Abstract
Objectives
DNA methylation is known to play a critical role in regulating development of placental morphology and physiology. The methylation of genes mediated by glucocorticoid hormones may be particularly vulnerable to intrauterine stress in the placenta. However little is known about DNA methylation of stress-related genes within a healthy placenta, and particularly whether methylation occurs uniformly across different regions of the placenta, which is a critical question for researchers seeking to analyze methylation patterns. We examined DNA methylation across four regions of the placenta to evaluate methylation levels of stress-related genes within a healthy placenta, and to evaluate whether methylation patterns vary by sampling location.
Study Design
We evaluated levels of DNA methylation of three stress-related genes: NR3C1, BDNF, and 11B-HSD2 and of the repetitive element, LINE-1, in four different sample locations of 20 healthy placentas.
Main Outcome Measures
Pyrosequencing was used to quantify levels of methylation at CpG sites within the promoter regions of each of the three stress-related genes, and global methylation of LINE-1.
Results
Very low levels of methylation were found across all three stress-related genes; no gene showed a median methylation level greater than 4.20% across placental regions. Variation in methylation between placental regions for stress-related genes and for LINE-1 was minimal.
Conclusions
Our data suggest that these frequently studied stress-related genes have low levels of methylation in healthy placenta tissue. Minimal variation between sites suggests that sampling location does not affect DNA methylation analyses of these genes or of LINE-1 repetitive elements.
Keywords: placenta, glucocorticoid receptor, BDNF, 11B-HSD2, LINE-1, stress
Introduction
A large body of epidemiologic and experimental evidence suggests that exposures to early life adverse influences may have long-term consequences for health and disease [1]. During certain critical periods of fetal development, nutritional, disease-related, and psychological stressors are hypothesized to alter the structure, function, or organization of cells and tissues in ways that may subsequently affect adult disease susceptibility [2]. While the mechanisms behind fetal programming are still relatively unclear, epigenetic modifications may be one process through which environmental exposures could lead to long-term changes in gene expression. DNA methylation is the best characterized epigenetic modification, in which methyl groups are added to cytosine nucleotides. These epigenetic mechanisms may be particularly important in the placenta for fetal programming, as the placenta is located at the interface of maternal and embryonic environments, and thus may be especially vulnerable to hormonal influences from both the mother and fetus [3].
Due to the growing interest in the exposure to stress during early development, and its influence on health over the life course, methylation of stress-related genes has been the focus of many recent studies in the rodent brain [4], human cord blood, and maternal blood [5, 6]. Here we define stress-related genes as those that are mediated by exposure to glucocorticoids, a class of steroid hormones that are elevated in response to stress. We focus on three stress-related genes, including NR3C1, which encodes the glucocorticoid receptor; BDNF, which influences neuronal development; and 11B-HSD2, which encodes an enzyme to regulate the transmission of cortisol to the fetus at the maternal-fetal boundary [7].
NR3C1 and BDNF have been targeted in the fetal programming literature to be particularly affected by stress in early life in rodents [4, 8], but have not yet been well examined in human placenta tissue. For example, within the hippocampus of rodent pups, studies have shown that receiving poor maternal care is associated with increased methylation of NR3C1 and BDNF [4, 8]. Regulation of the 11B-HSD2 gene is particularly important in the placenta, as it, prevents excess fetal exposure to cortisol [9], which can have negative long term effects on growth, metabolism, and mental health [10]. Elevated promoter methylation of 11B-HSD2 has been found in peripheral blood leukocytes of patients treated with glucocorticoids [11]. Researchers have also found that acute, but not chronic, exposure to stress during pregnancy upregulates placental 11B-HSD2 activity [12], but studies have not yet reported on methylation in placenta. The long interspersed nuclear element-1 (LINE-1) is a repetitive element that comprises 20% of the human genome, and is often highly methylated in somatic tissues [13]. Levels of LINE-1 are informative about potential genome-wide disturbances in methylation, but in studies of fetal programming, they have primarily been studied in cord and maternal blood [5, 14]. LINE-1 methylation likely differs between placenta and blood, as placenta is a proliferating tissue similar to cancerous tumors, which often show global deregulation [15].
