A minimal mathematical model based on stochastic attachment and detachment of kinetochore microtubules accurately reproduces both normal and abnormal chromosome segregation in fission yeast.
Abstract
In fission yeast, erroneous attachments of spindle microtubules to kinetochores are frequent in early mitosis. Most are corrected before anaphase onset by a mechanism involving the protein kinase Aurora B, which destabilizes kinetochore microtubules (ktMTs) in the absence of tension between sister chromatids. In this paper, we describe a minimal mathematical model of fission yeast chromosome segregation based on the stochastic attachment and detachment of ktMTs. The model accurately reproduces the timing of correct chromosome biorientation and segregation seen in fission yeast. Prevention of attachment defects requires both appropriate kinetochore orientation and an Aurora B–like activity. The model also reproduces abnormal chromosome segregation behavior (caused by, for example, inhibition of Aurora B). It predicts that, in metaphase, merotelic attachment is prevented by a kinetochore orientation effect and corrected by an Aurora B–like activity, whereas in anaphase, it is corrected through unbalanced forces applied to the kinetochore. These unbalanced forces are sufficient to prevent aneuploidy.
Introduction
The fidelity of chromosome attachment to the mitotic spindle is crucial to preventing the formation of aneuploid cells. To this end, kinetochores, protein structures that assemble at the centromeres of each pair of sister chromatids, must attach to microtubules from opposite spindle poles (chromosome biorientation) before the onset of anaphase. Precisely how the spindle captures and faithfully biorients all chromosomes within the short time between prophase and anaphase onset remains a fundamental question in biology. The bipolar mitotic spindle assembles during prometaphase by a “search and capture” process in which dynamically unstable microtubules make associations with kinetochores (Kirschner and Mitchison, 1986). The search and capture of kinetochores by microtubules is a common feature of mitosis in eukaryotes, which was initially visualized in newt lung cells (Hayden et al., 1990; Rieder and Alexander, 1990) and subsequently characterized in budding and fission yeasts (Tanaka et al., 2005, 2007; Franco et al., 2007; Gachet et al., 2008). After capture, chromosomes align at the metaphase plate, which forms equidistantly between the two centrosomes because of forces generated by kinetochore-bound mitotic motors (kinesins and dynein; Kops et al., 2010). Once the chromosomes are correctly bioriented, sister chromatids separate and move simultaneously toward the poles (Uhlmann et al., 1999; Oliveira et al., 2010). The correct back to back arrangement of sister kinetochores (kinetochore geometry) is crucial to prevent chromosome loss through defects, such as merotelic attachment, in which one kinetochore is attached to both poles (Gregan et al., 2007; Courtheoux et al., 2009; Sakuno et al., 2009; Gregan et al., 2011). Sister chromatid cohesion may thus define correct kinetochore geometry.
In the symmetrically dividing fission yeast Schizosaccharomyces pombe, segregation of the three mitotic chromosomes occurs within the nuclear envelope (so-called closed mitosis). The six kinetochores each contain attachment sites for up to four microtubules (Ding et al., 1993) emanating from the centrosome, also called the spindle pole body (SPB). The two SPBs also nucleate microtubules that interdigitate at the spindle center. Mitosis in S. pombe consists of three phases (Nabeshima et al., 1998; Tatebe et al., 2001). During phase 1, a short (<2.0 µm) spindle is formed. In phase 2 (prometaphase/metaphase/anaphase A), the spindle maintains roughly the same length, and the kinetochores make frequent, rapid oscillations between the poles. At the end of phase 2, the kinetochores congress to the spindle midzone; the sister chromatids then separate and move toward the SPBs during anaphase A (Tournier et al., 2004). In phase 3 (anaphase B), the spindle elongates along the longitudinal axis of the cell.
The spindle assembly checkpoint (SAC) controls the timing of anaphase onset to prevent chromosome loss as the result of incorrect attachments (Rieder et al., 1995; Rudner and Murray, 1996; Cleveland et al., 2003). Components of the SAC were first identified in the budding yeast Saccharomyces cerevisiae (Hoyt et al., 1991; Li and Murray, 1991), but structural and functional homologues of mitotic checkpoint proteins have since been identified in all other eukaryotes examined, including fission yeast (He et al., 1997; Bernard et al., 1998; Millband and Hardwick, 2002). In response to microtubule-disrupting agents, checkpoint proteins translocate to unattached kinetochores and delay the onset of anaphase. Aurora B kinases are also essential for accurate chromosome segregation (Lampson and Cheeseman, 2011). This kinase was first identified in S. cerevisiae (as Ipl1) in a screen for mutants that display an increase in ploidy (Chan and Botstein, 1993). Aurora B phosphorylates kinetochore substrates in the absence of tension. This destabilizes incorrect attachments and allows reorientation of the kinetochore toward the correct spindle pole (Cimini et al., 2006; Kelly and Funabiki, 2009).
Subunits of the monopolin complex also help to suppress merotelic attachment in S. pombe (Corbett et al., 2010; Rumpf et al., 2010; Gregan et al., 2011). Merotelic attachment occurs frequently during the early stages of mitosis but is not detected by the SAC (Gregan et al., 2011). Instead, it is corrected before anaphase onset by a mechanism dependent on Aurora B (Tanaka et al., 2002; Cimini et al., 2006; Knowlton et al., 2006). This attachment defect can also be corrected after anaphase onset through the forces produced by spindle elongation in both fission yeast and higher eukaryotes (Cimini et al., 2004; Courtheoux et al., 2009). Although the precise mechanism for this anaphase correction of merotelic attachment is unknown, earlier studies suggest that an imbalance of the forces exerted by microtubules on the kinetochore might be involved. In S. pombe, failure to correct merotely leads to the persistence of a chromosome at the site of cell cleavage (Courtheoux et al., 2009) and the so-called “cut” phenotype (the formation of the septum between daughter cells in the absence of normal nuclear division; Hirano et al., 1986).
To explore the possible mechanisms required to prevent or correct erroneous kinetochore attachments in metaphase and anaphase, we have designed a biophysical model of mitosis based on data obtained by video microscopy of living fission yeast cells. Although mathematical and computational models of mitosis have been developed over the past few years, none have combined a mechanical description of chromosome segregation with the correction of attachment defects (Brust-Mascher et al., 2004; Gardner et al., 2005; Civelekoglu-Scholey et al., 2006; Courtheoux et al., 2009; Paul et al., 2009). Our new model faithfully reproduces fission yeast chromosome oscillations and segregation from prophase to anaphase B, and it predicts the attachment state of all three chromosomes with time. It also predicts that the duration of phase 1 and 2 (prometaphase/metaphase/anaphase A) is long enough to allow proper biorientation to occur through a stochastic succession of attachment and detachment events biased toward biorientation. Finally, the model reproduces abnormal behavior of chromosome segregation after inhibition of Aurora B and predicts that merotelic attachments can also be corrected after anaphase onset by an imbalance of forces exerted by microtubules on the merotelic kinetochore. We conclude that two independent mechanisms, kinetochore orientation and Aurora-like activity, are required to prevent the appearance of merotelic attachment and the cut phenotype after anaphase onset.
Results
A simplified model of the mitotic spindle predicts anaphase A onset when the cohesin link between sister chromatids is removed
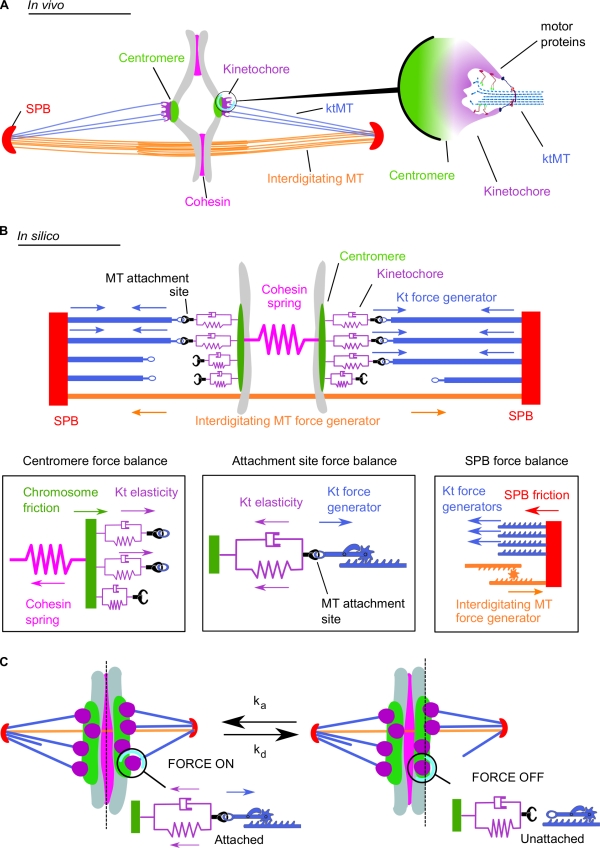
To model chromosome segregation in fission yeast, we took into account the interactions between several elements of the mitotic spindle (Fig. 1 A): (a) the SPBs, (b) the chromosomes, further divided into the centromeres and the kinetochores, (c) the kinetochore microtubules (ktMTs), linking SPBs to the microtubule attachment sites on the kinetochore, (d) interdigitating microtubules between the spindle poles but without contact to the chromosomes, and finally, (e) the cohesins, forming a link between sister chromatids before anaphase. The main assumptions (or approximations) made in this model are listed in the following section. We considered that the haploid mitotic fission yeast cell contains three chromosome pairs, i.e., a total of six kinetochores, each of which possesses up to four microtubule attachment sites (Fig. 1 A; Ding et al., 1993). We assumed a balance of opposing forces acting on spindle components (Fig. 1 B and Video 1; Civelekoglu-Scholey et al., 2006; Courtheoux et al., 2009) and made the approximation that attached ktMTs would always pull the chromosome toward the spindle pole. In the model, this pulling force is driven by a single force generator (kinetochore force generator; Fig. 1 B, blue arrows). This is a simplification for the variety of molecular motors present both on the microtubules and on the kinetochore in vivo (Fig. 1 A, right; Peskin et al., 1993; Maiato and Lince-Faria, 2010; McIntosh et al., 2010; Tanenbaum and Medema, 2010). The force is applied precisely at the microtubule attachment site, which is linked to the centromere by a spring and a dashpot, reflecting the elasticity of the kinetochore (Fig. 1 B and Video 1). The position or the state of the microtubule plus ends is not explicitly modeled. The pulling forces transmitted to the centromere are countered by a cohesion force mediated by cohesin protein complexes in vivo and modeled by a Hookean spring in Fig. 1 B (pink). The kinetochore force generator (Fig. 1 B, blue arrows) also results in an inward force on the SPBs that is balanced by an outward force created in vivo by antiparallel sliding of interdigitating microtubules and approximated in silico by a second single force generator (interdigitating microtubule force generator; Fig. 1 B, orange arrows).
