Hectd1 is a ubiquitin ligase that targets the chaperone Hsp90. In the absence of Hectd1 ubiquitin ligase activity, Hsp90 secretion is elevated, resulting in abnormal behavior of cranial mesenchyme cells.
Abstract
Hectd1 mutant mouse embryos exhibit the neural tube defect exencephaly associated with abnormal cranial mesenchyme. Cellular rearrangements in cranial mesenchyme are essential during neurulation for elevation of the neural folds. Here we investigate the molecular basis of the abnormal behavior of Hectd1 mutant cranial mesenchyme. We demonstrate that Hectd1 is a functional ubiquitin ligase and that one of its substrates is Hsp90, a chaperone protein with both intra- and extracellular clients. Extracellular Hsp90 enhances migration of multiple cell types. In mutant cranial mesenchyme cells, both secretion of Hsp90 and emigration of cells from cranial mesenchyme explants were enhanced. Importantly, we show that this enhanced emigration was highly dependent on the excess Hsp90 secreted from mutant cells. Together, our data set forth a model whereby increased secretion of Hsp90 in the cranial mesenchyme of Hectd1 mutants is responsible, at least in part, for the altered organization and behavior of these cells and provides a potential molecular mechanism underlying the neural tube defect.
Introduction
Hectd1 mutant embryos show exencephaly (Zohn et al., 2007), a neural tube defect that occurs when the anterior neural tube fails to close completely during embryonic development. These congenital malformations represent some of the most common birth defects in humans (Zohn and Sarkar, 2008). Proper closure of the neural tube is dependent on the orchestration of several complex cellular processes in both the neural tissue and surrounding epithelium and mesenchyme (Copp et al., 2003; Copp, 2005). Importantly, elevation of the anterior neural folds is mediated by expansion of the extracellular matrix, which in turn results in increased spacing of the cells in the mesoderm-derived cranial mesenchyme (CM; Morriss and Solursh, 1978a,b; Schoenwolf and Fisher, 1983; Morris-Wiman and Brinkley, 1990a,b,c). Defects in expansion of the CM are associated with exencephaly in rodent models of neural tube defects. For example, treatment of rat embryos during neurulation with hyaluronidase prevents expansion of the extracellular matrix, resulting in collapse of the CM and delayed neural tube closure (Morriss-Kay et al., 1986). Furthermore, exencephaly in Twist mutants is associated with reduced CM (Chen and Behringer, 1995), whereas we have shown that exencephaly in Hectd1 mutant embryos is associated with denser mesoderm-derived CM (Zohn et al., 2007).
Hectd1 encodes a novel homologous to E6-AP C-terminal (HECT)-domain containing E3 ubiquitin (Ub) ligase (Zohn et al., 2007). Ubiquitination involves a three-enzyme cascade (E1, E2, and E3), resulting in the conjugation of Ub as single (monoubiquitination; Ub) or multiple (multi- or polyubiquitination; Ubn) moieties onto either lysine (K) residues or the N terminus of substrate proteins (Pickart, 2001). The functional significance of Ubn is dependent on which K residues in Ub are used to form the chain. Conjugation onto K48 of Ub generally targets a substrate for proteasomal degradation, whereas K63-linked Ub chains regulate a multitude of processes including intracellular protein localization and trafficking (Mukhopadhyay and Riezman, 2007).
Heat shock proteins (Hsps) constitute a family of abundantly expressed protein chaperones that regulate numerous cellular functions, including protein folding and cell migration. Although the majority of studies on Hsp90 demonstrate intracellular functions, recent data establish that Hsp90 is secreted from the cell, stimulating enhanced motility (Eustace and Jay, 2004; Tsutsumi and Neckers, 2007). Although the full extent of extracellular client proteins remains unknown, Hsp90 is thought to exert its effect on cell motility in the extracellular space through its chaperone activity on cell surface receptors and extracellular matrix components (Tsutsumi and Neckers, 2007; Li et al., 2012). Furthermore, in spite of their clear importance in regulating migration outside the cell, our current understandings of the mechanisms controlling Hsp90 secretion remain fragmentary. Cell stressors such as treatment with proteasome inhibitors, heat shock, and hypoxia stimulate Hsp translocation to the membrane and/or secretion (Liao et al., 2000; Verschuure et al., 2002; Clayton et al., 2005; Li et al., 2007). Hsp90 does not have a signal sequence and is released from the cell in exosomes, secretory vesicles targeted for fusion with the plasma membrane (Hegmans et al., 2004; Cheng et al., 2008; McCready et al., 2010). Sorting of proteins into secretory vesicles is typically regulated by phosphorylation and/or ubiquitination (Bonifacino and Traub, 2003; Urbé, 2005). Accordingly, phosphorylation by protein kinase A (PKA) is required for both membrane translocation and secretion of Hsp90 (Wang et al., 2009). In contrast, the role of ubiquitination in Hsp90 secretion has not been explored.
Neural tube defects observed in Hectd1 mutants illustrate its critical importance during embryonic development; however, the key substrates regulated by Hectd1 remain unknown. Our previous and present studies show that Hectd1 is important for the organization of the CM, which facilitates elevation of the cranial neural folds and neural tube closure (Zohn et al., 2007). Here we find, for the first time, that Hectd1 functions as a Ub ligase in vivo and binds to Hsp90, resulting in its K63-linked Ubn. We demonstrate that Hectd1-dependent Ubn of Hsp90 influences its intracellular localization and negatively regulates its secretion, as in the absence of Hectd1 Ub ligase activity, extracellular Hsp90 levels are increased. Furthermore, we show that the disorganized CM in Hectd1 mutant embryos, a potential underlying cause of exencephaly, is due at least in part to the abnormal behavior of CM cells stimulated by the excess Hsp90 secreted from mutant cells.
Results
Exencephaly in Hectd1 mutant embryos is associated with abnormal emigration of CM cells from explants
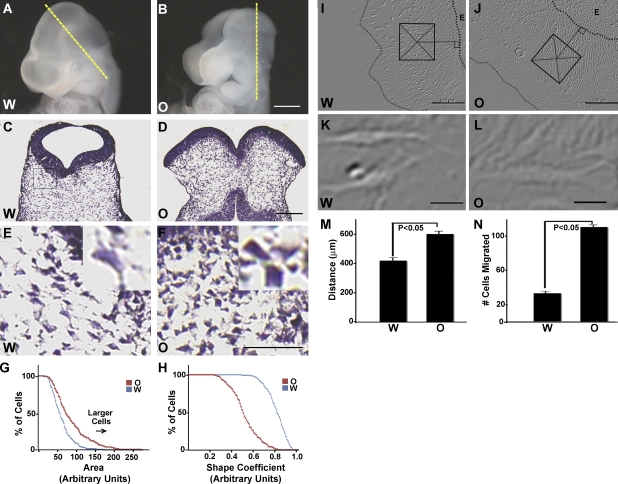
Our previous studies pointed to a failure of CM expansion as a potential underlying cause of exencephaly in Hectd1 mutants (Fig. 1, A–F; also see Fig. 2 A for diagrams of Hectd1 mutant alleles used in this study; Zohn et al., 2007). In addition to the increased cell density of mesoderm-derived CM cells in Hectd1 mutants previously noted (Zohn et al., 2007), Hectd1 mutant CM cells exhibit an altered morphology when compared with control cells (Fig. 1, E and F). Hectd1 mutant CM cells appeared larger than in wild-type CM. To quantitate these differences, cell area (Fig. 1 G) and cell perimeter were measured in hematoxylin and eosin–stained CM sections of embryonic day 9.5 (E9.5) wild-type and Hectd1 mutant embryos, and these values were used to calculate a shape coefficient (Fig. 1 H). These analyses revealed that CM cells in Hectd1openmind (Hectd1O) mutants are larger and have a significantly different shape in Hectd1wildtype (Hectd1W) embryos (Fig. 1, E–H).
Figure 1.
Exencephaly is associated with altered behavior of CM cells in Hectd1 mutant embryos during neurulation. (A and B) Hectd1W (A) and exencephalic Hectd1O (B) embryos at E9.5. (C–F) Hematoxylin and eosin (H&E) staining of transverse sections of E9.5 embryo heads taken at the level of the yellow broken lines in A and B revealing increased density and altered cell morphology of the CM in Hectd1O (D and F) compared with the Hectd1W (C and E) heads. E and F show higher magnification images of the boxes in C and D. Insets in E and F represent high-magnification images of individual cells. (G and H) Morphometric measurements reveal that Hectd1O mutant CM cells are larger and have a different shape coefficient then Hectd1W cells (n = 130 cells). (I–N) The mean migratory distance (M) from the circumference of the explant (broken lines) to the migratory front and the mean cell number (N) in a square area (200 µm2, box in I and J) perpendicular and mid-distant between the migratory front and explant circumference are increased in Hectd1O (J) compared with Hectd1W (I) CM explants. Error bars represent the mean ± SEM (SEM) of five independent experiments performed in triplicate. The shapes of cells migrating on fibronectin away from the explant do not appear to differ between Hectd1O (L) and Hectd1W (K). All data are representative of three independent experiments unless otherwise indicated. W, Hectd1W; O, Hectd1O. Bars: (B) 300 µm; (D) 125 µm; (F) 50 µm; (I and J) 200 µm; (K and L) 20 µm.