So far, only one study has analyzed DNA methylation levels of one of these three stress-related genes (NR3C1) in human placenta tissue [16], despite the fact that DNA methylation is critical to the regulation of placental growth and function [17]. This study of 480 placentas observed low levels of NR3C1 methylation (~3–4%) in placenta tissue from babies born small or average for gestational age (SGA, AGA), but slightly higher levels (~5–6%) in placentas of babies large for gestational age (LGA) [16]. It is unclear whether this small but significant methylation difference has any long term health consequences, and thus further study in placenta tissue is warranted.
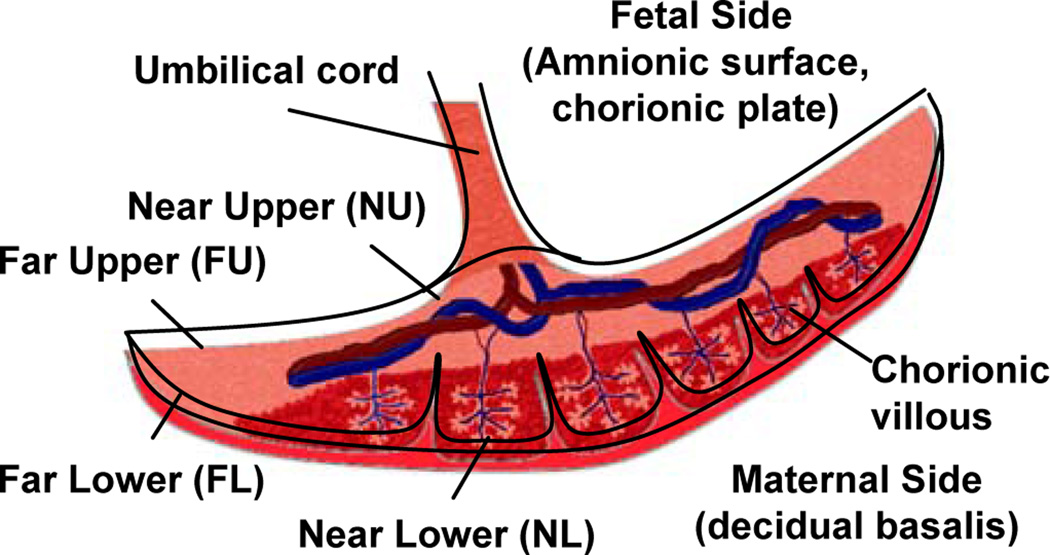
When analyzing methylation levels of genes in a complex organ such as the placenta, which contains a mosaic of many different cell and tissue types, it is important to understand the degree of intra-organ variability. Methylation of stress-related genes may be affected by stress in different ways within the upper side (generally maternally-derived region) versus the lower side (generally fetally-derived region) of the placenta, as each may differ in cellular composition and underlying genome (Figure 1). The physical location of cells within the placenta can also affect exposure to environmental factors, such as hormones or oxygen, or affect the proportion of undifferentiated cells at a particular placental site [18]. All of these sources of variation could contribute to different epigenetic profiles across the placenta, and must be considered when determining a sampling strategy.
Figure 1.
Diagram of placenta. This diagram demonstrates the four regions of the placenta sampled in this study, including Near Upper (NU) and Far Upper (FU) on the fetal side and the Near Lower (NL) and Far Lower (FL) regions on the maternal side.
Knowledge of the normal variation in methylation across a healthy organ is also useful for characterizing and predicting abnormal organ development, which is particularly important for an organ such as placenta, which exhibits an extremely wide range of normal variation in size and structure [18]. However, only a few prior studies have investigated intra-placental variation in DNA methylation, [18–20] most of which focused only on the chorionic villi, and to our knowledge, none of which have considered stress-related genes. Thus it is not clear whether and how methylation patterns may vary across placental regions among stress-related genes previously identified as important in studies of fetal programming.
In the current study, we evaluated the variation and distribution of DNA methylation within placenta tissues of 20 healthy individuals of three stress-related genes, as well as for the LINE-1 repetitive element. These data can help us understand the levels of methylation at key stress-related genes, and the effect of sampling location on methylation patterns in placenta tissue.
Methods
Study population
The placenta samples investigated in this study were selected from the Epigenetic Birth Cohort, which comprises 1941 mother-child dyads collected at the Brigham and Women’s Hospital in Boston. Collection of data and biospecimens in this cohort has been previously published [21]. The study protocol was approved by the Institutional Review Boards of BWH and Harvard School of Public Health.
