RAD23 proteins facilitate lesion recognition by XPC but then dissociate from the XPC complex and do not participate in the downstream DNA repair process.
Abstract
The recognition of helix-distorting deoxyribonucleic acid (DNA) lesions by the global genome nucleotide excision repair subpathway is performed by the XPC–RAD23–CEN2 complex. Although it has been established that Rad23 homologs are essential to protect XPC from proteasomal degradation, it is unclear whether RAD23 proteins have a direct role in the recognition of DNA damage. In this paper, we show that the association of XPC with ultraviolet-induced lesions was impaired in the absence of RAD23 proteins. Furthermore, we show that RAD23 proteins rapidly dissociated from XPC upon binding to damaged DNA. Our data suggest that RAD23 proteins facilitate lesion recognition by XPC but do not participate in the downstream DNA repair process.
Introduction
Nucleotide excision repair (NER) is a versatile DNA repair mechanism that repairs a variety of helix-disturbing lesions including those induced by the UV component of sunlight (Hoeijmakers, 2001). Two NER subpathways exist that differ in their mechanism of lesion recognition. Stalling of RNA polymerase II at lesions in transcribed regions initiates transcription-coupled NER (Fousteri et al., 2006). The Xeroderma Pigmentosum protein C (XPC) complex binds to lesions located anywhere in the genome and initiates global genome NER (GG-NER). After lesion recognition, the two subpathways funnel into a common mechanism that involves DNA unwinding, coordinated excision of a 25–30-nucleotide region containing the lesion (Staresincic et al., 2009), single-strand gap filling by the replication enzymes (Ogi et al., 2010; Overmeer et al., 2010), and finally sealing of the nick by a ligase (Moser et al., 2007).
Damage recognition is a crucial NER-initiating step and likely rate-determining parameter (Luijsterburg et al., 2010). Lesion discrimination in mammalian GG-NER is achieved by an intricate mechanism involving the UV–damaged DNA binding (DDB) and XPC complexes (Sugasawa et al., 2009). Xeroderma pigmentosum patients that carry mutations in the XPC gene are highly susceptible to develop tumors on sunlight-exposed areas of the skin (Cleaver, 2005), a feature recapitulated in Xpc knockout mice (Cheo et al., 1997; Sands et al., 1995). XPC purified from HeLa cell extracts was found to copurify with RAD23B and to a lesser extent with RAD23A (Masutani et al., 1994), which are two mammalian paralogs of the yeast Rad23 NER protein. As a third binding partner, the CEN2 protein has been identified (Araki et al., 2001; Nishi et al., 2005). The interaction between RAD23 and XPC is evolutionarily conserved, as this interaction was also observed in yeast, arguing for an important role in driving NER (Guzder et al., 1998). Yeast rad23 mutants as well as mouse embryonic fibroblasts (MEFs) deficient in both Rad23a and Rad23b are hypersensitive to UV light (Ng et al., 2003; Watkins and Smerdon, 1985), a finding that could be recapitulated in human knockdown cells (Renaud et al., 2011). However, cells lacking RAD23A or RAD23B (single knockouts) do not display increased UV sensitivity, suggesting that they have redundant functions in NER (Ng et al., 2003).
It has been shown that loss of RAD23 function in both yeast (Lommel et al., 2002) and mammalian cells (Ng et al., 2003) leads to severely reduced steady-state levels of Rad4/XPC. Thus, it was suggested that the major function of the RAD23 proteins is to stabilize XPC by protecting it from degradation. However, the affinity of purified XPC for damaged DNA significantly increases in a cell-free assay after adding purified RAD23B or RAD23A (Sugasawa et al., 1996). Moreover, overexpression of Rad4 in yeast only partly suppresses the UV sensitivity of rad23 mutant cells (Xie et al., 2004), suggesting additional roles for the RAD23 proteins in NER besides stabilizing XPC. Despite insight into the structural requirements for XPC to bind to damaged DNA (Min and Pavletich, 2007), the molecular mechanisms underlying RAD23-dependent regulation of DNA damage recognition by XPC are currently poorly understood. Here, we demonstrate that the mammalian RAD23 proteins play a direct role in damage recognition by enhancing the binding of XPC to DNA damage in living cells in addition to stabilizing XPC. Remarkably, however, RAD23B quickly dissociates from XPC after binding to damage, suggesting that it does not participate in the downstream NER complex assembly.
