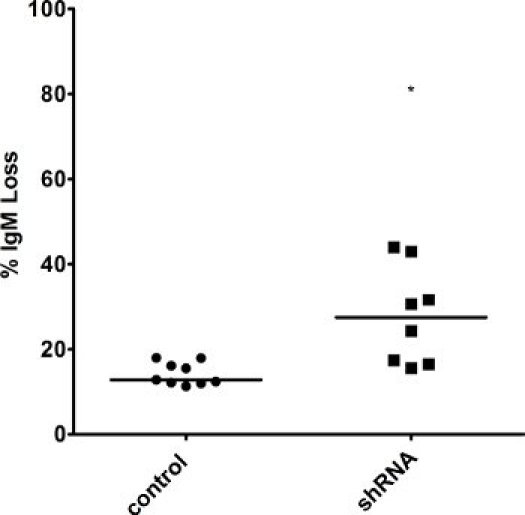 Figure 5. Increased IgM loss in AIDhi cells with knocked-down levels of a repair factor. Surface IgM in several clones was measured by FACS at the 3 week time point, and the percentage of IgM loss was calculated in cells still overexpressing AID. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student′s t-test.
Figure 5. Increased IgM loss in AIDhi cells with knocked-down levels of a repair factor. Surface IgM in several clones was measured by FACS at the 3 week time point, and the percentage of IgM loss was calculated in cells still overexpressing AID. The median value is indicated. * indicates a p value of <0.01, calculated as a one-tailed Student′s t-test.
 Table 1. Primer sequences for the Ig genes19: the constant region Cμ, the heavy chain V region (VH), and the light chain V region (VL).
Table 1. Primer sequences for the Ig genes19: the constant region Cμ, the heavy chain V region (VH), and the light chain V region (VL).
Discussion
As discussed previously, cell line models for antibody diversification have become a popular starting point to identify novel proteins that influence different steps during antibody diversification. We present here a method for using viral infection to either knock-down or overexpress proteins in the Ramos B-cell line and then examine the impact on SHM.
For these studies, we utilize both WT Ramos and AIDhi Ramos cells. WT cells can be used to study factors involved in targeting or expression of AID as well as repair of AID-generated lesions, while AIDhi cells exclude factors that affect AID expression.
It should also be noted that, SHM is measured here by assaying for loss of IgM from a population of cells that are initially IgM+. Alternatively, it is possible to start with IgM- cells and look for a gain of IgM+ cells in a reversion assay23.
By using different selection markers (e.g. puromycin, hygromycin, GFP, or Thy1.1), we can also perform multiple infections at the same time with the same protocol. However, we have found that when using two different antibiotic selection markers, the sensitivities of the cells change. It will be necessary to perform additional kill curves to optimize the conditions for selecting with two antibiotics at once. We can easily generate double knock-downs of two different factors or a knock-down of one factor plus an overexpression of another, etc.
We have also adapted this protocol for other B-cell-line models as well. For instance, with CH12F3-2 cells (a mouse B-cell line used to study CSR) 26,27, the same infection steps can be used to infect 1 x 106 CH12F3-2 cells seeded on the day of infection (variation of step 4.1). These cells are selected with 1 μg/mL of puromycin. Similarly, the same protocol can also be used for chicken DT40 cells - a cell line model for AID-initiated gene conversion.
Disclosures
No conflicts of interest declared.
Acknowledgments
The pMSCV-AID-I-Thy1.1 and pKat2 vectors were a kind gift from D. G. Schatz and the pVSV-G, pRSV-Rev, and pMDLg/pRRE vectors were a kind gift from B. R. Cullen.
References
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Peled JU. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr. Opin. Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Bardwell PD. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 2004;5:224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung QH. Increased hypermutation at G and C nucleotides in immunoglobulin variable genes from mice deficient in the MSH2 mismatch repair protein. J. Exp. Med. 1998;187:1745–1751. doi: 10.1084/jem.187.11.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Ehrenstein MR, Neuberger MS, Milstein C. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 1998;9:135–141. doi: 10.1016/s1074-7613(00)80595-6. [DOI] [PubMed] [Google Scholar]
- Rada C. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K. DNA polymerase eta is a limiting factor for A:T mutations in Ig genes and contributes to antibody affinity maturation. Eur. J. Immunol. 2008;38:2796–2805. doi: 10.1002/eji.200838502. [DOI] [PubMed] [Google Scholar]
- Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic. Acids. Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- Luo G. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7376–7381. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu U. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat. Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- Cumbers SJ. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nat. Biotechnol. 2002;20:1129–1134. doi: 10.1038/nbt752. [DOI] [PubMed] [Google Scholar]
- Papavasiliou FN, Schatz DG. Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- Parsa JY. AID mutates a non-immunoglobulin transgene independent of chromosomal position. Mol. Immunol. 2007;44:567–575. doi: 10.1016/j.molimm.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang W. Clonal instability of V region hypermutation in the Ramos Burkitt's lymphoma cell line. Int. Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- Ukai A. Induction of a:T mutations is dependent on cellular environment but independent of mutation frequency and target gene location. J. Immunol. 2008;181:7835–7842. doi: 10.4049/jimmunol.181.11.7835. [DOI] [PubMed] [Google Scholar]
- Dull T. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M. High frequency class switching of an IgM+ B lymphoma clone CH12F3 to IgA+ cells. Int. Immunol. 1996;8:193–201. doi: 10.1093/intimm/8.2.193. [DOI] [PubMed] [Google Scholar]
- Muramatsu M. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]


