Abstract
Androgen receptor activity drives incurable castrate-resistant prostate cancer. All approved antiandrogens inhibit androgen receptor-driven transcription, and in addition the second-generation antiandrogen MDV3100 inhibits ligand-activated androgen receptor nuclear translocation, via an unknown mechanism. Here, we report methoxychalcones that lock the heat shock protein 90-androgen receptor complex in the cytoplasm in an androgen-non-responsive state, thus demonstrating a novel chemical scaffold for antiandrogen development and a unique mechanism of antiandrogen activity.
Keywords: chalcone, antiandrogen, androgen receptor translocation, prostate cancer, Hsp90
Prostate cancer is one of the most common cancers in men and the second leading cause of cancer death in men in the United States. Although hormonal therapy is typically effective initially, resistance often develops and results in a state known as castration-resistant prostate cancer (CRPC).1 Recently it has been discovered that even in patients with castrate levels of androgen, prostate cancer remains dependent on androgen receptor (AR). Several AR reactivation mechanisms have been reported including overexpression of AR or AR coactivators, mutation of AR, indirect activation of AR by growth factors or cytokines, and aberrant phosphorylation of AR.1–5
Targeting AR signaling is a rational approach to the treatment of CRPC. The AR consists of an N-terminal regulatory domain (NTD), a DNA-binding domain (DBD), a hinge region and a C-terminal ligand-binding domain (LBD).4,5 In the absence of androgen, AR is held in the cytoplasm by heat shock protein 90 (Hsp90). Androgen binding to the LBD of AR results in a change of AR conformation and release of AR from Hsp90. AR translocates to the nucleus, binds to androgen response elements and AR target gene expression is activated.1,5 Prostate–specific antigen (PSA) and transmembrane protease serine 2 (TMPRSS2) are representative AR target genes. Several strategies have been reported to target AR in either a direct or indirect manner.6–13 These approaches include AR antagonists for reducing AR target gene expression,6–8 cytochrome P-450 CYP17A1 inhibitors for blocking androgen synthesis,14,15 and Hsp90 inhibitors for degrading AR protein.16 Recently, the antiandrogen MDV3100 has shown promising results in preclinical and clinical trials.17 MDV3100 inhibits AR translocation to the nucleus and binding of AR to androgen response elements. Chart 1 shows structures of androgen receptor agonists and antiandrogens.
Chart 1.
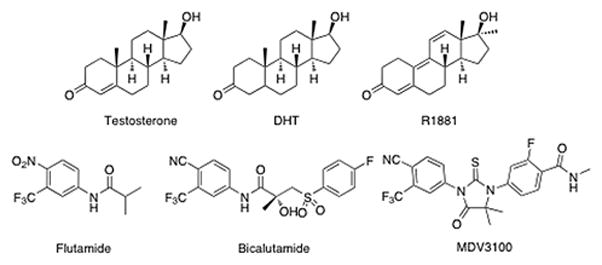
Structures of androgen receptor modulators.
Chalcones (trans-1,3-diphenyl-2-propene-1-ones) are α,β-unsaturated carbonyl compounds with two aromatic rings. They are intermediates in the biosyntheses of flavonoids and isoflavonoids and are abundant in plants. They are known to exhibit several biological activities such as anti-oxidant, anti-inflammatory, antiviral, antibacterial, antimalarial and anticancer activities.18–21 We describe here a novel mechanism of action of chalcone derivatives in the AR signaling pathway for the treatment of prostate cancer.