Results and discussion
RAD23A and RAD23B immobilize XPC on DNA damage in living cells
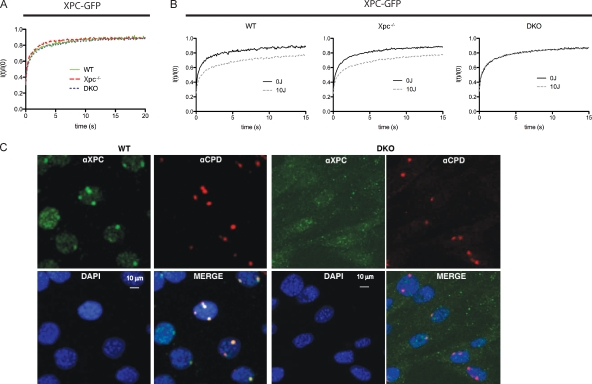
The finding that purified XPC has a lower affinity for damaged DNA in the absence of RAD23A or RAD23B in vitro (Sugasawa et al., 1996) prompted us to assess the impact of RAD23A and RAD23B on the binding of XPC to damaged DNA in living cells. Toward that aim, GFP-tagged XPC (XPC-GFP; Hoogstraten et al., 2008) was transiently expressed in either Rad23a/b double knockout (DKO) cells, Xpc−/− MEFs, or wild-type (WT) MEFs, and the mobility of XPC-GFP was assessed by FRAP analysis (Houtsmuller and Vermeulen, 2001). For the FRAP analysis, cells were selected that express XPC-GFP at similar amounts as within a previously described human XPC cell line (Hoogstraten et al., 2008) that stably expresses XPC-GFP at near physiological levels, comparable with endogenous XPC in WT cells. The mobility of XPC-GFP (Fig. 1 A) in the absence of DNA damage was similar in all three different MEF cells lines (WT, Xpc−/−, and DKO) and was identical to the mobility of XPC-GFP expressed in human XPC cells (Hoogstraten et al., 2008). We previously observed that the relatively slow mobility of XPC-GFP is a result of the continuous nonspecific association with and dissociation from chromosomal DNA (Hoogstraten et al., 2008). Thus, our mobility measurements show that nonspecific binding of XPC to DNA is not affected by the absence of RAD23 proteins. After UV irradiation (16 J/m2), resulting in a relatively high concentration of DNA lesions in the genome, the mobility of XPC-GFP was significantly reduced in both Xpc−/− and WT cells (Fig. 1 B). This UV-induced decrease in XPC-GFP mobility was also observed for other NER proteins and was thought to be a result of the transient incorporation of repair proteins into chromatin-associated repair complexes (Vermeulen, 2011). Strikingly, however, we did not detect any changes in the mobility of XPC-GFP after UV irradiation in Rad23a/b DKO cells (Fig. 1 B), suggesting that the RAD23 proteins are essential for the binding of XPC to UV-induced DNA damage in living cells. The reduced ability of XPC-GFP to bind to damaged DNA was confirmed in situ by immunostaining of XPC on local UV-irradiated areas (Fig. 1 C; Moné et al., 2001). In WT cells, a clear colocalization of endogenous XPC with UV-induced DNA lesions detected by an anti–cyclobutane pyrimidine dimers (CPDs) antibody was observed, whereas XPC failed to accumulate at DNA lesions in DKO cells.
Figure 1.
RAD23A and RAD23B immobilize XPC on DNA damage in living cells. (A and B) FRAP analysis of XPC-GFP in the absence and presence of UV damage. The relative fluorescence recovery (It/I0) immediately after bleaching is plotted against the time (seconds). (A) In the absence of DNA damage, no apparent difference in the mobility rate of XPC-GFP could be detected when expressed in either WT, Xpc knockout, or Rad23a/b DKO cells. (B) After UV treatment, part of the XPC-GFP is immobilized (incomplete fluorescence recovery) when expressed in WT cells or in Xpc−/− MEFs. However, when expressed in DKO cells, no immobilization upon UV treatment was observed. Mobilities were measured between 30 and 60 min after UV-C (16 J/m2) treatment. (C) Immunofluorescence analysis of XPC (green channel) at local UV-induced DNA-damaged areas, identified by antibodies that recognize the main UV photoproduct CPD (red channel) in different genetic backgrounds. Nuclei are counterstained by DAPI (blue channel), and the bottom right panel is a merge of all three channels. Cells were fixed 45 min after local UV-C irradiation. (left) Endogenous XPC accumulates at local damaged sites, as is indicated by the presence of the CPDs (red) in WT cells. (right) In Rad23a/b DKO cells, no XPC is found at the local UV damage. Note that the image settings are changed (increased background) to compensate for reduced XPC levels in DKO cells.
Our data indicate that the Rad23 homologs are not only critical for XPC stabilization but are also essential for the efficient binding of XPC to DNA lesions in living cells, as we show that the UV-induced immobilization of XPC is impaired in the absence of the RAD23 proteins even when XPC is expressed at levels comparable with those in WT cells. Importantly, loss of RAD23 proteins did not affect the nonspecific association to and dissociation from nondamaged chromosomal DNA of XPC in living cells (Hoogstraten et al., 2008). As XPC still binds to nondamaged DNA in the absence of RAD23 proteins both in vitro and in living cells (relative slow mobility as determined by FRAP), we propose that RAD23A and RAD23B assist in the formation of a stable anchorage of XPC during the early steps of NER.
RAD23A and RAD23B do not accumulate at local DNA damage
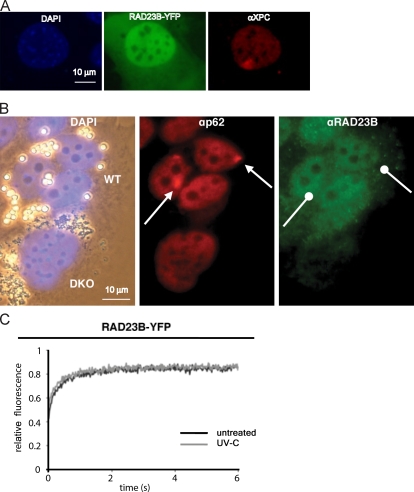
To measure the dynamic interaction of RAD23B with the NER machinery, we tagged mouse RAD23B (mRAD23B) at its C terminus with the YFP and a FLAG tag (RAD23B-YFP-FLAG). The fusion protein was stably expressed in the Rad23a/b DKO MEFs. Clones were selected by virtue of their ability to rescue the UV-sensitive phenotype of the Rad23a/b DKO cells (Fig. S1 A). Both endogenous mRAD23B and the mRAD23B-YFP-FLAG were homogeneously expressed in the cytoplasm and the nucleus, with less abundant amounts in nucleoli (Fig. 2, A and B). Immunofluorescence analysis showed that mRAD23B-YFP also restored the reduced endogenous mXPC levels to WT amounts (Fig. S1 B). Both the increase in mXPC levels as well as the rescue of the UV hypersensitivity upon expression in DKO cells indicate that the mRAD23B-YFP-FLAG protein is fully functional in GG-NER. 30 min after local UV irradiation of mHR23B-YFP cells, a clear accumulation of endogenous mXPC at damaged sites was observed, again showing the functionality of the RAD23B-YFP fusion protein. Despite the proven functionality, no accumulation of mRAD23B-YFP-FLAG could be detected at sites of local UV irradiation (Fig. 2 A). In line with these findings, none of the antibodies against endogenous mRAD23B revealed accumulation at locally damaged areas in which endogenous XPC clearly accumulated (Fig. 2 B and not depicted). Likewise, transiently expressed mRAD23A-GFP2-MYC also failed to accumulate at local UV damage in DKO cells (unpublished data). We subsequently applied FRAP after UV irradiation on cells expressing mRAD23B-YFP-FLAG. In contrast to all other NER factors tested thus far (Vermeulen, 2011), no immobilization of mRAD23B-YFP-FLAG could be detected upon UV exposure (Fig. 2 C).