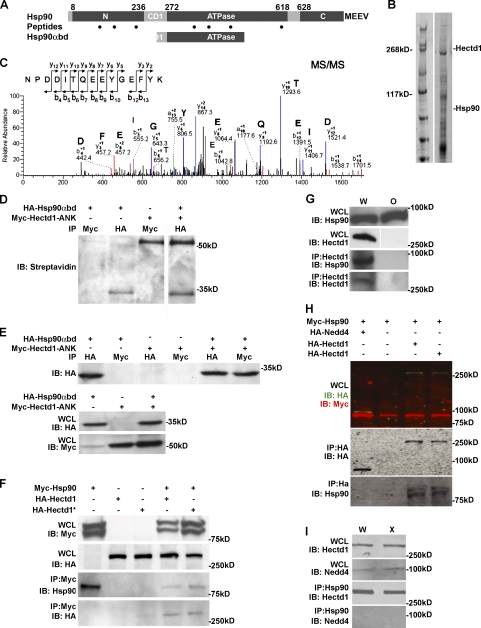
Figure 2.
Hectd1 functions as a Ub ligase. (A) Diagrammatic sketch of the Hectd1 alleles and plasmid constructs used in this study: Hectd1W (W; wild type), Hectd1O (O; opm; openmind), and Hectd1X (X; XC gene trap) are mouse alleles; Myc-Hectd1ANK, HA-Hectd1, and HA-Hectd1* are mammalian expression constructs. Hectd1O was generated in an ENU mutagenesis screen and harbors a missense mutation resulting in truncation of Hectd1. Hectd1X is a gene trap allele where the Ub ligase domain is disrupted by insertion of a β-geo (LacZ) cassette (Zohn et al., 2007). HA-Hectd1ANK consists of amino acids 1–551 of Hectd1 encompassing the ankyrin (ANK) domain. pCMVHA-Hectd1* is Ub ligase deficient because of mutation of the active site cysteine (C2579G). Other motifs present in Hectd1 include Mindbomb (mib) and Sad1/UNC (SUN) domains. The inverted Y denotes the paratope of the Hectd1 antibody that recognizes Hectd1W and Hectd1X but not Hectd1O proteins. (B) Reduced ubiquitination of Hectd1 and associated proteins in HEK293T cells expressing cysteine mutant Hectd1*. HA-Hectd1 immunoprecipitates were subjected to Western blot analyses to detect Hectd1 and mono- and polyubiquitinylated protein conjugates (FK2). (C) The appearance of HMW Hectd1 is dependent on its ligase activity in vivo. Hectd1 immunoprecipitates from E11.5 embryo heads of the indicated genotypes were probed with anti-Hectd1 antibody. (D) The conjugation of K63-linked Ub chains onto Hectd1 is dependent on its ligase activity. Hectd1 was immunoprecipitated from Hectd1W and Hectd1X CM cultures and immunoblotted with antibodies to detect K63-linked Ub chains (K63Ub) and Hectd1. (E and F) Reduction of total Ub proteins in Hectd1X compared with Hectd1W CM cultures. (E) Ub proteins were pulled down (PD) using Rad23 beads followed by immunoblotting to detect mono- and polyubiquitinylated protein conjugates (FK2). (F) Quantitation of normalized intensity of Western blots shown in E. Error bars represent the mean ± SEM of two independent experiments performed in triplicate. Statistical significance was determined by paired two-tailed Student’s t tests. W, Hectd1+/+; O, Hectd1opm/opm; X, Hectd1X/X; WCL, whole cell lysate; Con, control; IB, immunoblot; IP, immunoprecipitation; FK2, anti–mono- and polyubiquitinylated proteins; (Ub)n, Ubn; K63Ub, lysine 63 linked Ubn chains.
Mesoderm-derived CM cells are not generally considered a migratory population of cells. Rather, elevation of the neural folds is thought to be driven by the expansion of the extracellular matrix, displacing CM cells and leading to increases in the space between individual cells of the CM (Morriss and Solursh, 1978a,b; Schoenwolf and Fisher, 1983; Morris-Wiman and Brinkley, 1990a,b,c). Nevertheless, we have found that cells will migrate from CM explants when cultured on fibronectin-coated plates. To establish that the aberrant morphology observed in the CM of Hectd1 mutant embryos represents altered behavior of mutant cells, we performed a CM explant outgrowth assay. For these experiments, CM explants from E8.5 wild-type and Hectd1O embryos were microdissected and cultured on fibronectin-coated plates. We chose fibronectin as a substrate, as it is commonly used in these types of outgrowth assays, is generally associated with cell migration during embryonic development, and is a component of the extracellular matrix in the CM (Tuckett and Morriss-Kay, 1986; Zohn et al., 2006). Analyses of the mean number of cells emigrating from the explants demonstrated that Hectd1O mutant CM cells emerge from the explant at a significantly (P < 0.05) higher rate than wild-type cells (Fig. 1, I–L and N). In addition, Hectd1O mutant CM cells migrated significantly (P < 0.05) further from the explant than wild-type cells (Fig. 1 M). Differences in emigration of Hectd1O mutant cells could reflect differences in proliferation or death in the CM; however, our previous results indicate that these are not altered in Hectd1 mutant CM (Zohn et al., 2007). Alternatively, differences in emigration from the explant could indicate that Hectd1O mutant cells are differentially recruited from the explant than wild-type cells. Additionally, the observation that Hectd1O mutant cells migrated further suggests that mutant cells are likely more motile than their wild-type counterparts. In conclusion, these results demonstrate that the cells in Hectd1O mutant CM exhibit abnormal behaviors.
Hectd1 encodes a functional Ub ligase
A gene trap insertion disrupting the HECT domain of Hectd1 in the Hectd1XC (Hectd1X) allele phenocopies defects observed in Hectd1O-null mutants (Zohn et al., 2007), which indicates that the HECT domain is necessary for the biological activity of Hectd1. To establish that Hectd1 is a functional E3 Ub ligase, full-length HA-tagged constructs encoding wild-type Hectd1 (pCMV-HA-Hectd1) or a ligase incompetent version of Hectd1 (pCMV-HA-Hectd1*), in which the conserved active site cysteine2579 is mutated to glycine (Fig. 2 A), were transfected into human embryonic kidney 293T (HEK293T) cells. To assess the effect of the proteasome on the stability of proteins ubiquitinated by Hectd1, cultures were treated for 4 h before lysis with the cell-permeable proteasomal inhibitor MG132 (carbobenzoxy-leucyl-leucyl-leucinal). Western blots of Hectd1-immunoprecipitated lysates were probed with anti-Ub antibodies, revealing that cells expressing HA-Hectd1 contain significantly more Ub-conjugated proteins bound to Hectd1 than cells expressing cysteine mutant HA-Hectd1* (Fig. 2 B). These results demonstrate that Hectd1 exhibits Ub ligase activity. Furthermore, because Ub proteins are not stabilized by proteasomal inhibition, Hectd1 substrates are likely not targeted for proteasomal degradation.
We also examined the Ub ligase activity of Hectd1 in vivo (Fig. 2, C and D). For these experiments, we took advantage of our two mutant mouse lines (Fig. 2 A; Zohn et al., 2007). The Hectd1O allele contains a nonsense mutation resulting in truncation of Hectd1, whereas the Hectd1X allele contains a gene trap insertion disrupting the catalytic HECT domain but leaving the substrate interaction domains intact. Furthermore, wild-type (Hectd1W) and Hectd1X proteins but not Hectd1O were detected by our anti-Hectd1 antibody, demonstrating antibody specificity (Fig. 2, A and C; and Fig. S1). The presence of both native and high molecular weight (HMW) Hectd1 in immunoprecipitates of Hectd1 from Hectd1W but not Hectd1O E11.5 embryo head lysates confirms antibody specificity and that the HMW band is Hectd1 (Fig. 2 C). Significantly, this HMW band is not detected in Hectd1X mutant lysates, where the Ub ligase domain of Hectd1 is disrupted, indicating that the HMW band represents Hectd1 auto-ubiquitination (Fig. 2 C). Additionally, Hectd1 incorporates K63-linked Ubn chains in vivo that are absent in Hectd1X mutant CM cells (Fig. 2 D). Because conjugation of K63-linked Ubn chains onto substrate proteins generally serves nonproteolytic functions, this result further suggests that Hectd1-dependent Ubn is unlikely to target substrates to the proteasome for degradation.
Loss of Hectd1 affects the global cellular Ub protein pool
To determine the extent of cellular ubiquitination influenced by Hectd1, Ub proteins were enriched from lysates of CM cultures isolated from the cranial region of E12.5 Hectd1W and Hectd1X mutant embryos. To enrich for Ub proteins, cell lysates were incubated with the Ub-binding protein Rad23 bound to agarose beads (Chen and Madura, 2002; Hartmann-Petersen and Gordon, 2004). Western blotting of enriched proteins reveals a reduction in the total amount of Ub proteins in mutant samples (Fig. 2, E and F). These results indicate that Hectd1 likely catalyzes the ubiquitination of multiple and/or highly abundant proteins.