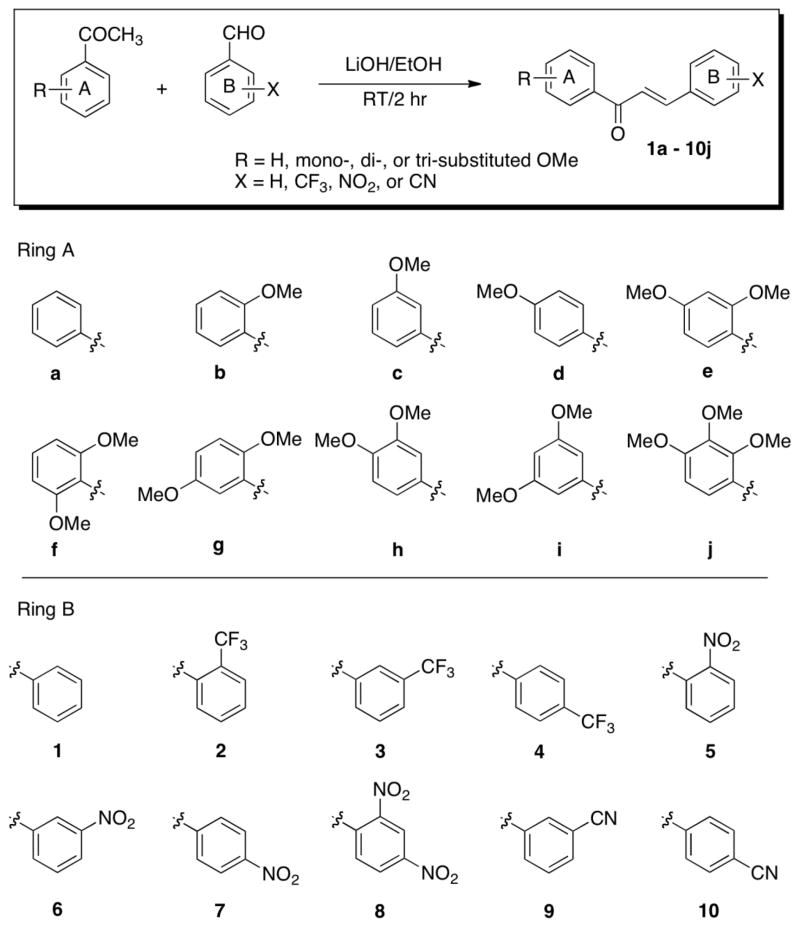
To determine the structure-activity relationship, 100 chalcone derivatives 1a–10j were synthesized in this study by condensation between substituted benzaldehyde and substituted acetophenone in the presence of LiOH in ethanol (Scheme 1). Once the synthesis of desired chalcones was achieved, their antiandrogen activity was screened using real-time reverse transcription-polymerase chain reaction (RT-PCR). The prostate cancer cell line LNCaP was treated with the test compounds at 2.5 μM for 20 hr and PSA mRNA levels were measured by real-time RT-PCR. The AR of LNCaP cells has a T877A point mutation in the ligand binding domain. This AR mutation, which has been observed in primary prostate cancer tissue of prostate cancer patients treated with the antiandrogen flutamide, has been shown to be associated with conversion of flutamide from an antagonist to an agonist.22,23 PSA is an androgen receptor target gene and inhibition of AR results in decreased PSA mRNA expression. We identified 24 potent compounds, which inhibited PSA mRNA expression over 80% at 2.5 μM (Table 1). Unsubstituted chalcones 1a–1j were found to be least active with maximum inhibition of only 73% in case of 1b. Also, the chalcone derivatives with cyano-substitution were inactive with the exception of compounds 10g and 10h, which showed 82 and 85% inhibition of PSA mRNA expression, respectively. As shown in Table 1, chalcone series b with an o-methoxy group, showed good activity when the B ring had a nitro- or trifluoromethyl- group. Among series b, compounds 2b and 5b showed the highest activity (over 94% inhibition of PSA mRNA expression) in which B ring had an o-trifluoromethyl or o-nitro group. It is also worth mentioning that the chalcone series f and j with two o-methoxy groups also showed significant inhibition, in particular, compounds 2f (87%), 6f (85%), 7f (90%), 8f (81%), 3j (81%), 6j (84%) and 7j (84%). Thus, in general, structure-activity relationship (SAR) studies revealed that chalcones with an o-methoxy group on A ring were highly active, suggesting that the o-methoxy substituted ring is important to enhanced activity.
Scheme 1.
Molecular structures of chalcones. Chalcones were prepared by the condensation of a substituted acetophenone and a substituted benzaldehyde in the presence of LiOH at room temperature.
Table 1.
