Abstract
Transcript profiling using a zebrafish heart cDNA library previously revealed abundant expressed sequence tags (ESTs) upregulated in zebrafish embryos treated with the aryl hydrocarbon receptor (AHR) agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Here, we identify those ESTs as LTR-containing retroelements termed EZR1 (Expressed-Zebrafish-Retroelement group 1). EZR1 is highly redundant in the genome and includes canonical long terminal repeats (LTRs) flanking an integrase-like open reading frame and a region similar to retroviral envelope protein genes. EZR1 sequences lack reverse transcriptase, RNase H, or protease, indicating retrotransposition would be nonautonomous. No AHR binding motifs were found in the EZR1 promoter region. A putative NF-κB-binding site was found, and TCDD-treated zebrafish embryos had significantly increased levels of nuclear protein(s) binding to this sequence. Protein–EZR1 DNA complex formation was partially competed by a mammalian consensus κB sequence, consistent with NF-κB-like activation contributing to increased protein binding to this site. Mobility of the TCDD-induced protein-EZR1 complex differed from that of authentic NF-κB protein bound to the consensus κB site. The results suggest that EZR1 is regulated by interaction with NF-κB or NF-κB-like protein(s) different from the NF-κB protein binding to the consensus κB site. The nature of the NF-κB-like protein and the relationship between EZR1 induction and cardiovascular toxicity caused by TCDD warrant further investigation.
Introduction
The common environmental contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is highly toxic to many vertebrate species.1 Early developmental stages are particularly susceptible to TCDD toxicity, and prominent effects in embryos are those involving the cardiovascular system.2,3 In an effort to understand the molecular basis of effects of TCDD on the developing cardiovascular system, we carried out gene expression profiling of zebrafish embryos exposed to TCDD.4 That profiling uncovered a prominent expressed sequence tag (EST) cluster, which we referred to as TR004. There were two notable observations regarding the TR004 EST cluster. First, the abundance of TR004 probes on a microarray, fabricated from a zebrafish heart cDNA library, suggested that the corresponding transcripts were highly expressed in cardiac tissue. Second, teratogenic doses of TCDD caused a robust induction of this set of closely related ESTs.4 Here, we characterize the EST sequences and identify the TR004 cluster as comprising a novel unorthodox LTR retroelement, EZR1 (Expressed Zebrafish Retroelement group 1), with broad distribution in the zebrafish genome and similarity to ERV4-DR1 (RepBase).
Retroelements, such as short and long interspersed repeat elements (SINEs and LINEs), retrotransposons, and endogenous retroviruses, are potentially mobile genetic elements that require an RNA intermediate for transposition. Such elements are integral components of eukaryotic genomes, composing as much as ∼40% of total genomic material in mammals.5,6 Amplification of retroelements can have immediate impacts on gene expression (i.e., insertional mutagenesis) and local chromosomal structure, as well as lasting influence on recombination and genome rearrangement events.7 Different classes of retroelements exhibit a variety of retrotransposition mechanisms, amplification rates, and fates in the host genome. Endogenous retroviruses and closely related retroelements comprise a distinct group defined by the presence of flanking long terminal repeats (LTRs) in host genomes. In the zebrafish genome, about 8% of all transposon-related repeats correspond to LTR elements.8
Our initial observations suggested that TCDD can regulate expression of the TR004 cluster. TCDD binds to and activates the aryl hydrocarbon receptor (AHR), and many of its toxic effects are dependent upon this activation.1 TCDD also has been shown to suppress the activation of the pleiotropic transcription factor, NF-κB, through its interaction with the AHR.9,10 On the other hand, TCDD and structurally similar polychlorinated biphenyls also may stimulate NF-κB activation, possibly through an AHR- and CYP1A1-dependent oxidative signal.11–14 Intriguingly, TCDD has been implicated in stimulating retroviral expression, involving both the AHR and NF-κB.15 Thus, we investigated the regulation of EZR1, examining the promoter region of EZR1 for possible regulatory elements, including those possibly involved in the induction by TCDD and those that could be involved in retrotransposon regulation. Analysis of the promoter region of the EZR1 retroelement failed to disclose any AHR response elements; however, a putative NF-κB binding site is present. Results from electrophoretic mobility shift assays (EMSA) support the hypothesis that activation of NF-κB is involved in the TCDD-induced upregulation of EZR1 expression.