Figure 1.
Schematic representation of the metaphase spindle. (A) In vivo. (left) The two SPBs are linked by overlapping interdigitated microtubules (MT). The chromosome (gray) is linked to the SPBs by its centromere regions. The four microtubule attachment sites located on each kinetochore (Kt) are connected to the SPBs by ktMTs. The two sister chromatids are held together by the cohesin complex. (right) Schematic depiction illustrating the in vivo structure of the ktMT attachment site. Several motors are present at the ktMT interface. (B) In silico. (top) The SPBs are linked by the interdigitated microtubule force generator. Each microtubule attachment site on the kinetochore is linked to the SPB through a ktMT (blue). The four microtubule binding sites (purple) are associated to the chromosomes by the centromere and represented by a spring and a dashpot (purple). Cohesin between the sister chromatids is modeled as a single spring linking both centromeres. (bottom) Schematic depiction illustrating the different forces applied in silico at the centromere level (left), the microtubule attachment site (middle), and at the SPB (right). (C) A simple stochastic process of microtubule attachment and detachment reproduces the directional instability. At any time, microtubule attachment sites (purple) attach with the frequency ka (left, force is on) or detach with the frequency kd (right, force is off). This attachment/detachment process leads to an imbalance of the forces applied on the chromosome and to chromosome dynamics within the spindle (also see Video 1).
The positions of the various elements in the spindle are obtained by solving a set of first-order linear-coupled differential equations at each time point (typically every second), from the start of phase 1 until the completion of phase 3. In the simulations, setting the cohesin spring constant to zero triggers poleward movement of the chromosomes, driven by the kinetochore force generator at the attachment sites. This reflects the degradation of the cohesin complex that triggers anaphase A in vivo (Uhlmann et al., 1999). Coincidently, the interdigitating microtubule force generator drives rapid elongation of the spindle (Video 1) also as observed in vivo (Loïodice et al., 2005).
Main simplifying assumptions of the model and consequences of these assumptions
The main simplifying assumptions made in the model are the following. First, we assume that the initial attachment of ktMT to kinetochore is random. Second, we model the ktMT plus end and its attachment site as a single element. Third, the unattached ktMT plus ends are not considered in the model. Fourth, we assume that ktMT attachment and detachment is stochastic. Fifth, we make the simplifying assumption that kinetochore force generator maintains a linear force–velocity relationship with constant values of maximum speed and stall force. Finally, we also assume that the midzone force generator always pushes. The direct consequences of these assumptions are that the plus end of a ktMT always follows the position of its attachment site (i.e., the ktMT grows and shrinks when the kinetochore moves poleward or antipoleward). In addition, the distribution of microtubule plus ends is assumed to be uniform within the spindle. Other consequences of our assumptions are that the dynamics of microtubules attached to sister kinetochores are coupled to one another and that the number of interdigitating microtubules or their length is not modeled. We also make the approximation that such changes do not affect the force generator responsible for the outward pushing of the SPBs and that a ktMT attachment exerts the same pulling force on a kinetochore regardless of the dynamic state of the microtubule.
Stochastic attachment of ktMTs mimics chromosome dynamics in mitosis
It has been suggested that attachment and detachment of individual microtubules to the kinetochore occurs throughout prometaphase and metaphase (Mitchison et al., 1986; Mitchison, 1989)—an assumption that we also made in our model. Microtubules interact in various ways with kinetochores (e.g., laterally or end on), and these types of attachment may require distinct motor proteins at the microtubule–kinetochore interface (Merdes and De Mey, 1990; Rieder and Alexander, 1990; Tanaka et al., 2005; Grishchuk and McIntosh, 2006; Gachet et al., 2008). In our model, these processes are simplified and represented by a single switch mechanism, which functions as follows: when a microtubule attaches to a kinetochore (with a Poisson rate ka), the pulling force is “on,” and when it detaches (with a detachment rate kd), the pulling force is “off” (Fig. 1 C). Varying distances between the sister kinetochores (“breathing”) and oscillatory movements of kinetochore pairs along the spindle can therefore be modeled by a series of stochastic events of attachment and detachment at each microtubule attachment site. For example, if four microtubules are attached to the left-hand side of a kinetochore pair and only one is attached to the right-hand side, the chromosome will move to the left. If one more attaches to the right, the interkinetochore distance increases, and the chromosome slows down. The plus ends of microtubules are thus following the kinetochore (poleward or antipoleward) without detaching.
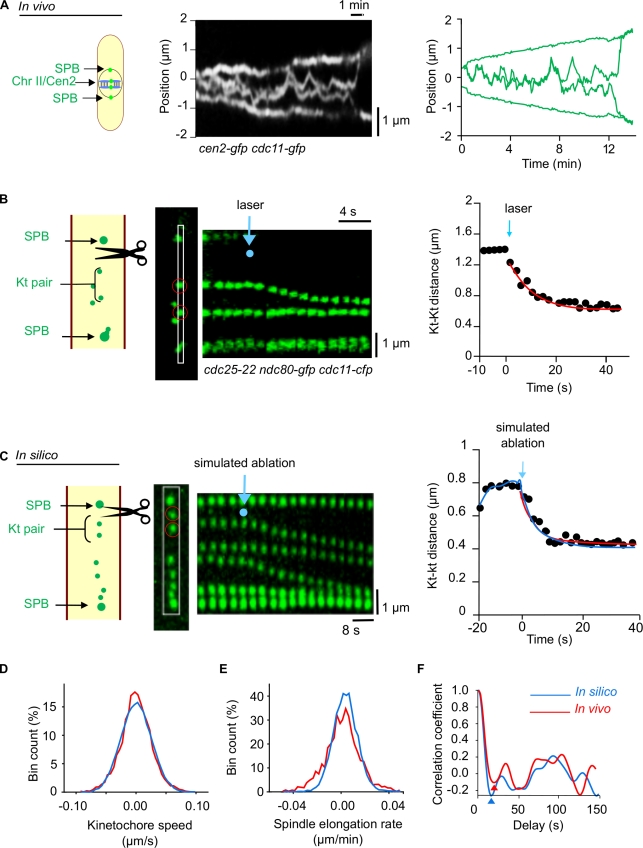
Modeling kinetochore dynamics involves 14 parameters, most of which were deducted from live-cell imaging using a strain that allows simultaneous observation of both the centromere of chromosome 2 (Cen2-GFP; Yamamoto and Hiraoka, 2003) and the SPBs (Cdc11-GFP; see Materials and methods; a link to the source code of the model is available on S. Tournier’s laboratory webpage at the Laboratoire de biologie cellulaire et moléculaire du contrôle de la prolifération; Fig. 2 A, Video 2, and Table 1, technical information). We first measured the timing of kinetochore oscillations between the SPBs to estimate the values for attachment (ka, force on) and detachment (kd, force off) rates. Using video microscopy, we detected centromere signals (cen2-gfp) and kinetochore signals (ndc80-cfp). Because the average distance between these two signals was very small (126 ± 10 nm, n = 22; Fig. S1), we assumed that centromere oscillations are very similar to kinetochore oscillations.
Figure 2.
Measurement of biological parameters to model chromosome segregation. (A, left) Kymograph of a cen2-gfp (centromere [Cen] chromosome [Chr] II) and cdc11-gfp (SPB) cell showing the in vivo oscillation of chromosome II between the SPBs (Video 2). (right) Tracked trajectories of the fluorescence signals from Cen2-GFP and Cdc11-GFP shown on the left. (B) In vivo experiment. (left) Laser ablation of the spindle apparatus in a metaphase cdc25-22 cell expressing a fluorescent marker protein is shown on the left for the kinetochore (Kt; Ndc80-GFP, green) and the SPB (Cdc11-GFP, green). Note that metaphase spindles in this mutant strain are longer than in wild-type cells. The positions of one kinetochore pair (red circles) are followed after laser ablation (the laser pulse is located at the blue dot) as illustrated on the kymograph constructed from the cropping outlined in white in the image. (right) Graphic representation showing the distance between the pair of kinetochores after laser ablation (time 0 = time of laser impact). The red line represents the best fit of an exponential relaxation curve. (C) In silico simulation. (left) Kymograph showing the dynamics of all three kinetochore pairs on a metaphase spindle of similar length to a cdc25-22 cell after elimination of the force generators (simulated laser ablation, blue dot). (right) Comparison of kinetochore relaxation seen in vivo (red trace) with that seen by modeling in silico (blue trace). (D–F) Further comparison of parameters of mitosis determined in vivo and modeled in silico. (D) Kinetochore speed distribution obtained after tracking of kinetochores in vivo and in silico. (E) Spindle elongation rate distribution for the in vivo and in silico trajectories. (F) Comparison of the autocorrelation function (mathematical tool to determine a repeat pattern) used to determine the attachment rate as described in the Materials and methods. Red and blue arrowheads show first minima of the autocorrelation functions for the in vivo and in silico autocorrelation functions, respectively.
Table 1.
Summary of the measures used in the study
| Description | Value | SD |
| Metaphase elongation rate | 0.001 µm/s | 0.2 × 10−3 µm/s |
| Anaphase elongation rate | 0.03 µm/s | 0.01 µm/s |
| Kinetochore poleward speed at anaphase A | 0.03 µm/s | 0.01 µm/s |
| Maximum metaphase kt–kt distance | 1.2 µm | 0.2 µm |
| Mean metaphase kt–kt distance | 0.4 µm | 0.2 µm |
| Resting kt–kt distance | 0.05 µm | 0.01 µm |
| Kt pair relaxation time | 10 s | 3 s |
| Merotelic kt relaxation time | 10 s | 3 s |
kt, kinetochore.