PSA mRNA level of LNCaP cells treated with the corresponding chalcone*
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| a | 0.78 | 0.13 | 0.41 | 0.44 | 0.14 | 0.24 | 0.21 | 0.64 | 0.67 | 0.61 |
| b | 0.37 | 0.05 | 0.09 | 0.52 | 0.06 | 0.10 | 0.08 | 0.09 | 0.31 | 0.26 |
| c | 0.43 | 0.32 | 0.67 | 0.59 | 0.30 | 0.21 | 0.21 | 0.38 | 0.58 | 0.36 |
| d | 0.68 | 0.46 | 0.56 | 0.71 | 0.22 | 0.35 | 0.32 | 0.43 | 0.71 | 0.71 |
| e | 0.63 | 0.35 | 0.36 | 0.51 | 0.44 | 0.33 | 0.30 | 0.39 | 0.34 | 0.29 |
| f | 0.81 | 0.13 | 0.29 | 0.27 | 0.29 | 0.15 | 0.10 | 0.19 | 0.32 | 0.35 |
| g | 0.47 | 0.23 | 0.48 | 0.44 | 0.16 | 0.11 | 0.33 | 0.24 | 0.24 | 0.18 |
| h | 0.74 | 0.40 | 0.20 | 0.20 | 0.31 | 0.16 | 0.06 | 0.75 | 0.81 | 0.15 |
| i | 0.52 | 0.31 | 0.48 | 1.61 | 0.26 | 0.80 | 0.20 | 0.40 | 0.60 | 0.51 |
| j | 0.50 | 0.48 | 0.19 | 0.62 | 0.23 | 0.16 | 0.16 | 0.36 | 0.35 | 0.30 |
PSA mRNA level was measured in LNCaP cells treated with the corresponding compound at 2.5 μM for 20 hr using real time RT-PCR. The PSA mRNA level is relative to DMSO-treated control cells in which PSA mRNA level is 1 (mean values, n = 3).
Further dose-dependent inhibition of AR target gene expression was achieved using compound 5b in LNCaP cells (Figure 1a). Chalcone 5b inhibited PSA and TMPRSS2 mRNA expression 50% at 0.60 μM and 0.75 μM, respectively. Importantly, no agonistic effect was observed in the range 5–0.05 μM. Compound 5b inhibited the growth of LNCaP cells after 3 days treatment with an IC50 3.4 μM (Figure 1b).
Figure 1.
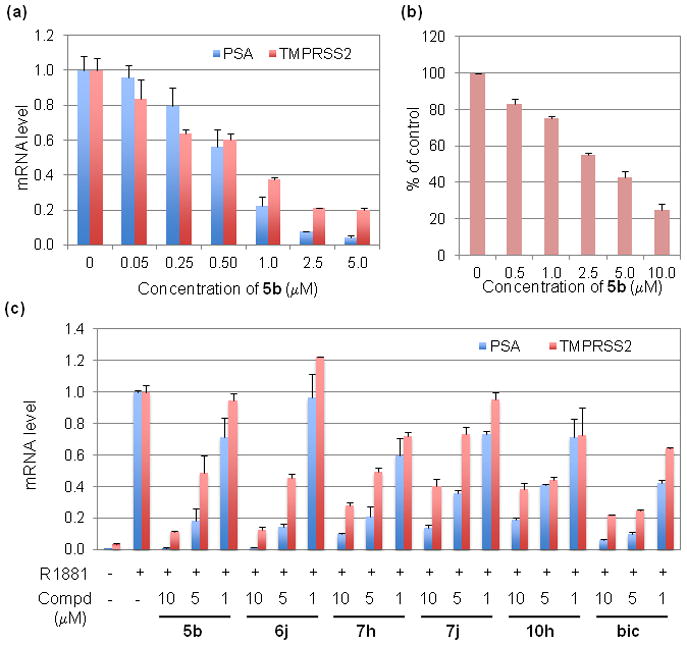
(a) Dose-dependent inhibition of PSA and TMPRSS2 mRNA expression by compound 5b at 20 hr. (b) Inhibition of LNCaP cell growth by compound 5b at day 3. (c) Inhibition of PSA and TMPRSS2 mRNA expression by compounds 5b, 6j, 7h, 7j, 10h, or bic in the presence of synthetic androgen R1881 at 20 hr. Test compounds were evaluated at 10, 5, or 1 μM and R1881 was used at 0.5 nM.
We then tested selected compounds for effect on AR target gene expression in the presence of the synthetic androgen R1881. LNCaP cells were incubated for 3 days in phenol red-free RPMI 1640 supplemented with 10% charcoal-stripped fetal bovine serum and 1% antimycotic-antibiotic solution, and then treated for 20 hr as indicated. The concentration of R1881 was 0.5 nM and that of the corresponding compounds was 10, 5 or 1 μM. As shown in Figure 1c, the AR target genes PSA and TMPRSS2 were induced about 500-fold and 30-fold, respectively, by R1881 compared with the DMSO control. R1881-induced gene expression was blocked by chalcones 5b, 6j, 7h, 7j, and 10h in a dose-dependent manner. Compound 5b (10 μM) inhibited PSA and TMPRSS2 mRNA expression 99% and 89% in the presence of 0.5 nM R1881 (Figure 1c). Although the activity of compound 6j was similar to that of compound 5b at high concentrations, it showed weak agonistic effect at 1 μM, which is not desirable. Thus we performed further analysis of the impact of compound 5b on androgen-regulated gene expression in LNCaP cells. We performed cDNA microarray analysis of LNCaP cells treated with 0.5 nM R1881 with/without 5 μM 5b for 20 hr, and Table 2 shows the fold-changes of gene expression compared to DMSO- treated control cells. Compound 5b inhibited expression of androgen-induced genes,24 including TMPRSS2, FK506 binding protein 5 (FKBP5), kallikrein-related peptidase 2 (KLK2), insulin-like growth factor 1 (IGF1) and chemokine (C-X-C motif) receptor 4 (CXCR4). The real time RT-PCR and cDNA microarray data demonstrate that compound 5b blocks AR target gene expression in LNCaP cells in the absence and presence of synthetic androgen R1881.