Figure 2.
RAD23A and RAD23B do not accumulate/immobilize at DNA damage and are not immobilized upon UV treatment. (A and B) Analysis of RAD23B-YFP and RAD23B at local damaged DNA spots. Cells were fixed between 30 and 60 min after local UV irradiation. (A) RAD23B-YFP (identified by the YFP fluorescence) does not accumulate at local damaged sites, whereas its complex partner XPC (identified by XPC antibodies) does. The nuclei are counterstained with DAPI (blue). (B) Comparative immunofluorescence on a mixture of WT and DKO MEFs, recognized by bead-labeled cells with small and large beads, respectively (DAPI-derived fluorescence mixed with transmitting light; left). Endogenous RAD23B (identified by RAD23B antibodies) fails to accumulate at local DNA damage (see arrows), as is indicated by the accumulation of the TFIIH subunit p62 (αp62). The specificity of the RAD23B antibody is illustrated by the virtual absence of staining in the DKO cells (small beads; left), whereas a clear signal is visible in WT cells (large beads; left). (C) FRAP analysis of RAD23B-YFP before and after UV-induced DNA damage. RAD23B-YFP does not immobilize after UV treatment. Mobilities were measured between 30 and 60 min after UV-C (16 J/m2) treatment.
The absence of UV-induced immobilization is in line with the lack of RAD23B accumulation at sites of local DNA damage. To verify these results in another cell type, we tagged the last exon of the mouse Rad23b gene with YFP-FLAG in a bacterial artificial chromosome (BAC) harboring the genomic mouse Rad23b locus, which was subsequently integrated in the genome of a WT mouse embryonic stem (ES) cell line. The expression level of tagged RAD23B is similar to endogenous RAD23B (Fig. S1 C). In this mouse ES cell line, genomically tagged RAD23B-YFP-FLAG also failed to accumulate at sites of local damage (unpublished data), confirming the data obtained in MEFs.
The XPC–RAD23B complex dissociates after binding to UV-induced DNA damage
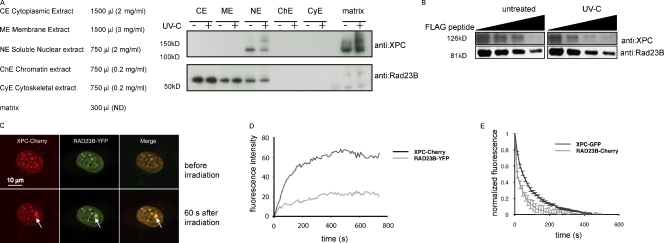
The previous data indicate that although RAD23 is required to efficiently load XPC onto damaged DNA, the protein itself does not stably bind damaged sites, which is in contrast to its complex partner XPC. This prompted us to test whether the XPC–RAD23B complex might be disrupted upon lesion binding of XPC. To this end, we used a biochemical fractionation approach and compared the complex composition isolated from irradiated and nonirradiated human U2-OS cells. The used fractionation protocol separates cells into a cytoplasmic, membrane, nuclear-soluble, chromatin, cytoskeletal, and matrix fraction. The latter fraction contains DNA-bound proteins that are resistant to micrococcal nuclease digestion and is devoid of membrane-bound proteins as a result of a separate membrane fractionation step. After fractionation of nonirradiated cells, we detected XPC in the nuclear-soluble fraction, the chromatin fraction (only detectable when equal protein concentrations are loaded), and the matrix fraction (Fig. 3 A and not depicted). As expected, we detected an increase of XPC in the DNA-bound fraction after UV exposure (Fig. 3 A). The presence of XPC in the matrix fraction before UV exposure is most likely caused by the presence of spontaneous/endogenous DNA damage. In contrast, RAD23B resides in all fractions except the matrix fraction (the presence in the chromatin and cytoskeletal fractions is only evident when equal protein concentrations are loaded; unpublished data), and UV irradiation did not appreciably affect the distribution in the different fractions. Importantly, very recently, a similar behavior of Rad23B and XPC during cellular fractionation was reported (Fei et al., 2011), underscoring our findings. Given that the matrix contains the damaged DNA-bound XPC pool, which is roughly half of the total amount of XPC molecules at a given time, we conclude that lesion-bound XPC is not, or at undetectable levels, in complex with RAD23B in vivo. We subsequently immunoprecipitated XPC from the nuclear, chromatin, and cytoskeletal fractions and probed for the presence of RAD23B. Considerably less RAD23B was coimmunoprecipitated with XPC after UV irradiation in the soluble nuclear fraction compared with nonirradiated cells (Fig. S2 A). To corroborate these findings, we incubated whole-cell extracts of mRAD23B-YFP-FLAG–expressing mES cells (described in Fig. S1 C), either mock or UV-C treated, with anti-FLAG antibody–coupled beads followed by elution with FLAG peptide. Equal amounts of RAD23B eluates were loaded, and blots were probed with anti-XPC antibodies. In agreement with our fractionation approach, a clear reduction in the stoichiometry of the XPC–RAD23B complex after UV treatment was apparent (Figs. 3 B and S2 [B and C]), showing that the XPC–RAD23 complex is disrupted upon UV irradiation.
Figure 3.