Identification of Hsp90α as a candidate Hectd1 substrate
HECT domain E3 Ub ligases physically interact with their substrates. To elucidate the pathways regulated by Hectd1, we conducted two independent proteomic screens to identify Hectd1 interactors and identified Hsp90 in both screens. We initially performed a yeast two-hybrid screen using the N-terminal 551 amino acids of Hectd1 containing the ankyrin domain (Hectd1ANK; Fig. 2 A) as bait to screen an E11.5 mouse cDNA library. Out of 4.5 million clones screened, 23 unique clones were identified, including two identical clones encompassing amino acids 241–459 of Hsp90α (Hsp90α binding domain [Hsp90αbd]). This region includes the charged domain 1 (CD1, amino acids 237–271) and ATPase domain (272–617), and is highly homologous to Hsp90β in primary amino acid sequence (red bar in Fig. 3 A). The second approach used liquid chromatography–mass spectrometry (LC-MS) to screen for proteins that coimmunoprecipitated with endogenous full-length Hectd1 from E12.5 mouse embryo heads. From this screen, seven peptide fragments of Hsp90 were found (dots in Fig. 3 A and Fig. 3, B and C). Identification of Hsp90 in both proteomic screens suggested that members of the Hsp90 superfamily may be substrates of Hectd1.
Figure 3.
Hectd1 physically interacts with Hsp90α. (A) Yeast two-hybrid screening of an E11.5 embryonic mouse cDNA library using Hectd1ANK as bait detected Hsp90α (Gene ID: Hsp90aa1). Two identical clones of Hsp90α consisting of amino acids 241–459 (Hsp90αbd) of the 732–amino acid Hsp90α protein partially overlap with the first charged domain (CD1, amino acids 236–272) and the ATPase domain (amino acids 272–618) of Hsp90α. Black dots indicate locations of Hsp90 peptide fragments found by LC-MS analysis: NPDDITQEEYGEFYK (300–315), TLTIVDTGIGMTK (88–100), KADLINNLGTIAKS (100–113), and GVVDSEDLPLNISR (387–400). Peptides common to Hsp90β include: KEDQTEYLEERR (190–201), RDNSTMGYMMAKK (620–632), and YIDQEELNK (284–292). The C-terminal amino acid sequence MEEVD of Hsp90α is essential for regulated secretion. (B and C) Liquid chromatography and tandem mass spectrometry (LC-MS/MS) proteomic screening of Hectd1-binding proteins from E10.5 Hectd1W embryo head lysates. Hectd1 was immunoprecipitated and associated proteins were resolved by 3–8% Tris-Acetate SDS-PAGE and visualized by Coomassie staining. (B) Individual bands from the Coomassie-stained gel were subjected to tryptic proteolysis, and the resulting peptides were analyzed by LC-MS/MS. (C) Representative MS/MS of a 1,293.54 D peptide. This and six other peptides (dots in A) with high XC scores (3.3–3.7) were identified as belonging to Hsp90α and Hsp90β when searched against the mouse Uniprot protein database using the Sequest algorithm as diagrammed in A. (D) Hsp90αbd binds to Hectd1ANK in rabbit reticulocyte lysates. In vitro translated, biotinylated Hsp90αbd and Hectd1ANK were bound and immunoprecipitated using the indicated antibodies and detected by Western blotting with streptavidin-HRP. (E and F) Hectd1 binds to Hsp90 in HEK293T cells. Cells were transfected and immunoprecipitated proteins were subjected to Western blot analyses as indicated. (E) Hsp90αbd binds to Hectd1ANK in HEK293T cells. (F) Full-length Hsp90 and Hectd1 bind in HEK293T cells. (G) Hsp90 binds to Hectd1 in the developing embryo. Hectd1 was immunoprecipitated from lysates prepared from E12.5 Hectd1W and Hectd1O embryo heads and subjected to Western blot analysis as indicated (n = 4). (H and I) Hsp90 binds to Hectd1 but not the related HECT domain containing Nedd4 Ub ligase. (H and I) HEK293T cells were transfected with HA-Nedd4 or HA-Hectd1 along with Myc-Hsp90. They were then HA immunoprocipitated and subjected to Western blotting (H), or Hsp90 (I) was immunoprecipitated from E12.5 Hectd1W and Hectd1O embryo head lysates followed by Western blotting to detect Nedd4 and Hectd1. All data are representative of three independent experiments unless otherwise indicated. W, Hectd1+/+; O, Hectd1opm/opm; X, Hectd1X/X; WCL, whole cell lysate; IB, immunoblot; IP, immunoprecipitation.
Hectd1 and Hsp90 physically interact
To explore the interaction between Hectd1 and Hsp90 (the Hsp90α isoform was used in all assays), we performed a series of in vitro and in vivo binding assays. Myc-Hectd1ANK and HA-Hsp90αbd (the fragment identified in the yeast two-hybrid screen) bind in an in vitro binding assay (Fig. 3 D) and when coexpressed in HEK293T cells (Fig. 3 E). Additionally, in HEK293T cells, full-length Myc-Hsp90 binds to both full-length wild-type Hectd1 (HA-Hectd1) and cysteine mutant Hectd1 (HA-Hectd1*; Figs. 2 A and 3 F). Furthermore, immunofluorescence analyses reveal that Myc-Hsp90 and HA-Hectd1 extensively colocalize when expressed in HEK293T cells (Fig. S2). Significantly, binding of endogenous proteins was detected in E12.5 head lysates from Hectd1W but not Hectd1O mutant embryos (Fig. 3 G). Intriguingly, binding was detected between endogenous Hsp90 and Hectd1 in the developing embryo under much more stringent conditions (e.g., radioimmunoprecipitation assay buffer vs. immunoprecipitation buffer with 0.1% Triton X-100; see Materials and methods) than when expressed in cell lines, which indicates that other factors likely facilitate this interaction in vivo. Importantly, demonstrating the specificity of binding in our assay conditions, Hectd1 but not Nedd4 (neural precursor cell expressed developmentally down-regulated protein 4), a related HECT domain Ub ligase, coimmunoprecipitated with Hsp90 in transfected HEK293T cells and head lysates from E12.5 Hectd1W and Hectd1X mutant embryos (Fig. 3, H and I).
Hectd1 is required for K63-linked Ubn of Hsp90
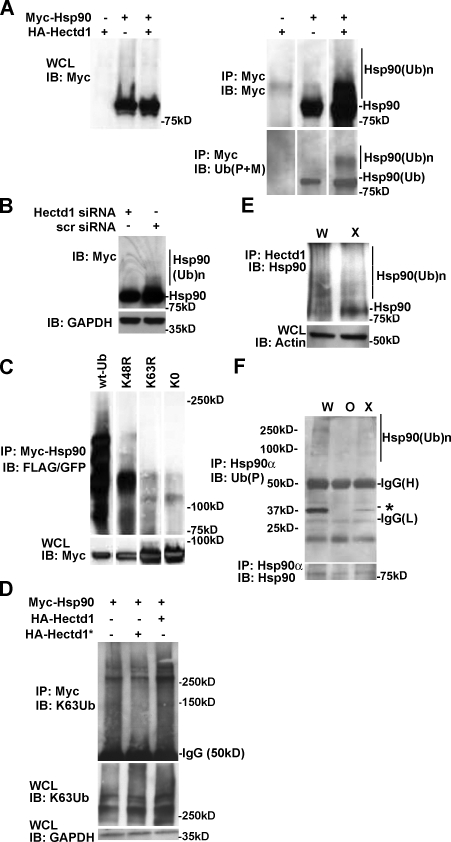
Next we tested whether the interaction of Hectd1 with Hsp90α results in the ubiquitination of Hsp90. Hsp90 coexpressed with Hectd1 in HEK293T cells resulted in the appearance of HMW Ub-Hsp90 species (Fig. 4 A). Furthermore, siRNA-mediated knockdown of Hectd1 in HEK293T cells reduced the appearance of HMW Myc-Hsp90 (Fig. 4 B). To discern the Ubn chain linkage specificity on Hsp90, HEK293T cells were transfected with Myc-Hsp90 along with wild-type Ub or Ub constructs where the critical lysine residues K48 or K63 were mutated to arginine (K48R-Ub and K63R-Ub, respectively). These mutant Ub proteins will incorporate into growing Ub chains, but if the mutated residue is utilized for chain elongation, the chain will fail to extend. In addition, a mutant Ub construct where all lysines in Ub are mutated to arginine (K0-Ub) was used as a negative control for chain elongation. Wild-type and K48R-Ub incorporated, whereas K0-Ub and K63R-Ub failed to incorporate into the Ubn chains on Hsp90 (Fig. 4 C). This result indicates that Ubn on Hsp90 utilizes K63 linkages. Similarly, incorporation of K63-Ubn is increased in HEK293T cells transfected with HA-Hectd1 but not ligase-deficient HA-Hectd1* (Fig. 2 D). Together, these observations indicate that the Hectd1-dependent appearance of HMW Hsp90 species is primarily caused by addition of K63 linkages.