Mitotic parameters, such as the speed of poleward chromosome movement, were either determined previously by video microscopy (Courtheoux et al., 2009) or obtained from the literature (Table 2). The mechanical characteristics of the cohesin bond (stiffness and friction coefficients between the sister kinetochores) were estimated by laser ablation of metaphase spindles. To do this, we took advantage of a temperature-sensitive fission yeast strain, cdc25-22, expressing fluorescent markers of kinetochores (Ndc80-GFP) and SPBs (Cdc11-GFP). These cells arrest in G2 after incubation for 4 h at the restrictive temperature of 36°C. Once released from G2 arrest, by return to the permissive temperature of 25°C, cells enter mitosis with a high degree of synchrony and relatively long metaphase spindles (<6 µm), which considerably facilitates laser ablation on one side of the spindle (Fig. 2 B and Video 3). After laser ablation of the spindle microtubules (both interdigitating and ktMTs), the stretched pairs of kinetochores immediately relaxed in a springlike behavior (Fig. 2 B). In silico, elimination of the force generator on one side of a stretched kinetochore pair immediately caused the sister kinetochores to come closer (Fig. 2 C, right), thus accurately reproducing the data measured in vivo (Fig. 2 C). The model also reproduced with reasonable accuracy both the dynamic characteristics of sister kinetochores with regard to oscillation, breathing, and speed (Fig. 2, D and F) and also spindle elongation during mitosis (Fig. 2 E).
Table 2.
Summary of the parameters used in this study
| Variable | Description | Value | Range |
| N | Number of chromosomes | 3 | 3 |
| Mk | Number of ktMTs per kinetochore | 4 | 4 |
| tA | Anaphase transition time | 720 s | 600–900 s |
| δt | Time step | 1 s | 0.1–3 |
| L0 | Spindle initial length | 0.3 µm | 0.1–5 µm |
| dc | Centromere pair rest length | 0.05 µm | 0.01–0.2 µm |
| κc | Cohesion spring constant | 90 pN/µm | 10–200 pN/µm |
| κk | Spring constant of the bond between attachment site and the centromere | 40 pN/µm | 10–200 pN/µm |
| μk | Damping coefficient for the attachment site | 900 pN.s/µm | 100–2,000 pN.s/µm |
| μc | Sister chromatid friction coefficient | 400 pN.s/µm | 100–2,000 pN.s/µm |
| μs | SPB friction coefficient | 500 pN.s/µm | 100–2,000 pN.s/µm |
| Fk | ktMT motor stall force | 10 pN | Fixed |
| Vk | ktMT motor maximum velocity | 0.03 µm/s | 0.01–0.06 µm/s |
| Fmz | Midzone motor stall force | 127 pN | 50–500 pN |
| Vmz | Midzone motor maximum velocity | 0.03 µm/s | 0.01–0.06 µm/s |
| ka | Attachment rate | 0.06/s | 0.01–0.1 /s |
| dα | Aurora B–like activity coefficient | 0.2 µm | 0.01–1 µm |
| β | Kinetochore orientation effect coefficient | 1 | 0–1 |
Chromosome biorientation requires a combination of a kinetochore orientation effect and Aurora B–like activity
Previous studies in wild-type fission yeast cells indicate that most erroneous attachments of spindle microtubules to kinetochores are detected and corrected before anaphase onset, which occurs on average 10–12 min after SPB separation (Nabeshima et al., 1998). In the model, if at least one kinetochore is completely unattached, the cohesin spring will persist until stochastic attachment corrects the defect. This behavior mimics the activity of the SAC in the sense that it prevents cells with a single unattached kinetochore from entering anaphase. We also introduced into the model two correction mechanisms.
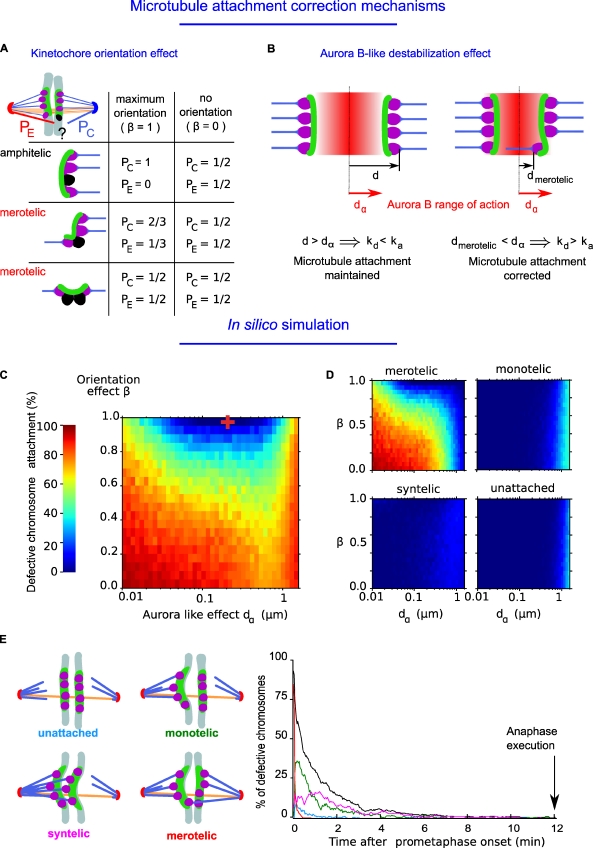
In the first (kinetochore orientation effect; Fig. 3 A), as previously assumed (Nicklas and Ward, 1994; Paul et al., 2009), a new ktMT attachment will be correct with a probability PC or erroneous with a probability PE that depends on the previous attachment state of the kinetochore to the poles. This is modulated with a parameter (β) spanning from 0 to 1. When β = 1 and the kinetochore is attached to a single spindle pole, the next attachment cannot be erroneous (Fig. 3 A, amphitelic, PC = 1 and PE = 0). However, when the kinetochore is attached to both poles, further attachment can be erroneous (Fig. 3 A, merotelic, PC < 1 and PE > 0). When β = 0, correct or erroneous attachments are equiprobable (PC = PE = 1/2). When β is between 0 and 1, correct attachment is favored (see Materials and methods).
Figure 3.
Relative contributions of kinetochore orientation effect and Aurora B–like activity to error correction. (A) Diagram illustrating the effect of a kinetochore orientation effect on the probability that an empty microtubule attachment site (in black) will be attached erroneously (in red, probability [PE]) or attached correctly (in blue, probability [PC]). As described in the Materials and methods, the kinetochore orientation effect is adjusted with a parameter β. Orientation effect is maximal when the parameter β = 1, and there is no orientation effect when β = 0. Several possibilities are shown, and the corresponding probabilities are indicated either for an amphitelic attachment (with a single unattached site) or a merotelic attachment (with one or two unattached sites). (B) Diagram illustrating the range of action of an Aurora B–like destabilization effect (dα; red gradient in between the two kinetochores) and its effect on the detachment rate (kd). When a merotelic kinetochore is localized in the range of action of the Aurora B–like destabilization effect, the distance from the position of the kinetochore binding site to the center of the kinetochore pair (dmerotelic) is lower than dα. Therefore, the probability for a microtubule to detach is high. Oppositely, if the position of the kinetochore binding site is not in the range of action of Aurora B–like destabilization effect, the distance (d) is higher than dα, and the probability for a microtubule to detach is low. (C) Simulations of the relative contributions of an Aurora B–like effect (dα) and kinetochore orientation effect (β) to the frequency of attachment defects. The percentage of chromosomes showing an attachment defect was measured 12 min after initiation of mitosis (average duration of metaphase, n = 45,100 simulations). The red cross indicates the optimum parameter values for Aurora B–like destabilization effect and kinetochore orientation effect at which there are the fewest chromosome attachment defects. (D) Simulations of the relative contributions of an Aurora B–like effect (dα) and kinetochore orientation effect (β) to the frequency of merotelic, monotelic, syntelic, and unattached. (E) Timing of correction of chromosome attachment defects (percentage) using the optimal value of correction mechanisms. The different types of erroneous attachments are shown: merotelic, monotelic, syntelic, unattached, and total defects (black).
The second correction mechanism mimics the role of Aurora B activity (Aurora B–like destabilization effect; Fig. 3 B). In silico, Aurora B activity modulates the probability of microtubule detachment as a function of the distance, d, between the microtubule attachment site and the center of the kinetochore pair (Fig. 3 B). Because a high value of d reflects tension at the kinetochore and increasing distance lowers the concentration of the kinase, the detachment rate should decrease with respect to d (Fig. 3 B). In the model, the detachment rate (kd) is inversely proportional to the distance (d):
the parameter dα is defined as the spatial range of Aurora B activity. Thus, when d is higher than dα, the attached state is favored (Fig. 3 B, left diagram). However, when d is lower than dα, the probability of microtubule detachment is increased, and correction takes place (Fig. 3 B, right diagram).
The kinetochore orientation effect (β) is likely to prevent the appearance of merotelic attachment, whereas the Aurora B–like destabilization effect (dα) will correct merotelic attachment. To quantify their respective contributions, we used the model to determine the percentage of chromosomes with defective kinetochore attachment when each value was altered (Fig. 3 C, n = 105 simulations). On a semilogarithmic plot, simulations reveal that the optimal condition for correct chromosome attachment is located at a combination of values of dα = 0.2 µm and of β = 1 (Fig. 3 C, represented by the red cross). The relative contribution of each type of attachment defect as a function of dα and β is shown in Fig. 3 D. As expected, when both parameters are low, the percentage of merotelic attachment is high (Fig. 3 D, red areas in the top left). In contrast, monotelic, unattached, and syntelic attachments are unaffected by changes in the value of the kinetochore orientation effect (β) but rather, only appear when the Aurora B–like destabilization effect (dα) is high (dα greater than the optimum; Fig. 3 D, pale blue areas in the three remaining simulations).
Using these optimum values, simulations reveal that the frequency of erroneous attachment decreases rapidly within the normal duration of phase 1 and 2 when kinetochores are randomly attached at time 0 (Fig. 3 E). Thus, our model demonstrates that a kinetochore orientation effect and fine tuning of an Aurora B–like activity are absolutely essential to prevent all types of attachment defects before anaphase onset (phase 3; 12 min).