The XPC–RAD23B complex dissociates upon UV-induced DNA damage. (A) Subcellular fractionation of human U2-OS with and without UV (60 min after 20 J/m2) treatment. The same volume was loaded on a gradient gel for each fraction. After Western blotting, membranes were stained with the indicated antibodies. (B) Immunoblot analysis for both XPC and RAD23B proteins of anti-FLAG–immunoprecipitated complexes from RAD23B-YFP-FLAG–expressing ES cells before and 60 min after UV-C (16 J/m2) treatment. Immunoprecipitated proteins were eluted with free FLAG peptide, and different fractions were collected, boiled, and separated on a gel. Upon elution with (free) Flag peptide, a lower amount of XPC coeluted when cells were treated with UV-C than from untreated cells. (C) Upon overexpression of XPC-Cherry (red channel), RAD23B-YFP does visibly accumulate at local damage (green channel), inflicted with a UV-C laser. Pictures of representative cells before (top row) and 60 s after (bottom row) UV-C are shown. Arrows indicate the spot of the local UV-C laser-induced DNA damage. (D) Relative accumulation of XPC-mCherry and RAD23B-YFP on local damage. Arbitrary fluorescence intensity units are plotted against the time (in seconds) after local damage infliction. (E) iFRAP, in which total fluorescence in the nuclei (except the fluorescence present at the local UV damage) is bleached and the subsequent loss in fluorescence of RAD23B-mCherry and XPC-GFP is monitored, which is a measure of the dissociation rate of these proteins from the local damage. RAD23B exhibits a quicker dissociation as compared with XPC, suggesting that RAD23B dissociates from XPC after binding of the complex to lesions. n = 10 cells. Error bars represent SEM.
This finding is rather unexpected, as previous studies indicated that the XPC–RAD23B complex is very stable and even resistant to high-salt conditions (Masutani et al., 1994). Moreover, within a recently solved structure of Rad4, the yeast ortholog of XPC, bound to nonpairing single-strand DNA opposite a CPD lesion, contained the Rad23 protein (Min and Pavletich, 2007). In this structure, it seems that the interaction between Rad4 and Rad23 might be maintained when Rad4 is bound to single-strand DNA. In addition, our current data indicate that RAD23B is required for efficient XPC binding to lesions (Fig. 1), suggesting that complex formation between these proteins is necessary during the early steps of NER. One way to explain this apparent discrepancy is that the intact XPC–RAD23B complex binds to lesions, and, soon after binding, RAD23B dissociates from the lesion-bound XPC. In this scenario, the intact XPC–RAD23 complex associates with DNA lesions, but the rapid dissociation and short chromatin dwell time of RAD23 make it difficult to detect RAD23 accumulation at lesions in FRAP and local damage experiments (Fig. 2). To test whether RAD23B-YFP can indeed be retained at damaged DNA, we overexpressed mCherry-tagged XPC at very high levels in the Rad23a/b DKO cells stably expressing RAD23B-YFP to shift the equilibrium between free RAD23B-YFP and RAD23B-YFP in complex with XPC. Only under these conditions were we indeed able to visualize RAD23B-YFP at damaged DNA induced by UV-C laser microirradiation (Fig. 3 C; Dinant et al., 2007).
Importantly, the relative amount of RAD23B-YFP that accumulated at damaged sites was significantly less than the amount of XPC that accumulated at the same damaged sites (Fig. 3 D), which indicates that unequal amounts of both proteins are retained at sites of DNA damage. To directly measure the dissociation rates of XPC and RAD23B from sites of DNA damage, we performed inverse FRAP (iFRAP; Dundr and Misteli, 2003) now using XPC-GFP and RAD23B-mCherry. In brief, we bleached both fluorescent tags at full laser power by scanning a selected region of interest comprising the entire nucleus except the locally damaged area for 4 s. Subsequently, we took images with time intervals of 15 s and measured the loss of fluorescence of both tagged proteins in the local damage, which reflects their dissociation rates. The observed dissociation rate of RAD23B-mCherry from damaged sites was significantly faster than the dissociation rate of XPC-GFP (Fig. 3 E), strongly indicating that after damage detection, RAD23B dissociates from the damage complex much earlier than XPC. As a control, to exclude possible artifacts as a result of different photophysical behavior of the fluorescent tags, we used the same protocol to measure the dissociation rates of mCherry-DDB1 and YFP-DDB2 (two subunits of an accessory GG-NER–initiating complex; Moser et al., 2005) that were simultaneously expressed in human fibroblasts (Fig. S2, D and E; Alekseev et al., 2008). As expected and unlike XPC and RAD23B, we found that the dissociation rates of DDB1 and DDB2 from damaged DNA were identical, suggesting that these proteins associate with and dissociate from DNA lesions as a complex, whereas the XPC–RAD23B complex is disrupted once XPC binds to damaged DNA.
The DNA-binding domain of XPC is localized between amino acid 607 and 742. Interestingly, the RAD23A- or RAD23B-binding region is between amino acid 496 and 734 (Uchida et al., 2002) and thus partially overlaps with the DNA-binding area. Therefore, it is tempting to speculate that the dissociation of RAD23B upon UV irradiation is necessary to make the DNA-binding domain of XPC (more) accessible. Recently, a study revealed that binding of XPC to a DNA lesion requires a two-stage discrimination mechanism, which initially involves a transient scanning mode followed by the more stable binding of XPC to initiate repair (Camenisch et al., 2009). As RAD23B is essential for retaining XPC at damaged DNA (the second stage) and seems to have no function in the initial scanning stage (Fig. 1), we suggest that RAD23B dissociates after having performed its function in recognition, now allowing the formation of a second, more stable damage-bound XPC complex.