Figure 4.
Hectd1 is required for K63-linked Ubn of Hsp90. (A–D) HEK293T cells were transfected, and lysates were subjected to immunoprecipitation and Western blot analysis as indicated. (A) Ubiquitination of Myc-Hsp90 increases with expression of HA-Hectd1 (n = 2). (B) siRNA-mediated knockdown of endogenous Hectd1 reduces the accumulation of HMW-Hsp90α (n = 2). (C) Hsp90α ubiquitination utilizes K63 linkages. (D) Hectd1-dependent polyubiquitination of Hsp90 occurs primarily through K63 linkages. (E) HMW Hsp90 species are reduced in Hectd1 mutant heads. E12.5 Hectd1W (W) and Hectd1X (X) embryos were cultured in the presence of 10 µM MG132 for 3 h before lysis and immunoprecipitation of Hectd1. Immunoprecipitates were subjected to Western blot analyses to detect Hsp90 that coimmunoprecipitated with Hectd1. (F) Hsp90 ubiquitination is reduced in CM cultures from Hectd1O (O) and Hectd1X (X) mutants compared with Hectd1W (W). Hsp90 was immunoprecipitated from E12.5 CM primary cultures in highly denaturing ubiquitination buffer plus 5% SDS and subjected to Western blot analyses as indicated. The appearance of a 30-kD ubiquitinated protein (asterisk) is reduced in Hectd1 mutant cells. All data are representative of three independent experiments unless otherwise indicated. Abbreviations: W, Hectd1+/+; O, Hectd1opm/opm; X, Hectd1X/X; WCL, whole cell lysate; IB, immunoblot; IP, immunoprecipitation; (Ub)n, Ubn; wt-Ub, wild-type Ub; K48R, mutant Ub lysine 48 arginine; K63R, mutant Ub lysine 48 arginine; K0, lysineless Ub.
To determine the in vivo relevance of Hectd1-dependent Hsp90 ubiquitination, we tested whether mutation of Hectd1 in the embryo would have an effect on endogenous Ubn-Hsp90 levels. To ensure stabilization of Ubn–protein conjugates in the embryo, we devised a short-term embryo culture protocol in the presence of the well-characterized cell-permeable proteasome inhibitor MG132. To determine if Hectd1 influences Ubn of Hsp90 in vivo, E11.5 wild-type (Hectd1W) and mutant (Hectd1X) embryos were cultured in the presence of MG132, followed by immunoprecipitation of Hectd1 and Western blotting to detect Hsp90. The gene trap insertion in Hectd1X disrupts the catalytic HECT domain but leaves the substrate interaction domains (Zohn et al., 2007) and the epitope recognized by our anti-Hectd1 antibody intact (Fig. 2 A). In Hectd1W lysates, HMW Hsp90 that coimmunoprecipitated with Hectd1 is detected as multiple bands (Fig. 4 E); however, in Hectd1X mutant lysates, HMW Hsp90 species are markedly diminished with a corresponding increase in low molecular weight Hsp90. Similarly, immunoprecipitation of Hsp90 from Hectd1W, Hectd1X, or Hectd1O primary CM cultures in a highly denaturing ubiquitination lysis buffer plus 5% SDS to minimize interaction of associated proteins demonstrates reduced ubiquitination of Hsp90 in Hectd1 mutant CM cells (Fig. 4 F). Interestingly, coimmunoprecipitation of Hsp90 with a 30kD Ubn-protein is dependent on Hectd1 activity (Fig. 4 F, asterisk). The binding of Hsp90 to this protein in highly denaturing conditions indicates a tight association or possibly a proteolytically processed Hsp90 fragment.
Hectd1-dependent Ubn influences the cellular localization and secretion of Hsp90
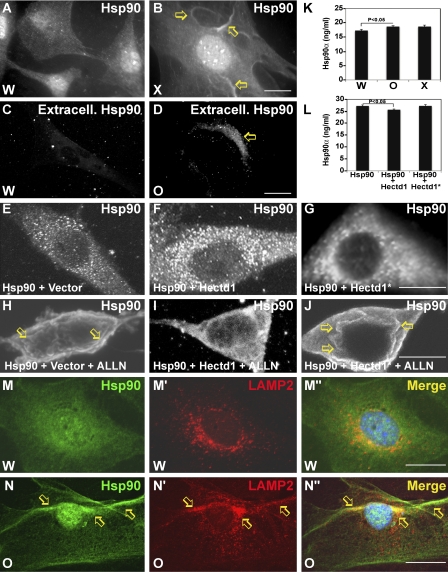
The conjugation of K63-linked Ubn chains onto substrate proteins typically does not target them to the proteasome for degradation but rather influences other processes such as intracellular localization/trafficking (Mukhopadhyay and Riezman, 2007). To determine if Hectd1-dependent K63-linked Ubn of Hsp90 regulates its intracellular localization, we examined the distribution of Hsp90 in primary CM cultures prepared from E10.5 Hectd1W and Hectd1X embryo heads. In Hectd1W CM cultures, Hsp90 is localized in the cytoplasm as well as in the nucleus (Fig. 5 A). In contrast, in Hectd1X mutant cells, Hsp90 was concentrated at the plasma membrane in ruffled-like structures (Fig. 5 B, arrows). To further investigate the plasma membrane localization of Hsp90, cells were immunostained with the primary anti-Hsp90 antibody before permeabilization with Triton X-100 to visualize only the Hsp90 localized to the outside of the plasma membrane. This experiment revealed extracellular localization of Hsp90 in Hectd1O mutant but not Hectd1W CM cells (Fig. 5, C and D). In addition, we used an experimental paradigm to stimulate Hsp90 translocation to the membrane by treating HEK293T cells with proteasomal inhibitors (Verschuure et al., 2002) to examine the role of Hectd1 in controlling the intracellular localization of Hsp90. Myc-Hsp90 is localized to the cytoplasm in HEK293T cells cotransfected with either HA-Hectd1 or HA-Hectd1* (Fig. 5, E–G). Treatment with the proteasomal inhibitor N-acetyl-leucyl-leucyl-norleucinal (ALLN; Fig. 5 H–J) results in Myc-Hsp90 localized to the membrane in cells transfected with Myc-Hsp90 alone or cotransfected with ligase-incompetent HA-Hectd1*. In contrast, cotransfection with HA-Hectd1 results in primarily cytosolic Myc-Hsp90.
Figure 5.
Hectd1 affects Hsp90 localization and secretion. (A and B) Hectd1 Ub ligase activity is required for normal cytoplasmic distribution of Hsp90. (A–D) Hectd1W (A and C) and Hectd1X (B and D) CM cells were subjected to immunofluorescence analyses to detect total Hsp90 (A and B) or extracellular Hsp90 (C and D). Arrows indicate membrane concentrated Hsp90 (B) or extracellular Hsp90 (D). (E–J) Hectd1-dependent ubiquitination prevents ALLN-induced translocation of Hsp90 to the membrane. HEK293T cells transfected as indicated were treated (H–J) or not treated (E–G) with ALLN and immunostained to detect Myc-Hsp90. Arrows indicate membrane-concentrated Hsp90. (K) Mutation of Hectd1 results in increased secretion of Hsp90. ELISA was used to detect Hsp90α in conditioned media from Hectd1W, Hectd1O, and Hectd1x CM cultures. (L) Expression of HA-Hectd1 but not Ub ligase–deficient HA-Hectd1* reduced secretion of Hsp90 in HEK293T cells. Statistical significance was determined by paired two-tailed Student’s t tests. Error bars represent the mean ± SEM (SEM) of three independent experiments performed in triplicate. (M–N″) Hsp90 (M and N) colocalizes (M″ and N″) with LAMP2 (M′ and N′) in the membrane (arrows) of Hectd1O CM cells. All data are representative of three independent experiments unless otherwise indicated. W, Hectd1+/+; O, Hectd1opm/opm; X, Hectd1X/X. Bars, 10 µm.
Plasma membrane localization of Hsp90 has been associated with secretion of Hsp chaperones through the exosome pathway (Hegmans et al., 2004; Cheng et al., 2008; McCready et al., 2010). Indeed, in Hectd1O mutant but not Hectd1W cells, Hsp90 colocalized extensively with LAMP2 (lysosomal-associated membrane protein 2), a protein that shuttles between lysosomes, endosomes, and the plasma membrane—organelles involved in exosomal secretion of Hsp90 (Fig. 5, M and N). Importantly, Hsp90 secretion is significantly (P < 0.05) increased in Hectd1X and Hectd1O mutants compared with Hectd1W CM cells as quantified by a sensitive ELISA assay of Hsp90 protein in the conditioned media of Hectd1X, Hectd1O, and Hectd1W CM cells (Fig. 5 K). Conversely, Hsp90 secretion was reduced in HEK293T cells cotransfected with Myc-Hsp90 along with HA-Hectd1 but not HA-Hectd1* (Fig. 5 L). Together, these results demonstrate that Hectd1-dependent Ubn of Hsp90 targets it away from the membrane and the secretory pathway.