We selected 20 samples from unrelated self-identified white women to represent a relatively homogeneous and healthy population, after excluding all pregnancies with any factors suspected to affect methylation patterns, i.e. twin births, prenatal smoking, drinking, use of medications, pregnancy complications (e.g. preterm birth, gestational diabetes, preeclampsia), or assisted reproduction. We also restricted our sample to pregnancies with gestational age between 37–41 weeks, and elective c-section deliveries, as variation in method of delivery can affect release of stress hormones that may influence methylation. Finally, we included only those with some pregnancy folate supplementation, as lack of any folate consumption may alter methylation patterns.. Sample characteristics are displayed in Table 1. Though our small sample of 20 individuals has limited power for detecting inter-individual differences, the four placental samples from each individual provides greater power for detecting intra-individual differences, which was the focus of this study.
Table 1.
Sample Characteristics.
| Variables (n=20) | Mean (range) |
|---|---|
| Mean Maternal Age, years (range) | 35.2 (29–42) |
| Sex of the baby, female | 65% |
| Mean Gestational age, weeks (range) | 38.8 (37.1–39.7) |
| Maternal pre-pregnancy BMI, kg/m2 (range) | 23.9 (20.0–30.9) |
| Mean birth weight, g (range) | 3551.6 (3185.0–4026.0) |
| Mean placental weight, g (range) | 866.9 (674.0–1156.0) |
| Average size for gestational age* | 85% |
| Large for gestational age* | 15% |
AGA and LGA were determined according to customized percentiles calculated with GROW software [32].
Sample collection and preparation
One sample (<1 cm3) was taken from each of four different regions of the placenta: 1) near the cord from the upper layer (Near Upper; NU), 2) at the placenta perimeter from the upper layer (Far Upper; FU), 3) near the cord from the lower layer (Near Lower; NL), and 4) at the placenta perimeter from the lower layer (Far Lower; FL) (Figure 1). For all samples, the cord insertion point was centrally located. Collected samples were snap-frozen and genomic DNA was isolated from placenta tissue using the QIAamp DNA Mini Kit (Qiagen).
DNA Methylation Assays
Bisulfite conversions were performed in duplicate on every sample using the EZ DNA Methylation Gold kit (Zymo Research, CA, USA) according to manufacturer’s protocol as described elsewhere [21]. Methylation was assayed in CpG islands within promoter regions of three genes, NR3C1, BDNF, and 11B-HSD2, and at the genome-wide repetitive element, LINE 1. Methylation assays were designed to contain sites previously shown to be regulated by methylation in NR3C1 and BDNF [5, 6], and a uniquely chosen CpG rich region in 11B-HSD2, using the Pyromark Assay Design 2.0 Tool (Qiagen), and for LINE-1 using the PyroMark LINE-1 kit (Qiagen). Primer sequencing information is available in Supplementary Table 1. Each assay was validated with a methylation scale (0-20-40-60-80-100% known methylation), described elsewhere [21]. Levels of methylation between technical replicates were tested for all genes.
Statistical Analyses
Methylation levels for each gene were calculated by taking an average across 5 CpG dinucleotides for NR3C1, 11 CpGs for BDNF, 14 CpGs for 11B-HSD2, and 3 CpGs for LINE-1, at each placental region. These levels were compared across all 20 placentas using Spearman correlations comparing each pair of regions, as levels of methylation were not normally distributed. Correlations between technical replicates were also calculated across all genes to test for reproducibility of our assay; e.g. NU fraction of NR3C1 showed a 0.6% average difference, and no pair of replicates differed by more than 2.6%. The Friedman rank sum test was used to test for intra-placental differences, while accounting for repeated measures. Post-hoc analysis was conducted with Wilcoxon signed rank tests. The Bonferroni method was used to correct for the six pairwise comparisons, corresponding to α-level=0.0083. All analyses were conducted using R software, v2.10. All statistical tests were two sided, and p-values of <0.05 were considered statistically significant.