RAD23B dissociation from chromatin-tethered XPC is triggered by UV-C
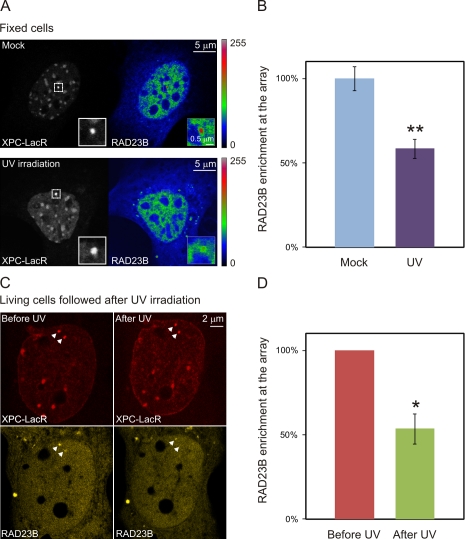
To gain more insight in the damage-induced dissociation of XPC and RAD23B, we used an in vivo targeting system for tethering proteins to chromatin (Soutoglou and Misteli, 2008). To visualize and target XPC, we generated an XPC-mCherry-LacR fusion protein, which was efficiently retained at an integrated 256x LacO array in NIH2/4 mouse cells (Soutoglou et al., 2007). Upon tethering of XPC-LacR to chromatin, clear accumulation of RAD23B-YFP could be detected, indicating that a complex between chromatin-bound XPC and RAD23B can be formed. UV-C irradiation (25 J/m2) triggered a marked reduction (50%) in the mean RAD23B-YFP intensity at the array upon XPC tethering compared with mock-treated control cells (Fig. 4, A and B). To confirm this observation in living cells, we measured the UV-induced change in the amount of RAD23B-YFP binding to chromatin-tethered XPC. Upon UV irradiation, a significant loss of RAD23-YFP from the LacO array was observed (Fig. 4, C and D). These results clearly demonstrate that UV irradiation triggers the dissociation of RAD23B from the chromatin-bound XPC protein.
Figure 4.
RAD23B dissociates from XPC after global UV treatment while it is tethered to DNA. (A) NIH2/4 cells containing a 256x LacO array were cotransfected with XPC-mCherry-LacR (grayscale images) and RAD23B-YFP (look-up table is shown next to the image) and subsequently mock treated (top) or exposed to UV light (25 J/m2; bottom) and fixed. Insets depict a magnified view of the area containing the integrated LacO array. (B) The intensity of the RAD23B-YFP signal at the XPC-LacR array in mock-treated or UV-irradiated cells was quantified (n = 40 for each condition from two independent experiments). (C) Living cells cotransfected with XPC-mCherry-LacR (red) and RAD23B-YFP (yellow) were monitored before and after UV exposure (25 J/m2). Arrowheads indicate the integrated LacO array. (D) The intensity of the RAD23B-YFP signal in the same cells before and after UV irradiation (n = 11 from three independent experiments). A single asterisk indicates significant differences (P < 0.05), whereas a double asterisk indicates highly significant differences (P < 0.005, based on an unpaired [B] or a paired [D] t test). Error bars indicate SDs.
The role of RAD23 in lesion recognition by XPC
In this study, we provide evidence that RAD23 proteins play an important role in recognition of UV-induced DNA damage but not in the formation of the downstream excision complex. Using biochemical and live-cell imaging approaches, we showed that although RAD23 is required to make XPC lesion binding competent, once XPC is bound to lesions, RAD23B rapidly dissociates from damage-bound XPC during the early steps of GG-NER.
The molecular triggers for the dissociation of RAD23 from XPC are currently unclear but may involve proteins in the XPC complex or additional NER proteins. For instance, the binding of XPC to single-strand DNA not only alters the structure of the DNA molecule by bending it (Janićijević et al., 2003) but also alters the conformation adopted by Rad4 (Min and Pavletich, 2007). Therefore, it is feasible that this altered conformation, which includes the interaction interface with Rad23, lowers the affinity of Rad4/XPC for Rad23/RAD23B. Moreover, it is also possible that the swift in vivo dissociation of RAD23B from UV-induced lesions is a result of conformational changes in the XPC complex that may also affect the binding of CEN2 to the XPC complex. As CEN2 is also implicated in regulating the XPC function within NER (Nishi et al., 2005), a role for this polypeptide in regulation RAD23B–XPC interaction when bound to lesions may not be excluded. However, our data do not provide further insight into the role of CEN2 in lesion recognition by XPC. Alternatively, but not necessarily mutually exclusive, up- or downstream NER factors, such as the UV-DDB or the TFIIH complexes, may play a role in the UV-induced RAD23B dissociation from the XPC complex. Given the fact that DDB2 mediates the ubiquitylation of XPC upon UV (Fig. 3 A; Sugasawa et al., 2005), it is also possible that this posttranslational modification of XPC triggers the RAD23B dissociation. As previously mentioned, XPC binds in two modes to DNA; only the longer immobilization leads to successful NER. As TFIIH is the immediate downstream factor of XPC required for lesion verification (Sugasawa et al., 2009), the scanning of TFIIH might disrupt the XPC–RAD23B complex. The possible role of the other NER factors in dissociating RAD23B from lesion-bound XPC may also provide an explanation for the apparent discrepancy with the structural data of the Rad4–Rad23–damaged DNA trimeric complex (Min and Pavletich, 2007). The interaction of Rad23 with lesion-bound Rad4 is apparently sufficiently stable to allow crystallization. Please note that the other up- and downstream NER factors as well as the UV-induced ubiquitylation were not present in the in vitro crystallization reaction (Min and Pavletich, 2007).
We propose a model in which XPC–RAD23B is the actual damage sensor. After recognition, RAD23B exposes the XPC damage–binding sites by dissociating from it, allowing XPC (without RAD23B) to then stably bind the DNA lesions. Given the intrinsic instability of the XPC protein, it is feasible that RAD23B delivers properly folded XPC to DNA lesions through the initial low-affinity binding of XPC and that the subsequent dissociation of RAD23B forces a more stable binding of XPC to the lesion, which concurrently also stabilizes the protein in the absence of RAD23B. Consequently, damage-bound XPC triggers downstream NER events, leading to successful repair.
Materials and methods
Cell culture
WT, Xpc−/−, and DKO MEFs (Ng et al., 2003) were cultured at 37°C in 5% CO2 in F10/DME culture medium (Invitrogen) supplemented with 15% FCS, 2 mM glutamate, and 50 µg/ml penicillin and streptomycin. NIH2/4 cells containing 256 copies of the LacO sequence stably integrated in their genome were previously described (Soutoglou et al., 2007). For transient expression experiments, medium was changed at least 1 d before the experiment to avoid toxicity of the transfection reagents. To obtain stably expressing mRAD23B-YFP-FLAG clones, cells were selected using 150 µg/ml hygromycin followed by (single cell) FACS sorting. Cells were differentially labeled with latex beads of different size by adding a suspension of beads to the culturing medium. Before the mixing, cells were thoroughly washed to remove free beads (Vermeulen et al., 1991).