Enhanced migration of Hectd1 mutant CM cells is associated with increased Hsp90 secretion
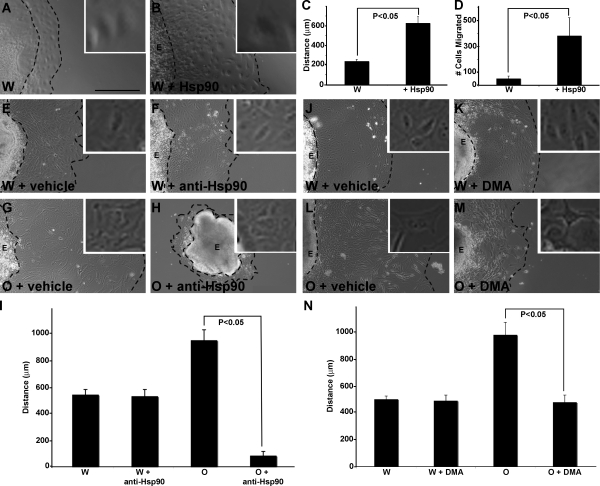
Secreted Hsp90 can enhance cell migration in numerous experimental paradigms (Tsutsumi and Neckers, 2007). Therefore, our observations that Hectd1 regulates Hsp90 secretion and that in Hectd1 mutant CM cells Hsp90 secretion is increased suggest a potential molecular mechanism underlying the abnormal behavior of mutant CM cells (Fig. 1). To establish that Hsp90 can stimulate emigration and migration of cells from CM explants, CM explants from wild-type embryos were microdissected, plated on fibronectin-coated plates, and incubated with recombinant Hsp90 protein. Treatment with Hsp90 significantly increased (P < 0.05) both the cells emigrating from the explants and the distance migrated (Fig. 6, A–D), which demonstrates that increased extracellular Hsp90 would be sufficient to cause the abnormal behavior of Hectd1 mutant CM cells in this assay. Furthermore, migration of Hectd1O mutant CM cells is highly dependent on secreted Hsp90. Treatment of Hectd1W and Hectd1O explants with a dose of anti-Hsp90 antibody that does not affect emigration of CM cells from Hectd1W explants significantly inhibited emigration of cells from Hectd1O mutant explants (Fig. 6, E–I). Similarly, treatment of explants with dimethyl amiloride (DMA), an inhibitor of secretion, reduced emigration of CM cells from Hectd1O mutant explants (Fig. 6, J–N). Together, these data demonstrate that enhanced emigration and migration of Hectd1 mutant CM cell explants is at least partly caused by increased secretion of Hsp90 in the absence of Hectd1 ubiquitination activity.
Figure 6.
Abnormal emigration of Hectd1 mutant CM from explants is highly dependent on Hsp90 activity. (A–D) CM explants were cultured in the presence (B) or absence (A) of 100 ng/ml Hsp90 protein. Migratory distance (C) and cell numbers (D) are increased in Hsp90-treated explants (n = 3). (E–N) CM explants from Hectd1W (E, F, J, and K) and Hectd1O (G, H, L, and M) embryos were incubated with DMSO/vehicle (E, G, J, and L), 25 µg/ml anti-Hsp90 antibody (F and H), or 200 µM DMA (K and M). The broken lines denote the edge of the explant (E) and the migration front. Insets show enlarged views of cells migrating from the explant. (I and N) Mean distance of cells migrated from explants treated in E–H and J–M, respectively. Error bars represent the mean ± SE of five independent experiments (unless otherwise indicated) performed in triplicate. Bar, 200 µm.
Discussion
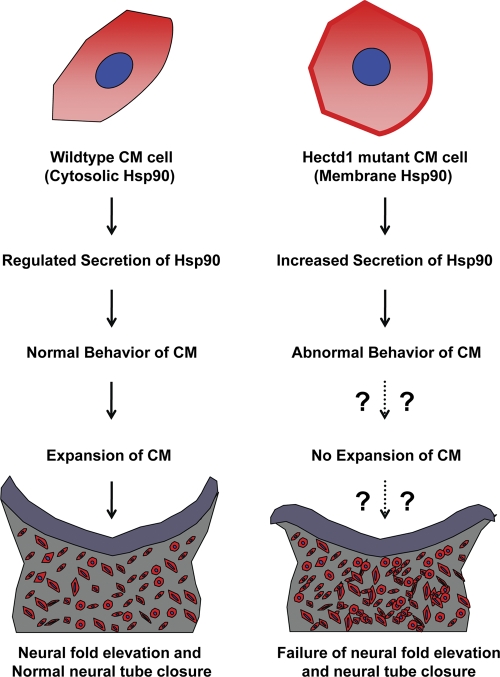
Here we demonstrate that Hectd1 is a functional Ub ligase and that Hectd1-dependent ubiquitination plays an essential role in controlling Hsp90 secretion by negatively regulating secretion of Hsp90. Our data indicate that Hectd1 binds to Hsp90 and is required for conjugation of K63-linked Ubn chains onto this cellular chaperone. Fig. 7 summarizes a model of how mutation of Hectd1 results in alterations in CM behavior that have emerged from this study. In the absence of Hectd1 Ub ligase activity, Hsp90 becomes concentrated at the cell surface of CM cells rather than its normal cytoplasmic distribution. Additionally, Hsp90 secretion is increased in Hectd1 mutant CM cells. This excessive secretion is at least partially responsible for the abnormal behavior of Hectd1 mutant CM cells. We propose that this abnormal CM behavior contributes to the failure of CM expansion found in Hectd1 mutant embryos. Because this CM defect is associated with failure of neural fold elevation (Fig. 1; Zohn et al., 2007), these results raise the distinct possibility that Hsp90 may be a critical Hectd1 substrate and that failure to regulate Hsp90 secretion in Hectd1 mutant CM may contribute to exencephaly in Hectd1 mutants.
Figure 7.
Proposed model depicting consequences of Hectd1-mediated Hsp90 K63 ubiquitination from the present study. In wild-type CM cells, K63-ubiquitinated Hsp90 (red) is localized to the cytosol. Hsp90 secretion is properly regulated in CM of wild-type embryos, resulting in normal behavior of the CM and possibly normal expansion of the CM required for neural fold elevation. In Hectd1 mutant CM cells, the lack of K63 ubiquitination of Hsp90 results in an increased pool of Hsp90 targeted for secretion and localized to the membrane (red). Excess secretion of Hsp90 leads to abnormal behavior of the CM, potentially contributing to failure of CM expansion, neural fold elevation, and neural tube closure.
Hectd1-dependent ubiquitination of Hsp90
Hsps have been shown to interact with other E3 Ub ligases, although a direct role in ubiquitination of these chaperones has not been demonstrated. The U-box E3 ligase, C terminus of Hsc70-interacting protein (CHIP) interacts with Hsp90, Hsp70, and Hsp40; however, instead of ubiquitinating these, CHIP acts as a quality controller by selectively targeting associated client proteins for degradation (McDonough and Patterson, 2003; Murata et al., 2003). In contrast, our data demonstrate that Hectd1 is required for K63-Ubn of Hsp90α itself, raising the possibility that Hectd1-dependent ubiquitination of Hsp90α serves an alternate purpose beyond that of quality control of client proteins. Although our data do not rule out the distinct possibility that Hectd1 also regulates the selective ubiquitination of Hsp90 client proteins, in this study we restricted ourselves to examining the Ub conjugation and function of Hsp90 itself. Accordingly, our data demonstrate that Hectd1-dependent K63-Ubn of Hsp90 controls its intracellular localization and secretion.
Hectd1-dependent ubiquitination of Hsp90 regulates it secretion
Little is known about the pathways regulating secretion of Hsp90. Phosphorylation of Hsp90α by PKA on Thr90 promotes its secretion (Wang et al., 2009). Furthermore, the four C-terminal amino acid residues of Hsp90α (EEVD) are critical for regulation of its secretion (Wang et al., 2009). This domain provides a point of interaction for tetratricopeptide repeat (TPR) domain–containing proteins preventing secretion by docking Hsp90α in the cytoplasm (Wang et al., 2009). Similarly, our data demonstrate that Hectd1 is a negative regulator of Hsp90α secretion. In contrast to the interaction of Hsp90α with TPR domain–containing proteins, our binding data demonstrate that Hectd1 interacts with Hsp90α through a region located between amino acids 241 and 617. It’s important to note that it is not simply the interaction of Hectd1 with Hsp90α that prevents its secretion; the Ub ligase activity of Hectd1 is also required. K63-linked Ub regulates trafficking of proteins in numerous systems (Mukhopadhyay and Riezman, 2007). It remains to be determined how Hectd1-dependent ubiquitination of Hsp90α prevents its secretion. One clue comes from our finding that a Hectd1-dependent 30-kD Ub protein coimmunoprecipitates with Hsp90α under highly denaturing conditions. This could either represent a tightly associated protein or a cleavage product of Hsp90α. It is possible that association of Hsp90α with this 30-kD Ub protein prevents membrane translocation. Alternatively, Hectd1-dependent K63-Ubn of Hsp90α may lead to cleavage of Hsp90α to a fragment that cannot be secreted. Precisely how Hectd1-dependent ubiquitination regulates Hsp90 secretion remains to be determined.