Results
Intra-placental variation in DNA methylation
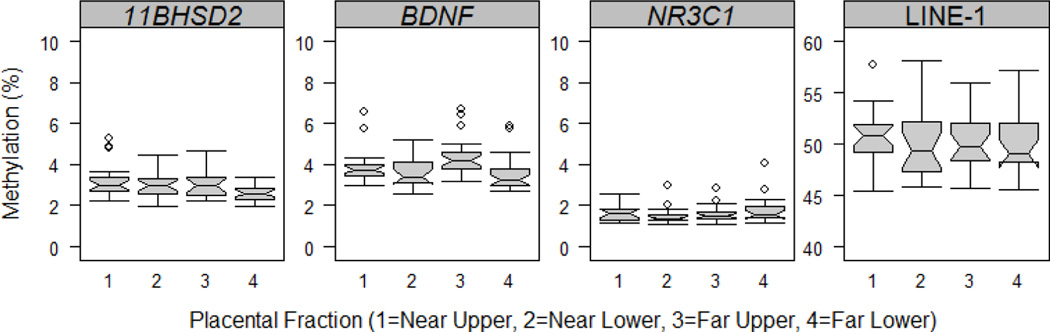
All three stress-related genes demonstrated low levels of methylation across all four placental regions (Figure 2, Supplemental Table 2). The lowest methylation levels were found for NR3C1, with medians ranging from 1.37% to 1.64% across the four placental regions, and the highest levels were found for BDNF, with medians ranging from 3.25% to 4.20% across all placental regions. These levels were within the same range as those found for the unmethylated DNA in the methylation scale (data not shown). Median methylation values for LINE-1 ranged between 49.10% and 50.85% across placental regions (Figure 2, Supplemental Table 2).
Figure 2.
Variation in methylation by placental region. Each boxplot displays the median and range of methylation values across the four placental regions for each of the three stress-related genes and the Line-1 repetitive element.
Methylation levels of stress-related genes were very similar across all four placental regions. For each of the three genes, no two samples differed by more than 4% across any of the four regions. No significant differences were found between any placental regions for NR3C1. At least one difference of statistical significance, but of limited magnitude, was found between regions for BDNF ( =16.7684, p<0.001) and for 11B-HSD2 ( =9.0706, P=0.028). After adjusting for multiple testing, significantly greater methylation was found in FU versus FL regions for both of these genes, as well as significantly greater methylation for FU versus NL regions for BDNF, and for NU versus FL regions of 11B-HSD2. No significant differences were found between placental regions of LINE-1.
After Bonferroni adjustment, we found no significant Spearman correlations between pairs of regions for NR3C1, one significant positive correlation for BDNF (between NU-FU, ρ=0.69, P=0.001), and one significant positive correlation for 11B-HSD2 (between NU-FU, ρ=0.74, P<0.001), (Table 2). The NU-FU regions showed the highest and most significant positive correlations across two of the genes: BDNF (ρ=0.69, P<0.001) and 11B-HSD2 (ρ=0.74, P<0.001). Methylation levels for LINE-1 were all significantly correlated (all ρ>0.60, all P≤0.005) between all six pairs of regions (Table 2).
Table 2.
Spearman correlations of methylation between placental regions
| NR3C1 | BDNF | 11B-HSD2 | LINE-1 | |||||
|---|---|---|---|---|---|---|---|---|
| Regions | ρ | P-value | ρ | P-value | ρ | P-value | ρ | P-value |
| NU-FU | 0.37 | 0.1218 | 0.69 | 0.0012* | 0.74 | <0.0001* | 0.70 | <0.0001* |
| NU-NL | 0.23 | 0.3212 | 0.27 | 0.2684 | 0.53 | 0.0208 | 0.76 | <0.0001* |
| NU-FL | 0.43 | 0.0689 | 0.14 | 0.5638 | 0.36 | 0.1425 | 0.68 | 0.0025* |
| NL-FU | 0.25 | 0.2872 | 0.37 | 0.1176 | 0.44 | 0.0641 | 0.61 | 0.0052* |
| NL-FL | 0.31 | 0.1979 | 0.56 | 0.0141 | 0.39 | 0.1205 | 0.77 | <0.0001* |
| FU-FL | 0.26 | 0.2818 | 0.42 | 0.0658 | 0.34 | 0.1709 | 0.72 | 0.001* |
NU=near upper, FU=far upper, NL=near lower, FL=far lower
Significant at Bonferonni-adjusted p-value of 0.0083.