Immunofluorescence
Cells were fixed at room temperature for 10 min in 2% PFA followed by a 5-min 0.1% Triton X-100 wash or for 10 min with ice-cold methanol (for RAD23B staining). Cells were washed in PBS containing 0.5% BSA and 0.15% glycine (PBS+). Primary antibody incubation (diluted in PBS+) was performed overnight at 4°C. After extensive washing with PBS+ (four times for 5 min), the cells were incubated with the secondary antibody for 60–90 min at room temperature followed by washing two times with PBS+ and three times with PBS at room temperature. Cells were preserved in VECTASHIELD Mounting Medium (Vector Laboratories) containing DAPI to visualize DNA. The antibodies used were anti-hXPC (rabbit polyclonal), anti-mRAD23B (rabbit polyclonal; a gift from K. Sugasawa, RIKEN, Saitama, Japan), anti-CPD (a gift from O. Nikaido, Kanazawa University, Kanazawa, Japan), anti-p62 (a gift from J.-M. Egly, Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France), and corresponding secondary antibodies coupled with Alexa Fluor 594 and Alexa Fluor 488 or Cy3 (Invitrogen and Jackson ImmunoResearch Laboratories, Inc.).
Fusion protein expression vectors
Both mRAD23A and mRAD23B cDNAs were cloned from mouse livers. mRAD23B was fused to a YFP-FLAG construct and an mCherry construct, and mRAD23A was fused to a GFP2-MYC construct. Mouse RAD23A and RAD23B were cloned in-frame into respective GFP2-MYC or YFP-FLAG mammalian expression vectors (Takara Bio Inc.). FLAG and MYC tags were inserted into the YFP-N3 or GFP2 vectors as double-stranded oligonucleotides into the SsB1–Not1 site. RAD23B was cloned from the RAD23B-YFP-FLAG construct into the mCherry expression vector. The XPC-GFP-HA-His6 construct was previously described (Hoogstraten et al., 2008; Ng et al., 2003). Briefly, full-length human XPC cDNA was cloned in-frame in the eukaryotic expression vector pEGFP-N3 (Takara Bio Inc.). An HA and 6xHis tag were added by insertion of a double-stranded oligonucleotide into the SspBI–NotI site. The LacR gene was fused to XPC-mCherry (Dinant et al., 2007; Soutoglou and Misteli, 2008).
Generation of mRAD23B-YFP-FLAG ES cells and culture
BAC clone RP23-302N23, spanning 200 kb of genomic mouse C57BL/6 sequence including the entire Rad23b locus, was ordered from the BACPAC Resources Center at Children’s Hospital Oakland Research Institute. A YFP-FLAG-LoxP cassette was inserted into the STOP codon of the Rad23b gene using RecA-mediated recombination in Escherichia coli (Imam et al., 2000). A kanamycin/neomycin gene driven by the dual bacterial/mammalian gb2/Pgk promoter and flanked by LoxP sites was inserted in the SacB gene of the BAC vector using λ-red–mediated recombination (Datsenko and Wanner, 2000). Two RAD23B-YFP-FLAG mouse 129 ES clones, F11 and B11, were selected for further experiments based on construct integrity (as analyzed by quantitative PCR, Southern blotting, and FISH) and YFP expression (as measured by flow cytometry). Both had a correct karyotype and only two and three tandem copies of the transgene, respectively, randomly integrated in a single locus in their genome. Mouse 129 ES cells were cultured on Petri dishes coated with 0.1% gelatin at 37°C in 5% CO2 in ES media (40% buffalo rat liver cell–conditioned DME, 40% DME, 15% FCS, 100 U/ml penicillin, 100 mg/ml streptomycin, nonessential amino acids, 1,000 U/ml of leukemia inhibitory factor, and 0.1 mM β-mercaptoethanol).
Protein analysis
Immunoblots for XPC–RAD23B were performed as previously described (Okuda et al., 2004). For immunoprecipitations, cells were washed with PBS and lysed in radioimmunoprecipitation assay (RIPA) plus buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 10% glycerol, protease inhibitors, and 0.1 mM DTT). Extracts were centrifuged, and supernatants were incubated with FLAG beads (M2; Sigma-Aldrich) for 3 h while rotating at 4°C. Beads were washed four times with RIPA plus buffer at 4°C for 10 min. The proteins were eluted from the FLAG beads by incubating the beads in RIPA plus buffer containing 0.2 mg/ml 3xFLAG peptide for 15 min on ice. The eluate was collected after centrifugation at 1,000 rpm for 1 min at 4°C, and the elution procedure was repeated three more times. Eluates were boiled in sample buffer followed by SDS-PAGE and immunoblotting using the indicated antibodies. The subcellular protein fractionation assay was performed using a subcellular protein fractionation kit (78840; Thermo Fisher Scientific). Subsequent immunoprecipitations were performed by adding XPC (rabbit polyclonal) antibodies and agarose beads (Sigma-Aldrich) to the extracts diluted in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 1 mM DTT, and complete protease inhibitors). Beads were collected and washed three times in lysis buffer and once in wash buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.1% NP-40, 0.05% deoxycholate, and complete protease inhibitors). SDS sample buffer was added, and samples were loaded on gradient gels (Invitrogen). Western blots were stained with XPC (ab6264; Abcam) and RAD23B (ab3835; Abcam) antibodies.