The model reproduces abnormal chromosome segregation behavior caused by Aurora B inhibition
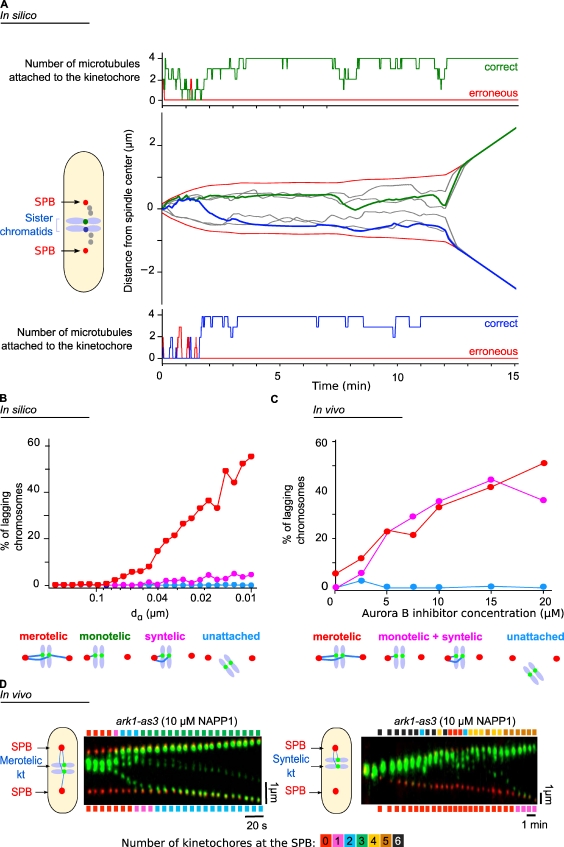
Our model accurately simulates the dynamics and breathing of kinetochores from prometaphase to anaphase B, and it predicts the level of occupancy of the attachment sites for any pair of sister kinetochores during mitotic progression. An example of a simulation is shown in Fig. 4 A and Video 4. In Fig. 4 A (center plot), the trajectories of the SPBs (Fig. 4 A, red) and the six kinetochores are shown during the interval from phase 1 (t = 0) to anaphase (t = 12 min). One pair is highlighted in green and blue, and the occupancy of the ktMT attachment sites for each is indicated (Fig. 4 A, green in the top plot corresponding to the green kinetochore and blue in the bottom plot corresponding to the blue kinetochore). Note that when an erroneous attachment occurs (Fig. 4 A, bottom, red line, between 1 and 2 min), it is rapidly corrected.
Figure 4.
Comparison of in silico and in vivo abnormal chromosome segregation behaviors in the case of Aurora B inhibition. (A) Simulations of the dynamics of the SPBs (black) and the three kinetochore pairs (gray and green/blue). In the center plot, the trajectories of the SPBs and innerkinetochores are shown during the interval from phase 1 (t = 0) to phase 3 (anaphase B). In the top and bottom plots, the occupancy of the ktMT attachment sites is indicated for two sister kinetochores (green in the top plot corresponding to the green kinetochore and blue in the bottom plot corresponding to the blue kinetochore). The red lines indicate the number of erroneous attachments, and the green/blue lines indicate the number of correct attachment. (B) Statistical analysis of the different chromosome attachment defects predicted by the model in silico when the Aurora B–like effect is reduced. (C) Quantification of the different types of attachment defects seen in vivo in the presence of increasing amounts of the inhibitor 1NAPP1. (D) Kymograph representations of the movements of the SPBs (Cdc11-cfp, red) and the kinetochores (kt; Ndc80-GFP, green) in a cell bearing an ATP analogue-sensitive mutation in the Aurora kinase gene, showing a stretched merotelic attachment (left) or syntelic attachment (right). The color bars above (and below) the kymographs give the number of kinetochores that have reached the upper (or lower) SPB at a given time, and the color code is detailed on the scale below the two kymographs.
To test the model further, we asked whether it would reproduce quantitatively and qualitatively the abnormal chromosome segregation observed in anaphase as a consequence of Aurora B inhibition in vivo (Hauf et al., 2007). We first performed simulations to identify the percentage of lagging chromosomes present after anaphase onset as a function of dα (Fig. 4 B). Decreasing dα (i.e., equivalent to inhibiting Aurora B) resulted in the maintenance of merotelic lagging chromosomes but very few syntelic attachments and no monotelic attachment (Fig. 4 B and see also Fig. S3 to visualize stretched ktMT attachment sites during merotely). We then tested the effect of inhibiting Aurora B in vivo by using conditional alleles of Aurora B sensitive to ATP analogues (ark1-as3, Shokat mutant; Hauf et al., 2007). With this strain, we can inhibit Aurora kinase activity simply by adding the ATP analogue 1NAPP1 to cells. To quantify the mitotic defects seen after Aurora inhibition, the temperature-sensitive double mutant cdc25-22 ark1-as3 was synchronized by incubation at 36°C to accumulate cells in G2 phase and then released into early mitosis by incubation at the permissive temperature of 25°C (>90% of cells were in phase 1) and adding various concentrations of 1NAPP1. Progress of the cells through mitosis was followed, and attachment defects in anaphase were quantified by live-cell imaging (Videos 5 and 6). We found that the percentage of merotelic attachments or the sum of monotelic and syntelic attachments increased with the degree of Aurora B inhibition (Fig. 4 C). When Aurora kinase was inhibited with 10 µM 1NAPP1, anaphase cells displayed single sister chromatids either stretched between the two poles (merotelic; Fig. 4 D, left) or sister chromatids attached to the same pole (syntelic or monotelic; Fig. 4 D, right). We conclude that our model qualitatively reproduces the abnormal chromosome segregation defects observed after Aurora inhibition.
Aurora activity and the kinetochore orientation effect generate unbalanced forces that trigger correction of merotelic attachment
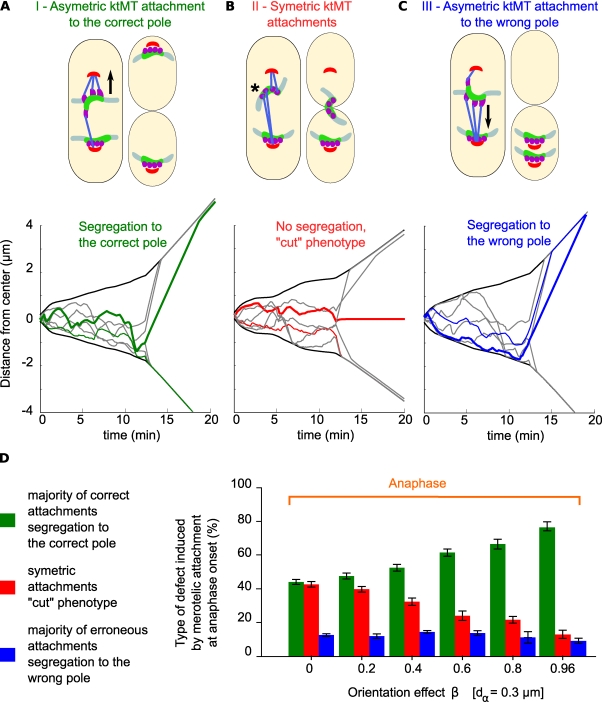
We previously showed that the vast majority of merotelic kinetochore attachments were resolved in anaphase, preventing the appearance of aneuploidy or the cut phenotype (Courtheoux et al., 2009). The precise mechanism for this anaphase correction is unknown, although previous studies suggest that an imbalance in the forces exerted by ktMTs might play a role (Cimini et al., 2004; Courtheoux et al., 2009). We performed simulations to test this hypothesis. We decreased β to increase the probability of merotelic attachment remaining in anaphase (β = 0.8 instead of the optimum) and then analyzed in the simulations the appearance of three major phenotypes (Fig. 5 A). In panel A, the merotelic (upper) kinetochore is attached asymmetrically and moves to the correct pole as shown by the dark green line in the simulation on the bottom (Fig. 5 A, category I, green). In Fig. 5 B, the merotelic (upper) kinetochore is attached symmetrically and fails to segregate, resulting in the cut phenotype as shown by the dark red line in the simulation on the bottom (Fig. 5 A, category II, red). In Fig. 5 C, the merotelic (upper) kinetochore is attached asymmetrically and moves to the wrong pole as shown by the dark blue line in the simulation on the bottom (Fig. 5 A, category III, blue).
Figure 5.
Unbalanced forces generated by an asymmetric attachment of microtubules promote segregation of merotely toward the correct pole in anaphase. (A, top) The merotelic (upper) kinetochore is attached asymmetrically and segregates to the correct pole (category I). (bottom) Example of a simulation corresponding to this category, the SPBs trajectories are in black. At anaphase onset (12 min), most kinetochores rapidly segregate to the poles (light green and gray traces), whereas the merotelic kinetochore (dark green trace) lags behind and regains the correct pole with a delay. (B, top) The merotelic (upper) kinetochore is attached symmetrically (asterisk) and fails to segregate, resulting in the cut phenotype as shown by the dark red line in the simulation below (category II). (C, top) The merotelic (upper) kinetochore is attached asymmetrically and slowly moves to the wrong pole as shown by the dark blue line in the simulation below (category III). Closed arrows indicate direction of chromosome movement. (D) Statistical analysis of the relative proportions of the three phenotypes generated by merotelic attachment in anaphase (n = 45,100 simulations) analyzed as a function of the kinetochore orientation parameter value β. Error bars represent the SD of the error rate for the 250 simulations used for each value of β.
In the absence of any influence from either dα or β, merotelic attachment will be unbiased. 50% of cells will show the cut phenotype (Fig. 5 A, category II), and the other 50% will show segregation to either the correct pole or to the wrong one (Fig. 5 A, categories I and III, 25% each). In the absence of β, but with an optimum value of dα, a small Aurora effect results in the reduction of category III from 25 to 10% (Fig. 5 C, blue bar, β = 0) and a corresponding increase in category I from 25 to 45% (Fig. 5 C, green bar, β = 0). Increasing β leads to no change in the value of category III, but the cut phenotype (Fig. 5 C, red bars) disappears progressively with a corresponding increase in category I. We conclude that unbalanced forces generated by an asymmetric attachment of microtubules are already present on merotelic kinetochores at anaphase onset to promote segregation toward the correct spindle pole. These unbalanced forces are generated rapidly in phase 1 by a combination of a (minor) Aurora-like destabilization effect and a (major) kinetochore orientation effect. This bias in kinetochore attachment, inherited from phase 1, is sufficient to explain the correction of merotely in anaphase.