We also examined if methylation levels differed significantly between individuals on the few characteristics that varied in our sample, such as birthweight, placenta weight:birthweight ratio, and fetal sex. No significant differences between groups across any of these variables were detected for any of the three stress-related genes (data not shown). Lower LINE-1 methylation levels were found in placentas of neonates with lower birthweight and lower placental weight/birthweight ratios in this study, and also in a previous study using a larger sample from this cohort [21].
Discussion
Methylation levels in stress-related genes and LINE-1
We found very low levels of methylation at all three stress-related genes, regardless of the sampling region of the placenta. These methylation levels were comparable to levels previously reported for these genes in cord blood and maternal blood. For example, similarly low levels of methylation have been found at BDNF (≤8%) across 11 CpG sites that correspond to those assayed in the current study [6] and ≤5% methylation across all five corresponding CpG sites in NR3C1 [5] in both cord and maternal blood. One prior study analyzed methylation of NR3C1 in placenta, and found similarly low (~3%) levels of methylation at five CpG sites corresponding to those in the current study. However, slightly higher levels of methylation (~6–6.5%) were identified at seven additional assayed sites only in placentas of LGA babies [16]. More research is needed to confirm this result and to determine if this small difference in methylation has significant functional consequences. No prior studies to date have reported levels of DNA methylation of 11B-HSD2 in human placenta, though reduced RNA expression in placenta has been associated with higher maternal anxiety [22], and with pregnancy complications that are not found in our dataset, such as preeclampsia [23] and growth restricted placentas [24].
Given the low levels of methylation, and very little variation we found in placenta and in previous reports in blood, we hypothesize that stress effects of these genes may not be as evident in these tissues compared to brain tissues, where variable methylation has been observed in rodent brains according to parental care [4] and in the brains of human suicide victims [25, 26].The low variation we observe in methylation may be related to the homogeneous sample of healthy mothers, or to the potential lack of variation in maternal stressors. Though we could not measure maternal stress in this sample, it is unlikely, that all subjects experienced the same degree of stress during pregnancy. Thus the highly similar methylation levels across all samples implies that methylation of these genes does not vary by maternal stress. Furthermore, the consistently low methylation across all samples indicates that these genes may be constitutively unmethylated in human placenta.
The median level of 50% methylation we observed for LINE-1 in the placenta were hypomethylated relative to other tissues, such as blood or brain, which usually have mean values of ~78–80% [14, 21], and ~66–69% [27], respectively. CpG sites in repetitive elements are usually highly methylated in order to silence expression and prevent transposition of these elements into other genomic regions. It is interesting to note that hypomethylation of LINE-1 is also found in cancer, where it is linked to chromosomal instability and the reactivation of transposable repetitive elements [15, 28].
Intra-Placental variation
Sampling region of the placenta did not appear to have a large effect on levels of methylation. In general, the intra-placental variation was very small, though statistically significant differences were found between some placental regions. The low correlation values for some regional comparisons could be a statistical artifact of small sample sizes and small inter-individual variation.. In contrast, all of the LINE-1 methylation levels, which showed higher values and a larger range of variation than the stress-related genes, were more highly correlated across placental regions.
The highest correlation between placental regions was generally found for the NU-FU comparisons. These two regions may be the most similar because the upper side of the placenta is more likely to contain only fetally-derived cells, whereas lower side could contain a mix of fetal and maternal cell types. This result is further supported by our finding of significant methylation differences only when comparing fetal side to maternal side. Thus, researchers interested in fetal effects may choose to sample consistently from the upper side of the placenta to avoid potential contamination from maternal decidua on the lower side. However, the differences in methylation between any two regions in this study were so small that we predict they will not have a biologically significant effect on fetal development, though further study is needed to confirm this hypothesis.
Our finding of low variation across the placenta is largely consistent with the results of Avila et al, who found consistent methylation patterns across four sample sites and four sample depths for genes related to trophoblast differentiation [18]. However, when they microdissected the placenta, they found different levels of DNA methylation in the amnion, chorion, trophoblast, and mesenchyme tissues, respectively, suggesting that microdissection is necessary when trying to isolate methylation levels in a particular cell or tissue type
The low intra-placental variability in these stress-related genes may not be generalizeable to other genes of interest. For example, high intra-placental variation was identified for differentially methylated regions associated with IGF2/H19 and IGF2R [20]. Methylation of imprinted genes may be more variable across the placenta due to stochastic variation in the number of trophoblast stem cell progenitors at the time when the imprints were established [20]. Additionally, genes affected by hypoxia [29] or cell differentiation [19] have shown greater variation across the placenta. Thus, future studies should evaluate each gene of interest in a similar manner across sampling sites. Researchers could also try to minimize any potential variation that might be found in other genes or other placental regions by sampling and pooling from multiple sites across the placenta, or by carefully sampling from a single site consistently throughout an entire population.