Hectd1-dependent ubiquitination of Hsp90 regulates CM cell behavior
Our data support the model that abnormal behavior of CM cells is mediated by increased secretion of Hsp90α in mutant cells (Fig. 7). Increased extracellular Hsp90α is sufficient to induce increased emigration from CM explants. Additionally, emigration and migration of cells from Hectd1 mutant CM explants is highly dependent on secretion of Hsp90α. How might abnormal behavior of the CM result in failure of neural fold elevation? Current models of cranial neural tube closure predict that a driving force for cranial neural fold elevation is the expansion of the extracellular matrix, displacing CM cells and resulting in expansion of the space between cells (Morriss and Solursh, 1978a,b; Schoenwolf and Fisher, 1983; Morris-Wiman and Brinkley, 1990a,b,c; Copp, 2005). Our data demonstrate that the abnormal behavior of Hectd1 mutant CM is related to the excess secretion of Hsp90. We propose that the abnormal behavior of the CM in Hectd1 mutants interferes with the expansion of the CM that occurs as the extracellular matrix expands, resulting in the dense CM found in Hectd1 mutants. The substrates of extracellular Hsp90 include cell surface receptors and extracellular matrix components and likely a host of other client proteins, which can regulate cell behavior (Tsutsumi and Neckers, 2007; Li et al., 2012; Sims et al., 2011). Thus, excess extracellular Hsp90 in the CM may influence expansion of the extracellular matrix, interaction of CM cells with the extracellular matrix, and/or motility of the CM cells. Our previous observation that the increased density of CM in Hectd1 mutants was not associated with alterations in cell proliferation and apoptosis (Zohn et al., 2007) supports the hypothesis that increased CM density is caused by effects on cell rearrangements. Whether this abnormal cellular behavior is truly responsible for the failed CM expansion and neural tube closure in Hectd1 mutants still remains to be determined. It is important to note that Hectd1 likely has multiple substrates, as demonstrated from our proteomic screens, and it is possible that these also contribute to the abnormal CM behavior and NTDs in Hectd1 mutants. In conclusion, our study establishes a novel Ub ligase–substrate interaction in vivo between Hectd1 and Hsp90α and provides a molecular mechanism for the abnormal migratory behavior of the CM in Hectd1 mutants.
Materials and methods
Mice genotyping and cell culture
Hectd1opm and Hectd1XC (Hectd1X) mouse lines have been described previously (Fig. 2 A; Kasarskis et al., 1998; Zohn et al., 2007). Hectd1opm represents an N-ethyl-N-nitrosourea (ENU)-induced allele, resulting in truncation of the Hectd1 protein at amino acid 144. Hectd1X results from a gene trap insertion into the HECT domain. Hectd1opm and Hectd1X were genotyped using amplification refractory mutation system PCR (ARMS-PCR; Ye et al., 2001) and gap PCR (Bandyopadhyay et al., 2004), respectively, and as shown (Fig. S3). For ARMS-PCR to genotype Hectd1opm, the primers used were: forward inner primer (A allele), 5′-AGGGGCAGTCTTTGAGGCTGGTGGATA-3′; reverse inner primer (T allele), 5′-TGTCACGAATGAAGGTNAGAACACAATACA-3′; forward outer primer, 5′-ACCTTGTGTCTCTGTTGTAGGTGCTGGA-3′; and reverse outer primer, 5′-GCTCCATTTTGCCACAGAGTCTCGATAC-3′. Mice carrying the Hectd1X gene trap allele were genotyped using gap PCR. The primers used were 52860334F, 5′-CACTGAAAGTGACGGGGCTTGGAAC-3′; 52859614R, 5′-ATAATAATAAAGCACTTAACTATTCCCGAG-3′; and B-Gal-Rev1, 5′-GACAGTATCGGCCTCAGGAAGATCG-3′. CM cultures were prepared according to a standard protocol for isolating mouse embryonic fibroblasts (MEFs) from E12.5 mouse embryo heads (Nagy et al., 2003). HEK293T and CM cells were cultured in DME (Invitrogen), supplemented with 10% FCS, 100 IU/ml penicillin G, and 100 µg/ml streptomycin sulfate (Invitrogen). Bright field images for whole embryos were obtained using a microscope (Discover V8; Carl Zeiss) using an Achromat S FWD 107 mm lens with a magnification of 0.63×. Images were captured using a camera (AxioCam ERc 5s; Carl Zeiss) and processed with Photoshop (8.0) and Illustrator (10.0; both from Adobe).
Mammalian expression constructs
pGBKT7-Hectd1ANK, pCMV-HA-Hectd1ANK, pCMV-HA-Hectd1, and pCMV-HA-Hectd1* were assembled by ligation of RT-PCR generated fragments and ESTs encoding fragments of mouse Hectd1 (Fig. 2). pCMV-Sport6-Hsp90aa1 expression plasmid was obtained from Invitrogen (Clone ID no. 6491307) and subcloned into pCMV-Myc (Takara Bio Inc.). EGFP-C1-UbKO was obtained from Addgene (Addgene plasmid 11934; Bergink et al., 2006). pCMV-FLAG-Ub, pCMV-FLAG-K63R-Ub, and pCMV-FLAG-K48R-Ub were provided by Drs. H. McLauchlan and P. Cohen (University of Dundee, Dundee, Scotland, UK; Windheim et al., 2008). Control, scrambled, and custom Hectd1 (5′-AGATAAAGGTGGTGATATA-3′) siRNA were purchased from Thermo Fisher Scientific. The effectiveness of the Hectd1 siRNA was demonstrated by transfection of HEK293T cells with control, scrambled, and Hectd1 constructs followed by Western blotting to detect Hectd1 (Fig. S4)
Proteomic screens
The Matchmaker Two-Hybrid System 3 was used according to manufacturer’s instructions to screen the Matchmaker Pretransformed E11.5 mouse cDNA library (Takara Bio Inc.) for proteins that bind to Hectd1ANK. For the LC-MS screen, Hectd1 was immunoprecipitated in immunoprecipitation buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, and 0.1% Triton X-100 with protease inhibitor cocktail) from lysates of wild-type E10.5 mouse; embryo heads were resolved on 3–8% Tris-acetate denaturing gels (Invitrogen) and processed (Jensen et al., 1999; Cargile et al., 2004; Essader et al., 2005). Peptide sequences were searched against the mouse Uniprot protein database (http://www.uniprot.org/) using the Sequest algorithm in the Bioworks Browser software package (Thermo Fisher Scientific).
Antibodies and immunofluorescence assay
Anti-peptide Hectd1 rabbit polyclonal antibody was generated (Biosynthesis Inc.) against mouse Hectd1 amino acids 487–505 (CPVNKGDDKKKDTNKDEE) and used for all experiments unless otherwise indicated. The specificity of the Hectd1 antibody was confirmed in an immunoblotting assay (Fig. S1). Serum from rabbits immunized with Hectd1 peptide was used for Western blot analysis or affinity purified using the RAP Affinity Purification kit (Invitrogen) for immunoprecipitation. For enrichment of Ub proteins that bind to Rad23, the ubiquitinated protein enrichment kit (EMD) was used according to the manufacturer’s instructions. Other antibodies used were: HSP90 (E289) antibody (4875; Cell Signaling Technology), anti-Hsp90 mouse monoclonal antibody (mAb; EMD-17D7, CA1023; EMD), HSP90α (ADI-SPS-771; Enzo Life Sciences), mAb to mono- and polyubiquitinylated protein conjugates FK2H (BML-PW0150; Enzo Life Sciences), FK1 mAb to polyubiquitinylated conjugates (BML-PW8805; Enzo Life Sciences), Hsp90β (RB-118-P0; Thermo Fisher Scientific), HECTD1 mAb clone 1E10 (H00025831-M03; Abnova), c-Myc mAb (631206; Takara Bio Inc.), polyclonal Myc-Tag antibody (2272; Cell Signaling Technology), HA-Tag polyclonal antibody (631207; Takara Bio Inc.), HA mAb clone 16B12 (MMS-101R; Covance), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 14C10), rabbit mAb (14C10; Cell Signaling Technology), anti-FLAG monoclonal (M2) antibody (F1804; Sigma-Aldrich), anti-actin mAb clone C4 (MAB1501R; Millipore), rabbit anti-GFP (A-11122; Invitrogen), goat anti–rabbit IgG (H+L)–HRP conjugate (1721019; Bio-Rad Laboratories), goat anti–mouse IgG (H+L)–HRP conjugate (1721011; Bio-Rad Laboratories), rabbit anti–goat IgG (H&L)–HRP conjugate (ab6741; Abcam), Hoechst 33342 (P5541; Promega), lysine 48 linkage-specific polyubiquitin (Ubn) antibody (4289; Cell Signaling Technology), Ubn (K63 linkage-specific) mAb (HWA4C4, BML-PW0600; Enzo Life Sciences), Alexa Fluor 555 goat anti–rat IgG (H+L; A21434; Invitrogen), Alexa Fluor 555 goat anti–rabbit IgG (H+L; A21429; Invitrogen), Alexa Fluor 488 goat anti–rabbit IgG (H+L; A11034, Invitrogen), Alexa Fluor 488 goat anti–mouse IgG (H+L; A11029; Invitrogen), and Alexa Fluor 546 donkey anti–goat IgG (H+L; A11056; Invitrogen). Because of the high homology between Hsp90 family members, we confirmed antibody specificity against Hsp90α and -β in transfected HEK293T lysates. Our results demonstrate that the Hsp90 antibody (no. 4877; Cell Signaling Technology) recognizes both α and β subtypes, whereas the Hsp90α (no. SPS-771; Enzo Life Sciences) uniquely recognizes Hsp90α. We did not identify an antibody that uniquely recognizes Hsp90β. To detect extracellular Hsp90α, live cells were treated with anti-Hsp90α antibody without permeabilization with Triton X-100 for 3 h at 37°C before fixation. In contrast, all other immunofluorescence experiments were performed as described previously (Zohn et al., 2006). In brief, cells were fixed for 20 min in 4% paraformaldehyde, washed in PBS, and permeabilized in PBS + 0.1% Triton X-100. Primary antibody incubations were performed overnight and secondary antibody incubations were performed for 20 min. Immunofluorescence images were obtained at 25°C with a fluoromount mounting medium (Sigma-Aldrich) using a confocal microscope (LSM-510; Carl Zeiss) with the 63× objective lens (Plan-Apochromat NA 1.4 oil, differential interference contrast). Digital images were acquired using AxioVision LE software (Carl Zeiss) and processed with Photoshop (8.0) and Illustrator (10.0).