Discussion
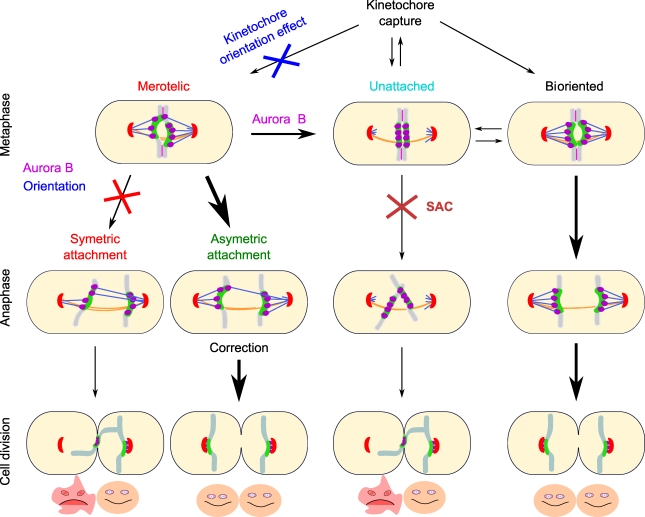
This study illustrates how a simplified force balance model, with stochastic attachment and detachment events and correction mechanisms (Aurora B and kinetochore orientation effect), can explain the segregation of chromosomes with a timing and accuracy similar to chromosome segregation in living wild-type fission yeast cells. The model reproduces the full dynamics of fission yeast chromosomes from phase 1 to anaphase B, with few free parameters. It also satisfies the requirements of the SAC by allowing correction of erroneous attachment before anaphase onset. Finally, the model reproduces the abnormal chromosome segregation behavior seen upon inhibition of Aurora B and corrects merotelic attachment in anaphase as previously observed in vivo (Courtheoux et al., 2009). The model thus identifies the critical parameters controlling both chromosome dynamics and the timing of correction of erroneous attachments in fission yeast (Fig. 6).
Figure 6.
Model summarizing the contributions of various mechanisms to correct microtubule attachment defects during cell division. Unattached kinetochores are captured during phase 1, and erroneous attachments, such as merotely, are produced when the kinetochore orientation effect is compromised. Aurora B activity corrects merotely by promoting microtubule destabilization, leading to unattached kinetochores. The spindle assembly checkpoint (SAC) prevents the segregation of unattached kinetochores. Aurora B–like activity and the kinetochore orientation effect also promote a reduction in the number of microtubules attached to the wrong pole, leading to unbalance forces at the merotelic kinetochore in anaphase. These unbalance forces are sufficient to promote correction in anaphase and proper cell division. On the contrary, if no correction occurs, the cytokinetic actin ring traps the merotelic chromosome during cell division, leading to aneuploidy. Therefore, a combination of Aurora B–like activity, the kinetochore orientation effect, and SAC promotes kinetochore biorientation and proper cell division.
It is now well established that kinetochore oscillations during mitosis can be affected by microtubule instability or microtubule attachment to kinetochores (Kops et al., 2010). Likewise, kinetochore alignment at the metaphase plate can be explained by various mechanisms, such as length-dependent microtubule destabilization (Varga et al., 2009) or the presence of a microtubule-stabilizing gradient at the spindle midzone (Gardner et al., 2008). What actually determines the directional instability of the kinetochore throughout mitosis, however, is still controversial. A direct modification of microtubule length or an alternative force applied to the chromosome might influence chromosome movement. Here, we provide evidence that a simple mechanism can generate chromosome oscillations during fission yeast mitosis. Our model predicts that simple forces applied stochastically on individual ktMT attachment sites are sufficient to produce the oscillations and movement of chromosomes observed in fission yeast mitosis. Therefore, yeast chromosome behavior can be modeled by a mechanism homologous to Ostergren’s traction force, except the magnitude depends on the number of microtubules, not their length (Ostergren et al., 1960; Hays et al., 1982).
In vivo experiments of vertebrate kinetochores have found multiple microtubule binding sites (20–30) that are gradually occupied by microtubules during early mitosis to reach a maximum after anaphase onset (McEwen et al., 1997). Interestingly, the SAC is only satisfied when ∼85% of microtubule attachment sites on the kinetochores are occupied, which might reflect the fact that attached microtubules are constantly turning over before anaphase onset (McEwen et al., 1997). However, in vertebrate, no correlation between the number of microtubules on sister kinetochores and the direction of chromosome movement has been observed, suggesting that our model is not compatible with orthomitosis (McEwen et al., 1997). There may thus be different mechanisms for chromosome movement in yeast versus higher eukaryotes. This hypothesis is supported by previous work showing that minus-end molecular motors are dispensable for poleward chromosome motion in fission yeast (Grishchuk and McIntosh, 2006).
Several in vivo processes have been deliberately omitted in the model for the sake of simplicity or to focus on S. pombe mitosis. For example, no chromokinesins are present in fission yeast (Wood et al., 2002), so there are no antipoleward ejection forces (Rieder et al., 1986). There is also no poleward flux of tubulin within the spindle in fission yeast (Mallavarapu et al., 1999) as opposed to higher eukaryotes (Mitchison, 1989). In contrast to a previous in vivo study performed in S. cerevisiae (Gardner et al., 2008), we assume that the force exerted on the kinetochore by ktMTs is independent of their length and that attachment and detachment rates are independent of the position within the spindle. Thus, our model does not reproduce chromosome congression at the metaphase plate. Such a feature could be easily implemented; however, it is unlikely to influence the correction of attachment defects. We also assume in our model that the detachment process is instantaneous and triggers a complete shutdown of the force at the kinetochore. Thus, we have neglected effects such as the penetration depth of the ktMT within the kinetochore plate (Hill sleeve model; Hill, 1985). Finally, we also made the first-order approximation that the force generators were following linear force velocity relationships, neglecting nonlinear effects from collective behavior of motors (Klumpp and Lipowsky, 2005; Guérin et al., 2010). These descriptions of both midzone and kinetochore force generators faithfully reproduce key aspects of the spindle dynamics, such as the increased rate of spindle elongation in phase 1, before kinetochore biorientation (this study), the observed dependency of the spindle elongation rate on the number of merotelic kinetochore at anaphase (Courtheoux et al., 2009), and the observed poleward movements of chromosomes at anaphase onset.
Evidence from studies of living cells suggests that lateral attachment of kinetochores to microtubules is common in eukaryotes (Hayden et al., 1990; Merdes and De Mey, 1990; Rieder and Alexander, 1990; Tanaka et al., 2005; Grishchuk and McIntosh, 2006; Franco et al., 2007; Gachet et al., 2008). In budding yeast (Kitamura et al., 2010), fly (Maiato et al., 2004b), and vertebrate cells (Khodjakov et al., 2003), microtubules can also be generated from kinetochores and facilitate kinetochore capture by the spindle apparatus. In fission yeast, unattached kinetochores are retrieved to the spindle pole by end-on attachment to the microtubule followed by microtubule depolymerization (Grishchuk and McIntosh, 2006; Gachet et al., 2008). This result suggests that end-on attachment to the microtubule is also promoted in a bipolar spindle. In our model, the forces are directly applied to individual kinetochore attachment sites, assuming that only end-on attachment of chromosomes is present; however, we cannot rule out that lateral attachment of kinetochores to microtubules may affect kinetochore dynamics in a different manner to end-on attachment.
Kinetochore geometry was originally defined as a back to back position of sister kinetochore (Tanaka, 2010). This implies that when one kinetochore attaches to microtubules from one spindle pole (monotelic attachment), the sister kinetochore can only associate with microtubules from the opposite pole. This would prevent the formation of aberrant ktMT attachments, such as syntelic attachment. A recent study suggests that the coorientation of sister ktMT attachment sites toward the same pole in meiosis requires the function of a protein complex called the monopolin complex (Monje-Casas et al., 2007). It has recently been proposed that subunits of this complex (Csm1/Lrs4) function as a molecular clamp, cross-linking kinetochore components to suppress merotelic attachment in S. pombe mitosis (Corbett et al., 2010; Rumpf et al., 2010; Gregan et al., 2011). These results suggest that the orientation of ktMT binding sites toward the same pole plays an important role in preventing merotely. Accordingly, our model predicts that a process favoring the orientation of ktMT binding sites toward the same pole is sufficient to prevent aberrant ktMT attachments, such as merotely, as observed in monopolin mutants (Gregan et al., 2007).
The implementation in the model of a control over ktMT attachment site orientation could prevent erroneous attachment but could not actively correct syntely or merotely. Only a control at the level of sister kinetochores could perform this function. Indeed, our model predicts that correct attachment also requires fine tuning of tension across sister kinetochores; either increasing or decreasing an Aurora B–like activity causes several kinds of defects, including merotelic and syntelic attachment. The model predicts that merotelic attachment is the most frequent attachment defect when Aurora B is inhibited, as confirmed in vivo, and demonstrates that merotelic attachment can lead to several phenotypes of aneuploidy. By classifying the different types of merotelic attachment according to the phenotype generated, we find that merotelic attachments segregating to the wrong pole leads to a syntelic phenotype, which is not detected by the SAC. The activity of Aurora B is therefore not only crucial to correct merotelic attachment in metaphase but also establishes the unbalanced force at the kinetochore that allows correction of merotelic attachment in anaphase.
As previously suggested by Nicklas and Ward (1994), our model reveals that the duration of prometaphase/metaphase in fission yeast is long enough to allow proper biorientation to occur by chance. Before anaphase, each chromosome pair needs to reach its equilibrium state of attachment (correct biorientation) by sorting each attachment site, through a stochastic succession of attachment and detachment events biased toward biorientation. What determines the timing of biorientation and consequently the duration of prometaphase in eukaryotic cells remains elusive. Our model provides the basis to understand this important question.
Materials and methods
In vivo methods
Cell culture.
Media, growth, maintenance of strains, and genetic methods were performed as previously reported (Moreno et al., 1991). Cells were grown at 25°C in yeast extract and centrifuged for 30 s at 3,000 g before mounting in an imaging chamber. The ark1-as3 mutant was provided by S. Hauf (Friedrich Miescher Laboratory, Tubingen, Germany). The various strains used in this study are listed in Table S1.