The strengths of our study are its focus on stress-related genes of interest to researchers of fetal programming, and in the use of a healthy and relatively homogenous population of placentas to examine normal levels of variation in methylation. To our knowledge, this is the first study to investigate methylation levels in these stress-related genes across different regions of the placenta. Our use of a methylation scale to test the performance of the assay and of technical replicates for all samples demonstrates the reproducibility of our assays.
Conclusions
Overall, our results indicate a consistent pattern of very low methylation across all sample regions in the three stress-related genes, and low levels of methylation for LINE-1 relative to other tissues. We have also identified that in a healthy sample, there is little intra-placental variation among these genes. This result suggests that sampling from any site will yield similar methylation levels, reducing the need to sample from multiple sites. Our findings provide a starting point for future studies to investigate the variation of these genes among a more heterogeneous population, including abnormal placental tissue where larger differences within or between placentas may be detected. Future studies should also investigate if these small differences in methylation values correlate with mRNA expression levels, and if differences in methylation or expression across sampling sites can be found in other genes of interest.
Supplementary Material
Acknowledgements
The authors would like to thank the women who participated in the study, as well as Joelle Perkins, Eliza Gardiner, Michelle Peters, and Seema Sannesy for help with data and specimen collection. We also thank Professor Drucilla Roberts for helpful advice about the placenta, and Professor Gail Adler for general guidance and discussion. The authors thank the Robert Wood Johnson Foundation Health & Society Scholars program for its financial support. Sample collection was funded by Public Health Research Grant 5R21CA128382 from the National Cancer Institute, National Institutes of Health (to K.B.M.). A.Binder was supported by Training Grant T32HD060454 in Reproductive, Perinatal and Pediatric Epidemiology from the National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amy L. Non, Email: amynon@hsph.harvard.edu.
Alexandra M. Binder, Email: abinder@hsph.harvard.edu.
Ludovic Barault, Email: Ludovic.barault@free.fr.
Rebecca C. Rancourt, Email: RRANCOURT@partners.org.
Karin B. Michels, Email: kmichels@rics.bwh.harvard.edu.
References
- 1.Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol. 2004;561(Pt 2):355–377. doi: 10.1113/jphysiol.2004.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab. 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn GA, Morgan CP, Bale TL. Sex-specificity in transgenerational epigenetic programming. Horm Behav. 2011;59(3):290–295. doi: 10.1016/j.yhbeh.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 5.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3(2):97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 6.Devlin AM, Brain U, Austin J, Oberlander TF. Prenatal exposure to maternal depressed mood and the MTHFR C677T variant affect SLC6A4 methylation in infants at birth. PLoS One. 2010;5(8):e12201. doi: 10.1371/journal.pone.0012201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seckl JR, Meaney MJ. Glucocorticoid programming. Ann N Y Acad Sci. 2004;1032:63–84. doi: 10.1196/annals.1314.006. [DOI] [PubMed] [Google Scholar]
- 8.Roth TL, Sweatt JD. Epigenetic marking of the BDNF gene by early-life adverse experiences. Hormones and behavior. 2011;59(3):315–320. doi: 10.1016/j.yhbeh.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benediktsson R, Calder AA, Edwards CR, Seckl JR. Placental 11 beta-hydroxysteroid dehydrogenase: a key regulator of fetal glucocorticoid exposure. Clin Endocrinol (Oxf.) 1997;46(2):161–166. doi: 10.1046/j.1365-2265.1997.1230939.x. [DOI] [PubMed] [Google Scholar]
- 10.Tzschoppe A, Struwe E, Blessing H, Fahlbusch F, Liebhaber G, Dorr HG, Rauh M, Rascher W, Goecke TW, Schild RL, Schleussner E, Scheler C, Hubler A, Dahlem P, Dotsch J. Placental 11beta-HSD2 gene expression at birth is inversely correlated with growth velocity in the first year of life after intrauterine growth restriction. Pediatr Res. 2009;65(6):647–653. doi: 10.1203/PDR.0b013e31819e7337. [DOI] [PubMed] [Google Scholar]