Confocal microscopy and photobleaching
Confocal laser-scanning microscopy images were obtained on a laser-scanning microscope (LSM 510 META; Carl Zeiss) equipped with a cell culture microscopy stage (set at 37°C for photobleaching studies and kept at room temperature for immunofluorescence on fixed material). Images were recorded using a 40× lens (NA 1.3; Carl Zeiss). GFP fluorescence imaging was recorded after excitation with a 488-nm argon laser, and emission light was captured behind a 515–540-nm band-pass filter, YFP fluorescence imaging was performed using a 514-nm argon laser, and a 535–560-nm band emission light was filtered by a 560–615-nm band-pass filter. Fluorescent redistribution after photobleaching was performed as previously described (Houtsmuller and Vermeulen, 2001). In brief, a narrow (∼1 µm) strip spanning the width of the nucleus was photobleached for 63 ms at 100% laser intensity. Recovery of fluorescence in the strip was subsequently monitored with 21-ms intervals at 1% laser intensity. Images obtained were analyzed using AIM software (Carl Zeiss). UV laser irradiation was performed as previously described (Dinant et al., 2007), and a 2-mW pulsed (7.8 kHz) diode–pumped solid-state laser emitting at 266 nm (Rapp OptoElectronic) was connected to a confocal microscope (LSM 510) with an Axiovert 200M housing adapted for UV by all-quartz optics. A special adaptor (ZSI-A200; Rapp OptoElectronic) to fit in the aperture slider position of an Axiovert 200 microscope was developed by Rapp OptoElectronic to focus the laser. For local UV-C irradiation experiments, cells were grown on 25-mm-diameter quartz coverslips (010191T-AB; SPI Supplies). Microscopy images were quantified in ImageJ software (National Institutes of Health). iFRAP analysis was performed by bleaching the entire nucleus except the region of interest (the local damage) at full laser power for 4 s, and images were recorded every 15 s.
Online supplemental material
Fig. S1 shows the biological activity of the YFP-tagged RAD23B by complementing the UV hypersensitivity and restoring the low XPC levels to WT levels in RAD23A/RAD23B DKO MEFs and shows that in ES cells, the BAC-derived RAD23B-YFP-FLAG is expressed to the same level as endogenous nontagged RAD23B. Fig. S2 shows that less RAD23B is coimmunoprecipitated with XPC after UV in soluble subcellular protein fractions (from the fractionation assay in Fig. 3 A), that the RAD23B dissociation from XPC is quantitatively confirmed and further corroborated by semiquantitative mass spectrometric analysis, and that the UV–DDB complex (other GG-NER initiation complex) does not dissociate after UV in contrast to XPC–RAD23B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201107050/DC1.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr. O. Nikaido, Dr. J.-M. Egly, and Dr. Sugasawa for kindly providing the anti-CPD antibody, the anti-p62 antibody, and the anti-RAD23B antibody, respectively. The authors thank T. Ammon for discussions and assistance.
This work was supported by the European Union FP6 IP DNA Repair project LSHG-CT-2005-512113; the ZonMW projects 912-03-012, 912-08-031, and 917-46-364; the Netherlands Organisation for Scientific Research project 014-90-001; and the European Science Foundation project ALW-805.47.193. C. Dinant is supported by the European Molecular Biology Organization long-term fellowship ALTF 681-2009. M.S. Luijsterburg is supported by a Netherlands Organisation for Scientific Research Rubicon fellowship, a European Molecular Biology Organization long-term fellowship, and a Federation of European Biochemical Societies long-term fellowship. N.P. Dantuma was supported by the Swedish Cancer Society and the Swedish Research Council. The authors declare that they have no conflict of interest.
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- CPD
- cyclobutane pyrimidine dimer
- DDB
- damaged DNA binding
- DKO
- double knockout
- ES
- embryonic stem
- GG-NER
- global genome NER
- iFRAP
- inverse FRAP
- MEF
- mouse embryonic fibroblast
- NER
- nucleotide excision repair
- RIPA
- radioimmunoprecipitation assay
- WT
- wild type
References
- Alekseev S., Luijsterburg M.S., Pines A., Geverts B., Mari P.O., Giglia-Mari G., Lans H., Houtsmuller A.B., Mullenders L.H., Hoeijmakers J.H., Vermeulen W. 2008. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol. Cell. Biol. 28:7402–7413 10.1128/MCB.01108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki M., Masutani C., Takemura M., Uchida A., Sugasawa K., Kondoh J., Ohkuma Y., Hanaoka F. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276:18665–18672 10.1074/jbc.M100855200 [DOI] [PubMed] [Google Scholar]
- Camenisch U., Träutlein D., Clement F.C., Fei J., Leitenstorfer A., Ferrando-May E., Naegeli H. 2009. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J. 28:2387–2399 10.1038/emboj.2009.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheo D.L., Ruven H.J., Meira L.B., Hammer R.E., Burns D.K., Tappe N.J., van Zeeland A.A., Mullenders L.H., Friedberg E.C. 1997. Characterization of defective nucleotide excision repair in XPC mutant mice. Mutat. Res. 374:1–9 10.1016/S0027-5107(97)00046-8 [DOI] [PubMed] [Google Scholar]
- Cleaver J.E. 2005. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer. 5:564–573 10.1038/nrc1652 [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 97:6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W. 2007. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 120:2731–2740 10.1242/jcs.004523 [DOI] [PubMed] [Google Scholar]
- Dundr M., Misteli T. 2003. Measuring dynamics of nuclear proteins by photobleaching. Curr. Protoc. Cell Biol. Chapter 13:Unit 13.5. [DOI] [PubMed] [Google Scholar]
- Fei J., Kaczmarek N., Luch A., Glas A., Carell T., Naegeli H. 2011. Regulation of nucleotide excision repair by UV-DDB: Prioritization of damage recognition to internucleosomal DNA. PLoS Biol. 9:e1001183 10.1371/journal.pbio.1001183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M., Vermeulen W., van Zeeland A.A., Mullenders L.H. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 23:471–482 10.1016/j.molcel.2006.06.029 [DOI] [PubMed] [Google Scholar]