Table 2.
Changes of gene expression in LNCaP cells*
| Symbol | Gene description | R1881 | R1881 + 5b |
|---|---|---|---|
| TMPRSS2 | transmembrane protease, serine 2 | 22.7 | 7.0 |
| FKBP5 | FK506 binding protein 5 | 29.7 | 5.3 |
| HPGD | hydroxyprostaglandin dehydrogenase 15-(NAD) | 70.8 | 13.9 |
| KLK2 | kallikrein-related peptidase 2 | 423.8 | 159.3 |
| NDRG1 | N-myc downstream regulated 1 | 8.6 | 3.9 |
| IGF1 | insulin-like growth factor 1 | 19.0 | 5.0 |
| EAF2 | ELL associated factor 2 | 20.5 | 1.6 |
| CXCR4 | chemokine (C-X-C motif) receptor 4 | 7.1 | 2.7 |
| CXCL5 | chemokine (C-X-C motif) ligand 5 | 15.1 | 2.6 |
| CXCL9 | chemokine (C-X-C motif) ligand 9 | 18.7 | 3.4 |
| KCNMA1 | potassium large conductance calcium-activated channel, subfamily M, alpha member 1 | 5.1 | 2.5 |
| PHLDB2 | pleckstrin homology-like domain, family B, member 2 | 6.8 | 2.8 |
| SOCS2 | suppressor of cytokine signaling 2 | 6.4 | 2.3 |
| SRMS | src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites | 10.3 | 0.5 |
LNCaP cells were treated with DMSO or 5b (5 μM) in the absence or presence of synthetic androgen R1881 (0.5 nM) for 20 hr, cDNA microarray was performed using the GeneChip Human Genome U133 Plus 2.0 Array. The values shown represent fold-changes compared to DMSO-treated control cells.
Immunocytochemistry was performed to determine if compound 5b inhibits AR translocation to the nucleus. LNCaP cells were grown on cover slips and then were treated with DMSO, 10 μM bicalutamide (bic) or 10 μM 5b for 3 hr, followed by treatment with 0.5 nM R1881 for 3 hr. After staining, AR protein distribution in the cells was identified by immunofluorescence and image analysis. Figure 2 shows the fluorescence images of LNCaP cells in which green and blue color represents AR and nuclear DNA, respectively. Before treatment with R1881, AR is located in both the cytoplasm and nucleus in LNCaP cells. After incubation with R1881, bright nuclear AR staining was observed, due to translocation of AR to the nucleus. The approved antiandrogen bicalutamide did not inhibit androgen-stimulated AR translocation to the nucleus, as shown in the fluorescence image with bright nuclear AR staining and very weak cytoplasmic AR staining, consistent with previous reports.17 In contrast, compound 5b inhibited AR translocation significantly, resulting in the presence of AR in the nucleus and cytoplasm (Figure 2). In addition to the immunofluorescence technique, AR protein levels in the nucleus and cytoplasm were measured by subcellular fractionation, followed by western blot with anti-AR antibody. LNCaP cells were treated for 20 hr in the presence of 0.5 nM R1881 with or without 10, 5 and 1 μM compound 5b. Nuclear and cytoplasmic fractions were isolated and the AR protein level was measured by western blot. As shown in Figure 3a, compound 5b inhibited R1881-induced AR nuclear translocation in a dose-dependent manner. The results are consistent with immunocytochemistry data and real time RT-PCR data showing inhibition of AR target gene expression in response to compound 5b. To demonstrate the purity of the two subcellular fractions, topoisomerase II and tubulin were used as markers of the nucleus and the cytoplasm, respectively. For nuclear translocation to occur, AR must be released from association with cytoplasmic Hsp90.1,4 Using co-immunoprecipitation (IP) analysis we found that R1881-induced release of AR from Hsp90 was inhibited by compound 5b (Figure 3b). Furthermore R1881-induced release of AR from the immunophilin FKBP52 was also inhibited, consistent with arrest of AR in a cytoplasmic Hsp90-AR complex,25 a mechanism of anti-AR activity distinct from all approved antiandrogens.