- 11.Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, Ravagnani V, Carletto A, Pattini P, Corrocher R, Olivieri O. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199(2):323–327. doi: 10.1016/j.atherosclerosis.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 12.Welberg LA, Thrivikraman KV, Plotsky PM. Chronic maternal stress inhibits the capacity to up-regulate placental 11beta-hydroxysteroid dehydrogenase type 2 activity. The Journal of endocrinology. 2005;186(3):R7–R12. doi: 10.1677/joe.1.06374. [DOI] [PubMed] [Google Scholar]
- 13.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33(21):6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kile ML, Baccarelli A, Tarantini L, Hoffman E, Wright RO, Christiani DC. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PLoS One. 2010;5(10):e13730. doi: 10.1371/journal.pone.0013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filiberto AC, Maccani MA, Koestler D, Wilhelm-Benartzi C, Avissar-Whiting M, Banister CE, Gagne LA, Marsit CJ. Birthweight is associated with DNA promoter methylation of the glucocorticoid receptor in human placenta. Epigenetics. 2011;6(5):566–572. doi: 10.4161/epi.6.5.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 17(3):397–417. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 18.Avila L, Yuen RK, Diego-Alvarez D, Penaherrera MS, Jiang R, Robinson WP. Evaluating DNA methylation and gene expression variability in the human term placenta. Placenta. 2010;31(12):1070–1077. doi: 10.1016/j.placenta.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. Cell specific patterns of methylation in the human placenta. Epigenetics. 2011;6(3):368–379. doi: 10.4161/epi.6.3.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turan N, Katari S, Gerson LF, Chalian R, Foster MW, Gaughan JP, Coutifaris C, Sapienza C. Inter- and intra-individual variation in allele-specific DNA methylation and gene expression in children conceived using assisted reproductive technology. PLoS Genet. 2010;6(7):e1001033. doi: 10.1371/journal.pgen.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories, and global DNA methylation of LINE-1 repetitive elements. PLoS One. 2011;6(9):e25254. doi: 10.1371/journal.pone.0025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Donnell KJ, Bugge Jensen A, Freeman L, Khalife N, O'Connor TG, Glover V. Maternal prenatal anxiety and downregulation of placental 11beta-HSD2. Psychoneuroendocrinology. 2011 doi: 10.1016/j.psyneuen.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Causevic M, Mohaupt M. 11beta-Hydroxysteroid dehydrogenase type 2 in pregnancy and preeclampsia. Mol Aspects Med. 2007;28(2):220–226. doi: 10.1016/j.mam.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Dy J, Guan H, Sampath-Kumar R, Richardson BS, Yang K. Placental 11beta-hydroxysteroid dehydrogenase type 2 is reduced in pregnancies complicated with idiopathic intrauterine growth Restriction: evidence that this is associated with an attenuated ratio of cortisone to cortisol in the umbilical artery. Placenta. 2008;29(2):193–200. doi: 10.1016/j.placenta.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 25.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nature neuroscience. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, Carli V, Sacchetti S, Lembo F, Angiolillo A, Jovanovic N, Pisanti F, Tomaiuolo R, Monticelli A, Balazic J, Roy A, Marusic A, Cocozza S, Fusco A, Bruni CB, Castaldo G, Chiariotti L. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67(3):258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 27.Ohka F, Natsume A, Motomura K, Kishida Y, Kondo Y, Abe T, Nakasu Y, Namba H, Wakai K, Fukui T, Momota H, Iwami K, Kinjo S, Ito M, Fujii M, Wakabayashi T. The Global DNA Methylation Surrogate LINE-1 Methylation Is Correlated with <italic>MGMT</italic> Promoter Methylation and Is a Better Prognostic Factor for Glioma. PLoS ONE. 2011;6(8):e23332. doi: 10.1371/journal.pone.0023332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt SM, Kraus FT, Roh CR, Elchalal U, Nelson DM, Sadovsky Y. The correlation between sampling site and gene expression in the term human placenta. Placenta. 2005;26(5):372–379. doi: 10.1016/j.placenta.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Gardosi J, Francis A. Customised Weight Centile Calculator – GROW-Centile v.5.12/6.2. 2009 Gestation Network, www.gestation.net.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.