Co-immunoprecipitation, ubiquitination assays, and ELISA
For in vitro binding assays, biotinylated peptides were synthesized using the TNT T7 Coupled Reticulocyte Lysate system and Transcend tRNA (Promega). Binding assays were performed on precleared lysates at 25°C for 1 h using the Matchmaker Co-IP kit (Takara Bio Inc.) and Western blots developed using the Transcend Chemiluminescent Non-Radioactive Translation Detection system (Promega). Binding assays in HEK293T cells were performed at 4°C for 18 h as described previously (Zohn et al., 2006). For lenient binding conditions, a mild immunoprecipitation buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, and 0.1% Triton X-100 with protease inhibitor cocktail) was used, and for stringent binding conditions, radioimmunoprecipitation assay buffer (no. 89901; Thermo Fisher Scientific) with protease inhibition cocktail (Roche) was used. For ubiquitination assays in HEK293T cells, 44 h after transfection, cells were treated with the proteasome inhibitor MG132 (20 µM; Sigma-Aldrich) for 4 h, washed in ice-cold PBS, and lysed in ubiquitination lysis buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, complete protease inhibitor cocktail, 100 µM MG132, 20 µM Ub aldehyde, and 100 µM N-ethylmalemide). Images of Western blot gels were acquired with a ChemiDoc XRS system (Bio-Rad Laboratories) and Quality One (4.6.3) software or using the Odyssey infrared imaging system (LI-COR Biosciences) and processed with Photoshop (8.0) and Illustrator (10.0). Scanned films were quantified using ImageJ software (http://rsb.info.nih.gov/ij/). To quantify ubiquitination assays, the whole lane from the well to the bottom of the gel was used for quantification. Statistically significant differences were assessed by a paired two-tailed Student’s t test. All statistical analyses were performed using MS Excel software (Microsoft). Differences were considered to be significant for values of P < 0.05.
Cell shape assay
10 embryos each of wild type and Hectd1 mutant at E9.5 were fixed and stained using the hematoxylin and eosin method as described previously (Zohn et al., 2007). Bright field images were acquired using a microscope (BX61; Olympus) at 10× and 40× magnification, with objective lenses of NA 0.4 and 0.85, respectively. Images were taken using a DP71 camera (Olympus). Digital images were acquired using DPController and DPManager software (Olympus) and processed with Photoshop (8.0) and Illustrator (10.0). Cells were imaged using bright field microscopy at 100× magnification. Cell perimeters (n = 130) were computed using NeuronJ (http://www.imagescience.org/meijering/software/neuronj/), and cell areas (n = 130) were computed using ImageJ (http://rsbweb.nih.gov/ij/). Cell shape was defined using the equation (4π × cell area/cell perimeter2), where >1 indicates a perfect circle and values <1 indicate a more irregular shape.
Cell migration assays and quantification
For explant migration assays, E8.5 Hectd1WT and Hectd1O embryos were collected and washed in 1× PBS, and explants were micro-dissected from the CM, taking care to remove the neural epithelium and surface ectoderm. Explants were cultured for 24 h on fibronectin-coated glass coverslips in DME containing 10% FBS with or without 25 µg/ml anti-Hsp90α antibody as described previously (Sidera et al., 2008). Explants were photographed using a microscope (CKX41; Olympus) with the 40×/NA 0.55 objective lens. Images were captured using a DP70 camera (Olympus) and processed with Photoshop (8.0) and Illustrator (10.0). The mean migratory distance from the circumference of the explant to the migratory front and the mean cell number in unit area (200 µm2) perpendicular and mid-distant between the migratory front and explant circumference (dotted lines) were quantitated. Results are expressed as the mean migratory distance (in micrometers) of 50 randomly chosen cells at the migrating front and the mean number of migrating cells in 10 randomly chosen squares of unit area (200 µm2) at a mid-distance between the explant circumference and the migrating front, perpendicular to the explant circumference. Inhibition of secretory mechanisms was achieved by treatment of explants for 16 h in 200 µM DMA, whereas control explants were treated with DMSO vehicle. Statistical significance of differences of means was tested with a paired two-tailed Student’s t test.
Online supplemental material
Fig. S1 shows the specificity of the anti-Hectd1 antibody used in this study. Fig. S2 shows colocalization of Hectd1 and Hsp90 in HEK293T cells. Fig. S3 shows the genotyping strategy to identify Hectd1opm and Hectd1X. Fig. S4 shows the effectiveness of the siRNA targeting Hectd1 in reducing protein expression when transfected in HEK293T cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201105101/DC1.
Acknowledgments
The authors would like to express their appreciation to Drs. Joshua Corbin, Jyoti Jaiswal, Judy Liu, and Vittorio Gallo for helpful discussions and critical reading of the manuscript, and Samer Nuwayhid for technical assistance. We are grateful to Dr. Lee Niswander for support in the early days of this project, members of the Dr. Mary Baylies laboratory for help with the yeast two-hybrid screen, members of the Center for Neuroscience Research for helpful suggestions, Drs Kristy Brown and Yetrib Hathout for help with MS instrumentation and data analysis, and Dr. Anastas Popratiloff, director of the Center for Microscopy and Image Analysis at the George Washington University, for help with confocal microscopy.
This work was partially supported by National Institutes of Health grant IDDRC P30HD40677 (to Y. Hathout and A. Popratiloff) and a Basil O’Conner Award from the March of Dimes, a Young Investigator Award from the Spina Bifida Association, and grant NICHD-R01-HD058629 (from the National Institute of Child Health and Human Development) to I.E. Zohn.