Figure 2.
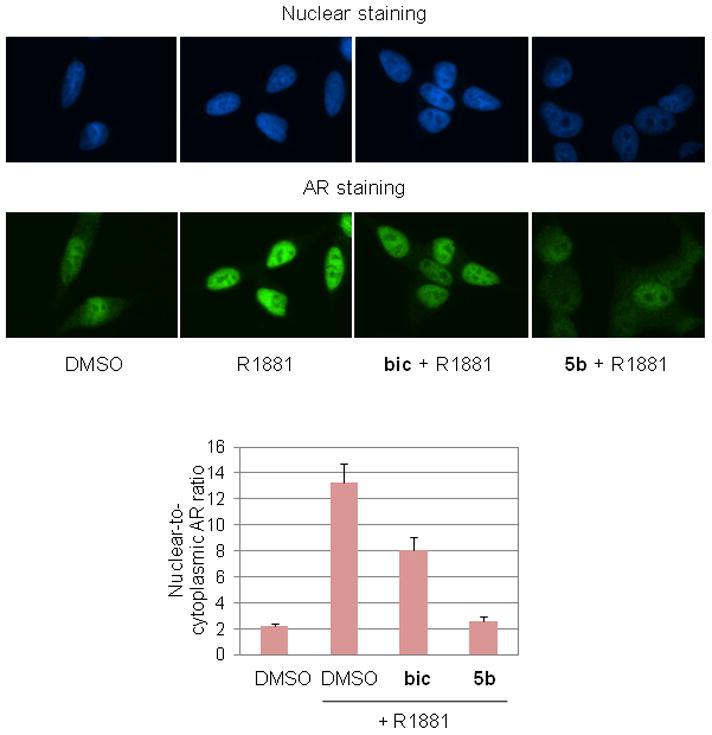
Inhibition of R1881-induced AR translocation to the nucleus. LNCaP cells were grown on cover slips and then were treated with DMSO, bic (10 μM) or 5b (10 μM) for 3 hr, followed by treatment with R1881 (0.5 nM) for 3 hr. Fluorescence images show nuclear DNA (blue color) or AR protein (green color) in LNCaP cells. Nuclear-to-cytoplasmic AR ratio was measured using Openlab software.
Figure 3.
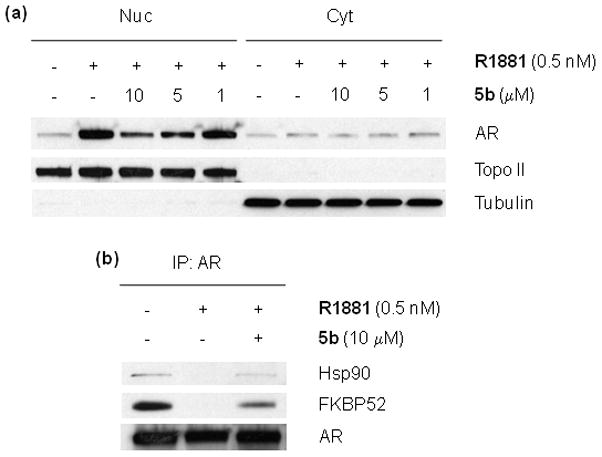
(a) AR protein level in nuclear and cytoplasmic fractions. (b) Co-immunoprecipitation (IP) was performed with anti-AR antibody and western blots were performed with anti-Hsp90 and anti-FKBP52 antibodies.
In conclusion, we have synthesized, screened and identified a series of antiandrogen chalcones in LNCaP cells. As demonstrated by real time RT-PCR, cDNA microarray, western blot and co-IP data, lead compound 5b, (E)-1-(2-methoxyphenyl)-3-(2-nitrophenyl)prop-2-en-1-one, inhibits AR translocation to the nucleus as well as AR target gene expression by locking the AR-Hsp90 complex in the cell cytoplasm in an androgen-nonresponsive state.