Live-cell imaging.
Live-cell analysis was performed in an imaging chamber (CoverWell PCI-2.5; Grace Bio-Labs, Inc.) filled with 1 ml of 1% agarose in minimal medium and sealed with a 22 × 22–mm glass coverslip. Time-lapse images were taken at 25°C. Exposure times were taken at 300–500 ms using a light source (HIGHlite; Roper Scientific) reduced to 30% to avoid phototoxicity and photobleaching. Images were visualized with a charge-coupled device camera (CoolSNAP HQ2; Roper Scientific and Princeton Instruments) fitted to an upright microscope (DM6000; Leica) with a 100×, 1.4 NA or a 63×, 1.4 NA objective and filters (Semrock) for GFP, CFP, or RFP. Images were recorded using the MetaMorph software package (Molecular Devices). Intensity and γ adjustments (threshold) were made using the Metamorph, ImageJ (National Institutes of Health), and Photoshop (Adobe) packages.
Laser ablation of mitotic cells.
Exponentially growing cultures of cdc25-22 cells at 25°C were arrested in G2 by incubation at 36°C for 4 h and then released into mitosis by rapid cooling to 25°C. 10 min after release, mitotic cells were followed by live microscopy at 25°C, and spindles were submitted to laser ablation. As previously described (Courtheoux et al., 2009), the system used to perform laser ablation is composed of a conventional inverted microscope (DMI6000 B; Leica) equipped with a heated stage covered with an incubation system, including a temperature controller. Mitotic spindle photoablation was achieved with a frequency-doubled neodymium-doped yttrium aluminum garnet–pulsed laser at a wavelength of 532 nm. The pulse duration was estimated as 600 ps with a repetition rate of 10 kHz (MicroChip, Sealed Green 532 nm; JDS Uniphase). An iLas head (Roper Scientific) comprised of a laser shutter and a galvanometer pair mirror was coupled to the microscope through the epifluorescence port and used to guide the laser beam within the field of view of the camera. The beam was focused by a high NA objective lens (HCS Plan Apochromat 100×, NA 1.4 oil immersion). Images were acquired with a cooled charge-coupled device camera (CoolSNAP HQ2). The system was controlled by MetaMorph software. The tight focusing of the laser beam to a beam waist of 1 µm allows the selective destruction of one SPB together with a small region of the spindle microtubules, whereas the kinetochores and the opposite SPB remain intact. Mitotic spindles were exposed to the pulsed beam for 25 ms.
In vivo data analysis
Multiparticle tracking.
Tracking of kinetochore trajectories was performed by using the particle tracker algorithm developed by Interactive Data Language (Crocker and Grier, 1996) and ported to MatLab by D. Blair (Georgetown University, Washington DC) and E. Dufresne (Yale University, New Haven, CT). In brief, this is a peak detection algorithm; localization of the fluorescent spot center with subpixel accuracy is achieved by finding the center of mass of the neighboring pixels. Trajectories are then reconstructed from the detected particles in each image by frame to frame distance minimization. After this automated reconstruction, the tracked trajectories are corrected through a graphical user interface written in the Python programming language.
To ensure that the Cen2 trajectories provided a faithful representation of kinetochore movement, we tracked a strain marked with the kinetochore marker Ndc80-GFP and the centromere marker Cen2-GFP (the Cen2-GFP strain corresponds to an insertion at a 5-kb distance from the centromere; Yamamoto and Hiraoka, 2003). In video frames in which both markers clearly belonged to the same kinetochore, the distance between the two dots was measured. From those images, we conclude that the average distance between Cen2 and Ndc80 was 126 ± 10 nm (Fig. S1). Cen2 is thus a good marker for the position of the chromosome 2 kinetochore.
Force balanced model of chromosome segregation
Spindle geometry.
In our model, the following elements of the spindle are taken into account: two SPBs, three centromere pairs, and four microtubule attachment sites per kinetochore. The spindle is defined at each time point by the speed and position of these elements as well as the attachment state of each microtubule attachment site. The attachment state can be 0 when no microtubule is attached to the site, 1 when a microtubule emanating from the correct SPB is attached, and −1 when the microtubule emanates from the erroneous SPB. When a microtubule is attached to the kinetochore, its plus end is assumed to follow the position of the attachment site and to grow and shrink when the kinetochore moves poleward or antipoleward. Although the molecular mechanisms responsible for this regulation are not fully understood, the ability of the microtubule plus end to remain attached to the kinetochore whether it moves poleward or antipoleward is generally accepted (Skibbens et al., 1993; Maiato et al., 2004a). We chose not to model the microtubules when they are not attached to the kinetochore, given the fact that, in this case, they exert no force. In fission yeast, kinetochore trajectories are parallel to the spindle axis. Therefore, the model describes the dynamics of these elements in one dimension (the pole to pole axis), with the axis origin located between the two poles.
Two classes of forces are considered: passive forces and active forces. Passive forces are the viscous drag applied to the SPBs and the kinetochores, the damped spring that links the attachment sites to the kinetochore, and a Hookean spring that links the kinetochore pairs. The active forces are pulling and pushing forces acting between the kinetochore and the SPB (the kinetochore force generator) as well as between both SPBs (the interdigitating microtubule force generator). In vivo, these forces arise from the activity of various molecular motors and from growing/shrinking microtubules. The activity of individual molecular motors has been characterized in vitro by optical tweezer experiments, and the importance of the collective behavior of individual motors has been demonstrated experimentally (Fallesen et al., 2011) and theoretically (Badoual et al., 2002; Klumpp and Lipowsky, 2005; Kunwar and Mogilner, 2010; Orlandi et al., 2010). The observed force–velocity relationships in such cases are often nonlinear, although they always exhibit an overall decreasing trend (high forces at low speeds and low forces at high speeds). The model makes the first-order approximation that active forces follow linear force–velocity relationships. The assumption that kinetochore force generators always pull the attachment site is in agreement with the fact that kinetochores always move toward, and not away from, the spindle pole during chromosome recapture experiments (Gachet et al., 2008).
Parameter definition.
When a microtubule is attached to both a kinetochore and one of the two SPBs, the microtubule exerts a force that depends linearly on the relative speed of the attachment site with respect to the SPB
in which vnm is the speed of the kinetochore attachment site, and vSPB is the speed of the SPB. The force is defined by two parameters: Vk is the motor maximum speed, and Fk is its stall force. Similarly, the midzone motors are modeled by a unique force–velocity relationship
in which is the speed of the left-hand side SPB (i.e., the SPB positioned on the negative part of the x axis), and Vmz and Fmz are, respectively, the midzone force generator’s maximum velocity and stall force. By definition of the reference frame with its origin equidistant of the SPBs, , so the previous relationship can be rewritten as
Thus, the midzone force generator always pushes the two poles apart.
In the model, microtubules can be attached to the kinetochore, in which case the pulling force is “on,” or detached, in which case the pulling force is “off.” The transition from one state to the other, we assume, is stochastic and instantaneous. Microtubules have a certain probability Pd to detach or Pa to attach to the kinetochore. These processes are Poissonian, as was already assumed in a study modeling the activity of the SAC (Doncic et al., 2005). In the time interval Δt, the probability that an attached ktMT will detach is given by Pd (Δt) = 1 − exp(−kdΔt), and similarly, the probability that a detached ktMT will attach is given by Pa (Δt) = 1 − exp(−kaΔt), in which kd and ka are the detachment and attachment rates. In vivo, attachment takes place when an attachment site encounters a free microtubule plus end undergoing dynamic instability. Here, the underlying free microtubule trajectory is implicit, and the stochastic nature of the attachment is a consequence of the random distribution of microtubule plus end within the spindle. We make the assumption that this distribution is uniform. Electron microscopy data of fission yeast spindles tend to strengthen this hypothesis (Ding et al., 1993). As a consequence of this approximation, ka is assumed to be constant. The detachment rate depends on the position of the attachment site, as will be detailed in the following paragraph.
Force balance model.
The force at the centromere is a balance between the Hookean spring between the sister chromatids, pulling them toward one another, the friction imposed by the viscous nucleoplasm opposing the centromere movement, and the ktMTs pulling toward the SPB. The force at the SPB is a balance between the ktMTs pulling toward the kinetochore (and thus the spindle center), the friction of the nucleoplasm, and the interdigitating microtubules pushing away from the spindle center.
The active forces at the midzone and the kinetochore are balanced by other components: (a) The friction forces on the kinetochores and the SPBs, described by Ff = −μv. (b) The bond between the centromere and the attachment sites, described by Fv = κk(xn − xnm − dk) − μk(vn − vnm), in which xn and vn are the position and speed of the n right-hand side centromere, and xnm is the position of the mth attachment site of the nth kinetochore. κk and μk are the spring constant and friction coefficient, respectively. (c) The cohesin bond between the centromeres, described by in which is the position of the n left-hand side centromere, κc is the cohesin spring constant, and dc is the centromere–centromere rest distance.
The force balance model is written as a system of coupled first-order differential equations, in which the sum of the forces applied to each element is equal to 0 (a link to the source code of the model is available on S. Tournier’s laboratory webpage at the Laboratoire de biologie cellulaire et moléculaire du contrôle de la proliferation). The simulation of full chromosome segregation relies on numerical resolution of the set of equations at each time step followed by Monte Carlo simulation of the stochastic attachment and detachment events according to the method previously proposed (Civelekoglu-Scholey et al., 2006). At anaphase onset, the cohesin spring constant is set to 0 to reflect the removal of the cohesin bond, and no further attachment or detachment processes take place.
Computer simulation.
A flow diagram of the simulation is given in Fig. S2. The simulations are implemented in Python; NumPy and SciPy libraries are used for the numerical aspects of the simulation. It is possible to simulate laser ablation. At a given time point, the force generators between one SPB and the other elements of the spindle are removed; all the attachment sites are detached and the midzone force generator is removed (by setting Fmz to 0). A graphical user interface developed in Qt (Nokia) allows observation of a given simulation and easy parameter exploration. Data analysis was also performed in Python with custom scripts. Graphical representations were obtained with the matplotlib library.