- Guzder S.N., Sung P., Prakash L., Prakash S. 1998. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J. Biol. Chem. 273:31541–31546 10.1074/jbc.273.47.31541 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 121:2850–2859 (published erratum appears in J. Cell Sci. 2008. 121:3991) 10.1242/jcs.031708 [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Vermeulen W. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 115:13–21 [DOI] [PubMed] [Google Scholar]
- Imam A.M., Patrinos G.P., de Krom M., Bottardi S., Janssens R.J., Katsantoni E., Wai A.W., Sherratt D.J., Grosveld F.G. 2000. Modification of human beta-globin locus PAC clones by homologous recombination in Escherichia coli. Nucleic Acids Res. 28:E65 10.1093/nar/28.12.e65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janićijević A., Sugasawa K., Shimizu Y., Hanaoka F., Wijgers N., Djurica M., Hoeijmakers J.H., Wyman C. 2003. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst.). 2:325–336 10.1016/S1568-7864(02)00222-7 [DOI] [PubMed] [Google Scholar]
- Lommel L., Ortolan T., Chen L., Madura K., Sweder K.S. 2002. Proteolysis of a nucleotide excision repair protein by the 26 S proteasome. Curr. Genet. 42:9–20 10.1007/s00294-002-0332-9 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., von Bornstaedt G., Gourdin A.M., Politi A.Z., Moné M.J., Warmerdam D.O., Goedhart J., Vermeulen W., van Driel R., Höfer T. 2010. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J. Cell Biol. 189:445–463 10.1083/jcb.200909175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Sugasawa K., Yanagisawa J., Sonoyama T., Ui M., Enomoto T., Takio K., Tanaka K., van der Spek P.J., Bootsma D., et al. 1994. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 13:1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.H., Pavletich N.P. 2007. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 449:570–575 10.1038/nature06155 [DOI] [PubMed] [Google Scholar]
- Moné M.J., Volker M., Nikaido O., Mullenders L.H., van Zeeland A.A., Verschure P.J., Manders E.M., van Driel R. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2:1013–1017 10.1093/embo-reports/kve224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J., Volker M., Kool H., Alekseev S., Vrieling H., Yasui A., van Zeeland A.A., Mullenders L.H. 2005. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amst.). 4:571–582 10.1016/j.dnarep.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Moser J., Kool H., Giakzidis I., Caldecott K., Mullenders L.H., Fousteri M.I. 2007. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol. Cell. 27:311–323 10.1016/j.molcel.2007.06.014 [DOI] [PubMed] [Google Scholar]
- Ng J.M., Vermeulen W., van der Horst G.T., Bergink S., Sugasawa K., Vrieling H., Hoeijmakers J.H. 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 17:1630–1645 10.1101/gad.260003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R., Okuda Y., Watanabe E., Mori T., Iwai S., Masutani C., Sugasawa K., Hanaoka F. 2005. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol. Cell. Biol. 25:5664–5674 10.1128/MCB.25.13.5664-5674.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 37:714–727 10.1016/j.molcel.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Okuda Y., Nishi R., Ng J.M., Vermeulen W., van der Horst G.T., Mori T., Hoeijmakers J.H., Hanaoka F., Sugasawa K. 2004. Relative levels of the two mammalian Rad23 homologs determine composition and stability of the xeroderma pigmentosum group C protein complex. DNA Repair (Amst.). 3:1285–1295 10.1016/j.dnarep.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Overmeer R.M., Gourdin A.M., Giglia-Mari A., Kool H., Houtsmuller A.B., Siegal G., Fousteri M.I., Mullenders L.H., Vermeulen W. 2010. Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30:4828–4839 10.1128/MCB.00285-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud E., Miccoli L., Zacal N., Biard D.S., Craescu C.T., Rainbow A.J., Angulo J.F. 2011. Differential contribution of XPC, RAD23A, RAD23B and CENTRIN 2 to the UV-response in human cells. DNA Repair (Amst.). 10:835–847 10.1016/j.dnarep.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Sands A.T., Abuin A., Sanchez A., Conti C.J., Bradley A. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature. 377:162–165 10.1038/377162a0 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E., Dorn J.F., Sengupta K., Jasin M., Nussenzweig A., Ried T., Danuser G., Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9:675–682 10.1038/ncb1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L., Fagbemi A.F., Enzlin J.H., Gourdin A.M., Wijgers N., Dunand-Sauthier I., Giglia-Mari G., Clarkson S.G., Vermeulen W., Schärer O.D. 2009. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 28:1111–1120 10.1038/emboj.2009.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Masutani C., Uchida A., Maekawa T., van der Spek P.J., Bootsma D., Hoeijmakers J.H., Hanaoka F. 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol. Cell. Biol. 16:4852–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., Hanaoka F. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 121:387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Akagi J., Nishi R., Iwai S., Hanaoka F. 2009. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol. Cell. 36:642–653 10.1016/j.molcel.2009.09.035 [DOI] [PubMed] [Google Scholar]
- Uchida A., Sugasawa K., Masutani C., Dohmae N., Araki M., Yokoi M., Ohkuma Y., Hanaoka F. 2002. The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair (Amst.). 1:449–461 10.1016/S1568-7864(02)00031-9 [DOI] [PubMed] [Google Scholar]
- Vermeulen W. 2011. Dynamics of mammalian NER proteins. DNA Repair (Amst.). 10:760–771 10.1016/j.dnarep.2011.04.015 [DOI] [PubMed] [Google Scholar]
- Vermeulen W., Stefanini M., Giliani S., Hoeijmakers J.H., Bootsma D. 1991. Xeroderma pigmentosum complementation group H falls into complementation group D. Mutat. Res. 255:201–208 [DOI] [PubMed] [Google Scholar]
- Watkins J.F., Smerdon M.J. 1985. Nucleosome rearrangement in vitro. 2. Formation of nucleosomes in newly repaired regions of DNA. Biochemistry. 24:7288–7295 10.1021/bi00346a040 [DOI] [PubMed] [Google Scholar]
- Xie Z., Liu S., Zhang Y., Wang Z. 2004. Roles of Rad23 protein in yeast nucleotide excision repair. Nucleic Acids Res. 32:5981–5990 10.1093/nar/gkh934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.