Footnotes
Abbreviations used in this paper:
- ALLN
- N-acetyl-leucyl-leucyl-norleucinal
- ANK
- ankyrin domain
- ARMS-PCR
- amplification refractory mutation system PCR
- CM
- cranial mesenchyme
- DMA
- dimethyl ameloride
- HECT
- homologous to the E6-AP C terminus
- Hectd1O
- Hectd1openmind
- Hectd1w
- Hectd1wildtype
- HectD1X
- Hectd1XC
- HEK293T
- human embryonic kidney 293
- HMW
- high molecular weight
- Hsp
- heat shock protein
- Hsp90αbd
- Hsp90α binding domain
- LC-MS
- liquid chromatography–mass spectrometry
- mAb
- monoclonal antibody
- Ub
- Ubiquitin
- Ubn
- polyubiquitin
References
- Bandyopadhyay A., Bandyopadhyay S., Basak J., Mondal B.C., Sarkar A.A., Majumdar S., Das M.K., Chandra S., Mukhopadhyay A., Sanghamita M., et al. 2004. Profile of beta-thalassemia in eastern India and its prenatal diagnosis. Prenat. Diagn. 24:992–996 10.1002/pd.1049 [DOI] [PubMed] [Google Scholar]
- Bergink S., Salomons F.A., Hoogstraten D., Groothuis T.A., de Waard H., Wu J., Yuan L., Citterio E., Houtsmuller A.B., Neefjes J., et al. 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 20:1343–1352 10.1101/gad.373706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., Traub L.M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Cargile B.J., Stephenson J.L., Jr 2004. An alternative to tandem mass spectrometry: isoelectric point and accurate mass for the identification of peptides. Anal. Chem. 76:267–275 10.1021/ac0352070 [DOI] [PubMed] [Google Scholar]
- Chen Z.F., Behringer R.R. 1995. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 9:686–699 10.1101/gad.9.6.686 [DOI] [PubMed] [Google Scholar]
- Chen L., Madura K. 2002. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 22:4902–4913 10.1128/MCB.22.13.4902-4913.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.F., Fan J., Fedesco M., Guan S., Li Y., Bandyopadhyay B., Bright A.M., Yerushalmi D., Liang M., Chen M., et al. 2008. Transforming growth factor alpha (TGFalpha)-stimulated secretion of HSP90alpha: using the receptor LRP-1/CD91 to promote human skin cell migration against a TGFbeta-rich environment during wound healing. Mol. Cell. Biol. 28:3344–3358 10.1128/MCB.01287-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A., Turkes A., Navabi H., Mason M.D., Tabi Z. 2005. Induction of heat shock proteins in B-cell exosomes. J. Cell Sci. 118:3631–3638 10.1242/jcs.02494 [DOI] [PubMed] [Google Scholar]
- Copp A.J. 2005. Neurulation in the cranial region—normal and abnormal. J. Anat. 207:623–635 10.1111/j.1469-7580.2005.00476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp A.J., Greene N.D., Murdoch J.N. 2003. The genetic basis of mammalian neurulation. Nat. Rev. Genet. 4:784–793 10.1038/nrg1181 [DOI] [PubMed] [Google Scholar]
- Essader A.S., Cargile B.J., Bundy J.L., Stephenson J.L., Jr 2005. A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 5:24–34 10.1002/pmic.200400888 [DOI] [PubMed] [Google Scholar]
- Eustace B.K., Jay D.G. 2004. Extracellular roles for the molecular chaperone, hsp90. Cell Cycle. 3:1098–1100 10.4161/cc.3.9.1088 [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R., Gordon C. 2004. Protein degradation: recognition of ubiquitinylated substrates. Curr. Biol. 14:R754–R756 10.1016/j.cub.2004.09.012 [DOI] [PubMed] [Google Scholar]
- Hegmans J.P., Bard M.P., Hemmes A., Luider T.M., Kleijmeer M.J., Prins J.B., Zitvogel L., Burgers S.A., Hoogsteden H.C., Lambrecht B.N. 2004. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 164:1807–1815 10.1016/S0002-9440(10)63739-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O.N., Wilm M., Shevchenko A., Mann M. 1999. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112:513–530 [DOI] [PubMed] [Google Scholar]
- Kasarskis A., Manova K., Anderson K.V. 1998. A phenotype-based screen for embryonic lethal mutations in the mouse. Proc. Natl. Acad. Sci. USA. 95:7485–7490 10.1073/pnas.95.13.7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li Y., Guan S., Fan J., Cheng C.F., Bright A.M., Chinn C., Chen M., Woodley D.T. 2007. Extracellular heat shock protein-90alpha: linking hypoxia to skin cell motility and wound healing. EMBO J. 26:1221–1233 10.1038/sj.emboj.7601579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sahu D., Tsen F. 2012. Secreted heat shock protein-90 (Hsp90) in wound healing and cancer. Biochim. Biophys. Acta. 1823:730–741 10.1016/j.bbamcr.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D.F., Jin Z.G., Baas A.S., Daum G., Gygi S.P., Aebersold R., Berk B.C. 2000. Purification and identification of secreted oxidative stress-induced factors from vascular smooth muscle cells. J. Biol. Chem. 275:189–196 10.1074/jbc.275.1.189 [DOI] [PubMed] [Google Scholar]
- McCready J., Sims J.D., Chan D., Jay D.G. 2010. Secretion of extracellular hsp90alpha via exosomes increases cancer cell motility: a role for plasminogen activation. BMC Cancer. 10:294 10.1186/1471-2407-10-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough H., Patterson C. 2003. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 8:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Wiman J., Brinkley L.L. 1990a. Changes in mesenchymal cell and hyaluronate distribution correlate with in vivo elevation of the mouse mesencephalic neural folds. Anat. Rec. 226:383–395 10.1002/ar.1092260316 [DOI] [PubMed] [Google Scholar]
- Morris-Wiman J., Brinkley L.L. 1990b. The role of the mesenchyme in mouse neural fold elevation. I. Patterns of mesenchymal cell distribution and proliferation in embryos developing in vitro. Am. J. Anat. 188:121–132 10.1002/aja.1001880203 [DOI] [PubMed] [Google Scholar]
- Morris-Wiman J., Brinkley L.L. 1990c. The role of the mesenchyme in mouse neural fold elevation. II. Patterns of hyaluronate synthesis and distribution in embryos developing in vitro. Am. J. Anat. 188:133–147 10.1002/aja.1001880204 [DOI] [PubMed] [Google Scholar]
- Morriss G.M., Solursh M. 1978a. Regional differences in mesenchymal cell morphology and glycosaminoglycans in early neural-fold stage rat embryos. J. Embryol. Exp. Morphol. 46:37–52 [PubMed] [Google Scholar]
- Morriss G.M., Solursh M. 1978b. The role of primary mesenchyme in normal and abnormal morphogenesis of mammalian neural folds. Zoon. 6:33–38 [Google Scholar]
- Morriss-Kay G.M., Tuckett F., Solursh M. 1986. The effects of Streptomyces hyaluronidase on tissue organization and cell cycle time in rat embryos. J. Embryol. Exp. Morphol. 98:59–70 [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 315:201–205 10.1126/science.1127085 [DOI] [PubMed] [Google Scholar]
- Murata S., Chiba T., Tanaka K. 2003. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int. J. Biochem. Cell Biol. 35:572–578 10.1016/S1357-2725(02)00394-1 [DOI] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R.R.2003. Manipulating the Mouse Embryo: A Laboratory Manual. Chapter 8, protocol 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. 371–375.
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Schoenwolf G.C., Fisher M. 1983. Analysis of the effects of Streptomyces hyaluronidase on formation of the neural tube. J. Embryol. Exp. Morphol. 73:1–15 [PubMed] [Google Scholar]
- Sidera K., Gaitanou M., Stellas D., Matsas R., Patsavoudi E. 2008. A critical role for HSP90 in cancer cell invasion involves interaction with the extracellular domain of HER-2. J. Biol. Chem. 283:2031–2041 10.1074/jbc.M701803200 [DOI] [PubMed] [Google Scholar]
- Sims J.D., McCready J., Jay D.G. 2011. Extracellular heat shock protein (Hsp)70 and Hsp90α assist in matrix metalloproteinase-2 activation and breast cancer cell migration and invasion. PLoS ONE. 6:e18848 10.1371/journal.pone.0018848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S., Neckers L. 2007. Extracellular heat shock protein 90: a role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 98:1536–1539 10.1111/j.1349-7006.2007.00561.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckett F., Morriss-Kay G.M. 1986. The distribution of fibronectin, laminin and entactin in the neurulating rat embryo studied by indirect immunofluorescence. J. Embryol. Exp. Morphol. 94:95–112 [PubMed] [Google Scholar]
- Urbé S. 2005. Ubiquitin and endocytic protein sorting. Essays Biochem. 41:81–98 10.1042/EB0410081 [DOI] [PubMed] [Google Scholar]
- Verschuure P., Croes Y., van den IJssel P.R., Quinlan R.A., de Jong W.W., Boelens W.C. 2002. Translocation of small heat shock proteins to the actin cytoskeleton upon proteasomal inhibition. J. Mol. Cell. Cardiol. 34:117–128 10.1006/jmcc.2001.1493 [DOI] [PubMed] [Google Scholar]
- Wang X., Song X., Zhuo W., Fu Y., Shi H., Liang Y., Tong M., Chang G., Luo Y. 2009. The regulatory mechanism of Hsp90alpha secretion and its function in tumor malignancy. Proc. Natl. Acad. Sci. USA. 106:21288–21293 10.1073/pnas.0908151106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windheim M., Stafford M., Peggie M., Cohen P. 2008. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol. Cell. Biol. 28:1783–1791 10.1128/MCB.02380-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Dhillon S., Ke X., Collins A.R., Day I.N. 2001. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 29:E88 10.1093/nar/29.17.e88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn I.E., Sarkar A.A. 2008. Modeling neural tube defects in the mouse. Curr. Top. Dev. Biol. 84:1–35 10.1016/S0070-2153(08)00601-7 [DOI] [PubMed] [Google Scholar]
- Zohn I.E., Li Y., Skolnik E.Y., Anderson K.V., Han J., Niswander L. 2006. p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell. 125:957–969 10.1016/j.cell.2006.03.048 [DOI] [PubMed] [Google Scholar]
- Zohn I.E., Anderson K.V., Niswander L. 2007. The Hectd1 ubiquitin ligase is required for development of the head mesenchyme and neural tube closure. Dev. Biol. 306:208–221 10.1016/j.ydbio.2007.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]