Supplementary Material
Acknowledgments
V.K. and S.V.M. would like to thank the National Cancer Institute (NCI) Developmental Therapeutics Program. This project has been funded in whole or in part with federal funds from the NCI, National Institutes of Health, to the Intramural Research Program (Y.S.K., A.I., S.L., L.N., and J.B.T.), and to DTP NCI-Frederick SAIC, under Contract No. HSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Feldman BJ, Feldman D. Nature Rev Cancer. 2001;1:34. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 2.Attar RM, Takimoto CH, Gottardis MM. Clin Cancer Res. 2009;15:3251. doi: 10.1158/1078-0432.CCR-08-1171. [DOI] [PubMed] [Google Scholar]
- 3.Attard G, Cooper CS, de Bono JS. Cancer Cell. 2009;16:458. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Knudsen KE, Scher HI. Clin Cancer Res. 2009;15:4792. doi: 10.1158/1078-0432.CCR-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dehm SM, Tindall DJ. Mol Endocrinol. 2007;21:2855. doi: 10.1210/me.2007-0223. [DOI] [PubMed] [Google Scholar]
- 6.Gao W, Bohl CE, Dalton JT. Chem Rev. 2005;105:3352. doi: 10.1021/cr020456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohler ML, Bohl CE, Jones A, Coss CC, Narayanan R, He Y, Hwang DJ, Dalton JT, Miller DD. J Med Chem. 2009;52:3597. doi: 10.1021/jm900280m. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Su L, Geng J, Liu J, Zhao G. ChemMedChem. 2010;5:1651. doi: 10.1002/cmdc.201000259. [DOI] [PubMed] [Google Scholar]
- 9.Knudsen KE, Penning TM. Trends Endocrinol Metab. 2010;21:315. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longo DL. N Engl J Med. 2010;363:479. doi: 10.1056/NEJMe1006300. [DOI] [PubMed] [Google Scholar]
- 11.Shepard DR, Raghavan D. Nature Rev Clin Oncol. 2010;7:13. doi: 10.1038/nrclinonc.2009.187. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Clegg NJ, Scher HI. Lancet Oncol. 2009;10:981. doi: 10.1016/S1470-2045(09)70229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Sawyers CL, Scher HI. Curr Opin Pharmacol. 2008;8:440. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labrie F. Nature Rev Urol. 2011;8:73. doi: 10.1038/nrurol.2010.231. [DOI] [PubMed] [Google Scholar]
- 15.Yap TA, Carden CP, Attard G, de Bono JS. Curr Opin Pharmacol. 2008;8:449. doi: 10.1016/j.coph.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Lattouf JB, Srinivasan R, Pinto PA, Linehan WM, Neckers L. Nature Clin Pract Urol. 2006;3:590. doi: 10.1038/ncpuro0604. [DOI] [PubMed] [Google Scholar]
- 17.Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, Higano CS, Beer TM, Hung DT, Scher HI, Jung ME, Sawyers CL. Science. 2009;324:787. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batovska DI, Todorova IT. Curr Clin Pharmacol. 2010;5:1. doi: 10.2174/157488410790410579. [DOI] [PubMed] [Google Scholar]
- 19.Dimmock JR, Elias DW, Beazely MA, Kandepu NM. Curr Med Chem. 1999;6:1125. [PubMed] [Google Scholar]
- 20.Go ML, Wu X, Liu XL. Curr Med Chem. 2005;12:481. doi: 10.2174/0929867053363153. [DOI] [PubMed] [Google Scholar]
- 21.Kontogiorgis C, Mantzanidou M, Hadjipavlou-Litina D. Mini Rev Med Chem. 2008;8:1224. doi: 10.2174/138955708786141034. [DOI] [PubMed] [Google Scholar]
- 22.Gottlieb B, Beitel LK, Wu JH, Trifiro M. Hum Mutat. 2004;23:527. doi: 10.1002/humu.20044. [DOI] [PubMed] [Google Scholar]
- 23.Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek SR, Jr, Weinmann R, Einspahr HM. Proc Natl Acad Sci USA. 2001;98:4904. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont KR, Tindall DJ. Adv Cancer Res. 2010;107:137. doi: 10.1016/S0065-230X(10)07005-3. [DOI] [PubMed] [Google Scholar]
- 25.De Leon JT, Iwai A, Feau C, Garcia Y, Balsiger HA, Storer CL, Suro RM, Garza KM, Lee S, Kim YS, Chen Y, Ning YM, Riggs DL, Fletterick RJ, Guy RK, Trepel JB, Neckers LM, Cox MB. Proc Natl Acad Sci USA. 2011;108:11878. doi: 10.1073/pnas.1105160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.