Modeling chromosome segregation with accuracy
In the model, microtubules attached to a kinetochore can bind to microtubules emanating from either of the two SPBs, leading to erroneous attachments. To reproduce the rare erroneous attachments observed in wild-type cells, we have included in the model a bias favoring correct attachment by modifying the attachment and detachment processes.
Kinetochore orientation effect on the microtubule binding site.
We assume that when one microtubule attaches a kinetochore to a SPB, further attachments from that kinetochore to the same SPB will be favored. When both SPBs are attached to the same kinetochore (i.e., the kinetochore is merotelic), the next attachment will be correct with the probability
or erroneous with the probability
in which nE is the number of erroneously attached sites, and nc is the number of correctly attached sites. This orientation effect is modulated by the factor β, such that Pc = PE = 1/2 when β = 0 (no effect), and Pc = nc/(nc + nE) when β = 1 (full effect).
Modeling Aurora B–like activity.
An evolutionarily conserved protein kinase, Aurora B, is known to destabilize ktMTs when they are not attached correctly. This destabilization is thought to depend on tension or kinetochore–kinetochore distance. In the model, the detachment rate of a microtubule is dependent on its distance from the center of the centromere pair. Mathematically, kd = ka(dα/d), with
with xn, , and xnm as described in the Force balance model section, and dα as an adjustable parameter, which is called the Aurora B–like activity parameter. When d is lower than dα, the detached state is more stable than the attached one, thus promoting correction. As this relation has only one adjustable parameter, it is easy to explore the role of this Aurora B–like activity on error correction by changing the value of dα. It has been shown in vitro that reconstituted kinetochore particles behave as catch bonds on the microtubule, so that the applied load stabilizes the attachment (Akiyoshi et al., 2010). In the absence of direct in vivo measurements for this phenomenon in fission yeast, we made the approximation that the ktMT detachment rate is solely dependent on this Aurora B–like positional effect (not on the applied force at the attachment site).
Parameter estimation
The stall force (Fk).
We use nondimensionalization to solve the system of differential equations. The stall force of the ktMT motor (Fk) is used as the unit force, thus all the other forces are calculated as multiples of Fk. Consequently, the determination of an absolute value for Fk was not required for this study. To allow comparison of the parameter values given by our model with previous studies, however, it was convenient to estimate Fk. It is difficult to measure this force in vivo in fission yeast. According to microneedle experiments performed on grasshopper spermatocytes (Nicklas, 1988), the maximum force exerted by one kinetochore fiber is ∼50 pN in anaphase A. Because kinetochore fibers in S. pombe contain about four microtubules, we estimate Fk as 10 pN (Table 2).
Chromosome number (N).
S. pombe contains three chromosomes, therefore N = 3. From electron microscopy (Ding et al., 1993), each kinetochore has four attachment sites for microtubules, therefore Mk = 4.
Intrakinetochore equilibrium distance (di).
In the absence of direct evidence from electron microscopy, we assume that the kinetochore and the centromere are closely apposed. Thus, we have fixed the equilibrium distance between the centromere and the attachment sites (dk) at 0 nm. A higher value for this parameter would only offset the attachment site trajectory with respect to the centromere.
Intercentromere distance (dc).
The equilibrium distance between the centromeres (dc) is set to 400 nm based on video microscopy of unattached centromere pairs (Gachet et al., 2008).
SPB friction coefficient (μs).
We have no rigorous way to estimate the SPB friction coefficient (μs). As the friction force is relatively small compared with the active forces, this value can be changed over one log without affecting the observed spindle dynamics. We therefore chose a value of 500 pN.s/µm, which is of the same order of magnitude as the value obtained for the friction coefficient of the kinetochore.
KtMT motor maximum speed (Vk).
Drag forces in anaphase A are much lower than the maximum force a ktMT can deliver (Nicklas, 1988). In our model, this means that the poleward speeds of the kinetochores are close to the ktMT motor’s maximum speed (Vk) because vnm − vSPB = Vk when the force is zero according to the linear force velocity relationship given (see previous paragraph). According to our own measurements, Vk = 0.03 µm/s.
Cohesin spring constant (κc).
The maximum distance between sister kinetochores in metaphase occurs when both kinetochores are saturated with microtubules and are immobile. In this case, the load on the spring equals MkFk, in which Mk is the maximum number of attachment sites. This force is balanced by the restoring force of the spring, κc(dmax − dc), in which κc is the cohesin spring constant. From these formulae, κc can be computed: κc = MkFk/(dmax − dc). According to our in vivo measurements, dmax is ∼1.0 µm, and thus, κc = κo ≈ 42 pN/µm when Fk = 10 pN.
Kinetochore spring constant (κk).
The spring constant associated with the structural bond between the inner and outer kinetochore plate can be determined when merotelic attachment occurs in anaphase. In this case, the kinetochore is stretched by the ktMTs emanating from both poles. The maximum stretching occurs when half the microtubules (Mk/2) are attached to one side, and half are attached to the other. We then have
and κk = 21 pN/µm with dk,max = 0.3 µm.
Friction coefficients.
Laser ablation experiments provide relaxation times for both inter- and intrakinetochore links. At first order, the relaxation time is given by the ratio of the spring constant to the friction coefficient τ = κ/μ. The friction coefficients for the sister chromatids and for the outer–inner bond are determined by using τ = 10 s in both cases. Thus, μk = 900 pN.s/µm and μk = 400 pN.s/µm.
Midzone motors maximum speed (Vmz).
The drag force opposing spindle elongation in anaphase B is weak compared with stall force of the midzone force generators. Thus, the anaphase spindle elongation rate can be used as an estimate of twice the midzone force generators’ maximum speed. This rate was measured as 0.06 µm/s, so Vmz = 0.03 µm/s.
Midzone motors stall force (Fmz).
In metaphase, the spindle elongates slowly as the attached ktMTs oppose the action of the midzone motors. The average elongation rate can be easily measured. The average force exerted by the ktMTs on the spindle poles is given by
in which, is the average SPB speed and equals half the metaphase elongation rate, and is the average occupancy of each attachment site. It can be shown that
and. The average distance between the kinetochore pair, , is measured experimentally. Neglecting friction, the force of the ktMTs is balanced by the force exerted by the midzone motors
From the two aforementioned relations, we have:
Three parameters thus remain to be determined: the attachment rate ka, the Aurora B–like parameter, dα, governing the detachment process, and the orientation parameter β. As discussed in the Results section (Fig. 3 C, red cross), the values of dα and β were determined from the minimum rate of misattachments at anaphase onset.
Attachment and detachment events trigger changes in kinetochore trajectories and contribute negatively to the trajectory’s autocorrelation function A, which is defined as follows:
with δt as the time step between two acquisitions, x(iδt) as the position of the kinetochore at the i time point, as the average of x, σ as its SD, and L as the total number of time points. The first minimum of this function (tmin) provides a good estimate of the directional instability time scale. We choose to set ka = 1/tmin. We verified that this estimate yielded accurate values for the first minimum of the autocorrelation function in silico (Fig. 2).
Comparison of the parameter values with previous studies
As already stated, the value of Fk is fixed to 10 pN, according to the values previously published and reviewed (Civelekoglu-Scholey and Scholey, 2010) or the values observed with purified kinetochore particles (Akiyoshi et al., 2010). Our value for the friction coefficient of the sister chromatid, μc, is around 500 pN.s/µm. For Drosophila melanogaster anaphase chromosomes, two studies use a value of 5 pN.s/µm (Marshall et al., 2001; Civelekoglu-Scholey et al., 2006). Our estimate is thus higher by a factor of 100. This higher value might be caused by an overestimation of Fk. It might also originate from a higher interaction of the kinetochore with spindle microtubules, e.g., through high affinity lateral interaction in phases 1 and 2. KtMT attachment and detachment frequencies are consistent with previous studies for microtubule rescue and catastrophe frequencies (in the range of 0.01–2/s; Gardner et al., 2005; Civelekoglu-Scholey et al., 2006). The elastic modulus for the kinetochore pair is consistent with previous studies (Civelekoglu-Scholey et al., 2006; Bouck and Bloom, 2007). Overall, and given the uncertainties for the magnitude of Fk, our parameter values are in good agreement with those obtained in previous studies, with the exception of the friction coefficients.
Online supplemental material
Fig. S1 illustrates the distribution of distances between the kinetochore signals and the centromere signals at the kinetochore. Fig. S2 shows a flow chart diagram of the simulation. Fig. S3 represents a simulation showing stretched ktMT attachment sites during merotely. Video 1 shows an animated representation of the model. Video 2 shows an example of fluorescent time-lapse imaging of chromosome 2 dynamics in a wild-type fission yeast cell. Video 3 shows a fluorescent time-lapse imaging of kinetochore dynamics after laser ablation of a fission yeast metaphase spindle. Video 4 represents an in silico simulation showing kinetochore dynamics during mitosis. Video 5 shows fluorescent time-lapse imaging of merotelic kinetochore dynamics after Aurora B inhibition. Video 6 shows fluorescent time-lapse imaging of syntelic kinetochore dynamics after Aurora B inhibition. Table S1 shows strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201107124/DC1.
Acknowledgments
We would like to thank S. Hauf for supplying the ark1-as3 mutant, J. Hyams and A. Merdes for critical reading of the manuscript, X. He for helpful discussions, R. Duteuil for mathematical tips, and the reviewers for spending time improving our paper.
G. Gay was supported by the Centre National de la Recherche Scientifique and the L’Agence Nationale de la Recherche. T. Courtheoux was supported by La ligue contre le cancer. This work was supported by the Association pour la Recherche sur le Cancer. The microscopy equipment was funded by the Centre National de la Recherche Scientifique, l’Association de la Recherche sur le Cancer and GlaxoSmithKline. This work is funded by the L’Agence Nationale de la Recherche (grant 2010 blanc 120601).
Footnotes
Abbreviations used in this paper:
- ktMT
- kinetochore microtubule
- SAC
- spindle assembly checkpoint
- SPB
- spindle pole body
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badoual M., Jülicher F., Prost J. 2002. Bidirectional cooperative motion of molecular motors. Proc. Natl. Acad. Sci. USA. 99:6696–6701 10.1073/pnas.102692399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Hardwick K., Javerzat J.P. 1998. Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143:1775–1787 10.1083/jcb.143.7.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K. 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17:741–748 10.1016/j.cub.2007.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher I., Civelekoglu-Scholey G., Kwon M., Mogilner A., Scholey J.M. 2004. Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc. Natl. Acad. Sci. USA. 101:15938–15943 10.1073/pnas.0407044101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.S., Botstein D. 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 135:677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Cameron L.A., Salmon E.D. 2004. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 14:2149–2155 10.1016/j.cub.2004.11.029 [DOI] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C.B., Salmon E.D. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Scholey J.M. 2010. Mitotic force generators and chromosome segregation. Cell. Mol. Life Sci. 67:2231–2250 10.1007/s00018-010-0326-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Sharp D.J., Mogilner A., Scholey J.M. 2006. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 90:3966–3982 10.1529/biophysj.105.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- Corbett K.D., Yip C.K., Ee L.S., Walz T., Amon A., Harrison S.C. 2010. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 142:556–567 10.1016/j.cell.2010.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtheoux T., Gay G., Gachet Y., Tournier S. 2009. Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J. Cell Biol. 187:399–412 10.1083/jcb.200902093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J.C., Grier D.G. 1996. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179:298–310 10.1006/jcis.1996.0217 [DOI] [Google Scholar]
- Ding R., McDonald K.L., McIntosh J.R. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A., Ben-Jacob E., Barkai N. 2005. Evaluating putative mechanisms of the mitotic spindle checkpoint. Proc. Natl. Acad. Sci. USA. 102:6332–6337 10.1073/pnas.0409142102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallesen T.L., Macosko J.C., Holzwarth G. 2011. Force-velocity relationship for multiple kinesin motors pulling a magnetic bead. Eur. Biophys. J. 40:1071–1079 10.1007/s00249-011-0724-1 [DOI] [PubMed] [Google Scholar]
- Franco A., Meadows J.C., Millar J.B. 2007. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J. Cell Sci. 120:3345–3351 10.1242/jcs.013698 [DOI] [PubMed] [Google Scholar]
- Gachet Y., Reyes C., Courthéoux T., Goldstone S., Gay G., Serrurier C., Tournier S. 2008. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol. Biol. Cell. 19:1646–1662 10.1091/mbc.E07-09-0910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Pearson C.G., Sprague B.L., Zarzar T.R., Bloom K., Salmon E.D., Odde D.J. 2005. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell. 16:3764–3775 10.1091/mbc.E05-04-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Bouck D.C., Paliulis L.V., Meehl J.B., O’Toole E.T., Haase J., Soubry A., Joglekar A.P., Winey M., Salmon E.D., et al. 2008. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 135:894–906 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Riedel C.G., Pidoux A.L., Katou Y., Rumpf C., Schleiffer A., Kearsey S.E., Shirahige K., Allshire R.C., Nasmyth K. 2007. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol. 17:1190–1200 10.1016/j.cub.2007.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Polakova S., Zhang L., Tolić-Nørrelykke I.M., Cimini D. 2011. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 21:374–381 10.1016/j.tcb.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., McIntosh J.R. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25:4888–4896 10.1038/sj.emboj.7601353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin T., Prost J., Martin P., Joanny J.F. 2010. Coordination and collective properties of molecular motors: theory. Curr. Opin. Cell Biol. 22:14–20 10.1016/j.ceb.2009.12.012 [DOI] [PubMed] [Google Scholar]
- Hauf S., Biswas A., Langegger M., Kawashima S.A., Tsukahara T., Watanabe Y. 2007. Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I. EMBO J. 26:4475–4486 10.1038/sj.emboj.7601880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden J.H., Bowser S.S., Rieder C.L. 1990. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J. Cell Biol. 111:1039–1045 10.1083/jcb.111.3.1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays T.S., Wise D., Salmon E.D. 1982. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J. Cell Biol. 93:374–389 10.1083/jcb.93.2.374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Patterson T.E., Sazer S. 1997. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 94:7965–7970 10.1073/pnas.94.15.7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T.L. 1985. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA. 82:4404–4408 10.1073/pnas.82.13.4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. 1986. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 5:2973–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Kelly A.E., Funabiki H. 2009. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr. Opin. Cell Biol. 21:51–58 10.1016/j.ceb.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Copenagle L., Gordon M.B., Compton D.A., Kapoor T.M. 2003. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160:671–683 10.1083/jcb.200208143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M., Mitchison T. 1986. Beyond self-assembly: from microtubules to morphogenesis. Cell. 45:329–342 10.1016/0092-8674(86)90318-1 [DOI] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C., Tanaka T.U. 2010. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell. 18:248–259 10.1016/j.devcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S., Lipowsky R. 2005. Cooperative cargo transport by several molecular motors. Proc. Natl. Acad. Sci. USA. 102:17284–17289 10.1073/pnas.0507363102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., Stukenberg P.T. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Kops G.J., Saurin A.T., Meraldi P. 2010. Finding the middle ground: how kinetochores power chromosome congression. Cell. Mol. Life Sci. 67:2145–2161 10.1007/s00018-010-0321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar A., Mogilner A. 2010. Robust transport by multiple motors with nonlinear force-velocity relations and stochastic load sharing. Phys. Biol. 7:16012 10.1088/1478-3975/7/1/016012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Cheeseman I.M. 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21:133–140 10.1016/j.tcb.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A.W. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531 10.1016/0092-8674(81)90015-5 [DOI] [PubMed] [Google Scholar]
- Loïodice I., Staub J., Setty T.G., Nguyen N.P., Paoletti A., Tran P.T. 2005. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell. 16:1756–1768 10.1091/mbc.E04-10-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Lince-Faria M. 2010. The perpetual movements of anaphase. Cell. Mol. Life Sci. 67:2251–2269 10.1007/s00018-010-0327-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., DeLuca J., Salmon E.D., Earnshaw W.C. 2004a. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477 10.1242/jcs.01536 [DOI] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., Khodjakov A. 2004b. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A., Sawin K., Mitchison T. 1999. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol. 9:1423–1426 10.1016/S0960-9822(00)80090-1 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Marko J.F., Agard D.A., Sedat J.W. 2001. Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr. Biol. 11:569–578 10.1016/S0960-9822(01)00180-4 [DOI] [PubMed] [Google Scholar]
- McEwen B.F., Heagle A.B., Cassels G.O., Buttle K.F., Rieder C.L. 1997. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137:1567–1580 10.1083/jcb.137.7.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Volkov V., Ataullakhanov F.I., Grishchuk E.L. 2010. Tubulin depolymerization may be an ancient biological motor. J. Cell Sci. 123:3425–3434 10.1242/jcs.067611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes A., De Mey J. 1990. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur. J. Cell Biol. 53:313–325 [PubMed] [Google Scholar]
- Millband D.N., Hardwick K.G. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22:2728–2742 10.1128/MCB.22.8.2728-2742.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J. 1989. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109:637–652 10.1083/jcb.109.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Evans L., Schulze E., Kirschner M. 1986. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 45:515–527 10.1016/0092-8674(86)90283-7 [DOI] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V.R., Lee B.H., Boselli M., Amon A. 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 128:477–490 10.1016/j.cell.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., Yanagida M. 1998. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 9:3211–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. 1988. The forces that move chromosomes in mitosis. Annu. Rev. Biophys. Biophys. Chem. 17:431–449 10.1146/annurev.bb.17.060188.002243 [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Ward S.C. 1994. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 126:1241–1253 10.1083/jcb.126.5.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R.A., Hamilton R.S., Pauli A., Davis I., Nasmyth K. 2010. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 12:185–192 10.1038/ncb2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi J.G., Blanch-Mercader C., Brugués J., Casademunt J. 2010. Cooperativity of self-organized Brownian motors pulling on soft cargoes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82:061903 10.1103/PhysRevE.82.061903 [DOI] [PubMed] [Google Scholar]
- Ostergren G., Mole-Bajer J., Bajer A. 1960. An interpretation of transport phenomena at mitosis. Ann. NY Acad. Sci. 90:381–408 10.1111/j.1749-6632.1960.tb23258.x [DOI] [PubMed] [Google Scholar]
- Paul R., Wollman R., Silkworth W.T., Nardi I.K., Cimini D., Mogilner A. 2009. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc. Natl. Acad. Sci. USA. 106:15708–15713 10.1073/pnas.0908261106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin C.S., Odell G.M., Oster G.F. 1993. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65:316–324 10.1016/S0006-3495(93)81035-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Alexander S.P. 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 110:81–95 10.1083/jcb.110.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Davison E.A., Jensen L.C., Cassimeris L., Salmon E.D. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 103:581–591 10.1083/jcb.103.2.581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948 10.1083/jcb.130.4.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner A.D., Murray A.W. 1996. The spindle assembly checkpoint. Curr. Opin. Cell Biol. 8:773–780 10.1016/S0955-0674(96)80077-9 [DOI] [PubMed] [Google Scholar]
- Rumpf C., Cipak L., Schleiffer A., Pidoux A., Mechtler K., Tolić-Nørrelykke I.M., Gregan J. 2010. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 9:3997–4004 10.4161/cc.9.19.13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuno T., Tada K., Watanabe Y. 2009. Kinetochore geometry defined by cohesion within the centromere. Nature. 458:852–858 10.1038/nature07876 [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Skeen V.P., Salmon E.D. 1993. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122:859–875 10.1083/jcb.122.4.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 434:987–994 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178:269–281 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U. 2010. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 29:4070–4082 10.1038/emboj.2010.294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., Medema R.H. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Tatebe H., Goshima G., Takeda K., Nakagawa T., Kinoshita K., Yanagida M. 2001. Fission yeast living mitosis visualized by GFP-tagged gene products. Micron. 32:67–74 10.1016/S0968-4328(00)00023-8 [DOI] [PubMed] [Google Scholar]
- Tournier S., Gachet Y., Buck V., Hyams J.S., Millar J.B. 2004. Disruption of astral microtubule contact with the cell cortex activates a Bub1, Bub3, and Mad3-dependent checkpoint in fission yeast. Mol. Biol. Cell. 15:3345–3356 10.1091/mbc.E04-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 400:37–42 10.1038/21831 [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., Diez S., Howard J. 2009. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 138:1174–1183 10.1016/j.cell.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature. 415:871–880 10.1038/nature724 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Hiraoka Y. 2003. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22:2284–2296 10.1093/emboj/cdg222 [DOI] [PMC free article] [PubMed] [Google Scholar]