Materials and Methods
Zebrafish embryo culture
All fish used in these experiments were from an inbred line of wild-type TL zebrafish (Danio rerio) maintained in the Fishman laboratory facility at Massachusetts General Hospital. To obtain embryos, trios of one male and two female mature fish were held in divided mating tanks overnight. To ensure that all embryos were synchronous to within two cell cycles, embryos were collected within 30 min after removing the barrier the following morning. Embryos were maintained in Tübingen E3 egg water at 28°C.
For expression analyses, 48 and 72 hours post fertilization (hpf), groups of 100 embryos were anesthetized on ice and placed in 1 mL 4% paraformaldehyde for 2–16 h at 4°C. Fixed embryos were rinsed three times with 1 mL methanol and subsequently stored in 1 mL methanol at −20°C. Additional embryos (72 hpf) were flash frozen in liquid nitrogen and stored at −80°C for subsequent RNA isolation. For EMSA analyses, 500 zebrafish embryos each were exposed to vehicle (0.05% DMSO) or 5 nM TCDD for 2 h starting at 2 hpf, and embryos were sampled 72 h later, at 74 hpf. Nuclear extracts were prepared directly following sampling.
5′ RACE and RT-PCR
Total RNA was prepared using TriZol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. cDNA was generated from total RNA using the SMART RACE kit (BD Biosciences Clontech, Mountain View, CA) according to the manufacturer's protocol. 5′ RACE PCR was performed using Advantage 2 Polymerase (BD Biosciences Clontech). For all other PCR, Taq polymerase (Epicentre Biotechnologies, Madison, WI) was used. Gene-specific PCR primer sequences (5′ to 3′) are listed in Table 1.
Table 1.
Gene-Specific Primers
| F1 | CCATGCAACCAGGATAAAACGAGC |
| R1 | GCCTGACAACACAGGATGGACAGG |
| R2 | CAGTCCCAATGTCCATAGCCACTTC |
| R3 | AGGTGCTCGTTTTATCCTGGTTGCATGG |
| R4 | GTTCTGGTTACAGCCACGACATCCGTCC |
DNA sequencing
Contaminating dNTPs and enzymes were removed from aliquots of PCR reactions using QiaQuick PCR Purification spin columns according to standard protocols (Qiagen,Valencia, CA). Purified PCR products were ligated into pGEM-T Easy vector (Promega, Madison, WI). JM109 cells were transformed with the plasmids by the standard heat-shock method and selected on agar containing ampicillin, IPTG, and X-gal (Promega). Plasmid DNA was prepared using QiaPrep Spin Mini-Prep columns according to the manufacturer's protocol (Qiagen). Inserts were amplified by PCR using universal SP6 and T7 primers, and PCR products were purified as above. DNA sequencing reactions were performed by the Massachusetts General Hospital DNA sequencing core facility. All sequenced clones of EZR1 have been deposited in GenBank (Accession Numbers GU133025–GU133046).
Radiation hybrid mapping
Mapping PCRs were performed using 5 μL genomic DNA from the Goodfellow T51 radiation hybrid panel16 and primers F1 and R2 (sequences above). PCR products were visualized by agarose gel electrophoresis and scored as present (1), absent (0), or ambiguous (2).
Sequence analysis
Nucleotide sequence alignments were produced using Muscle v3.7, with default settings.17 RAxML (v7.2.6) was used to perform a maximum likelihood analysis (GTRMIX model with GTR+gamma analysis of final tree18). Bootstrap confidence values were calculated from 100 replicates. This result was compared to the most probable tree morphology found by 100,000 generations (trees sampled every 100 generations) of an incrementally heated Metropolis-coupled Monte Carlo Markov Chain analysis (MrBayes v3.1, 4-by-4 nucleotide substitution model with rate variation according to determined gamma distribution).
Transcription factor binding motifs were identified by using MatInspector v2.2 to search the TransFac 4.0 database.19 EZR1 sequences were analyzed using GCG/Seqlab (Accelrys, San Diego, CA) and with LTR-FINDER using default settings.20 Zebrafish genomic sequence data used herein were produced by the Zebrafish Sequencing Group at the Sanger Institute and can be obtained freely from ftp://ftp.sanger.ac.uk/pub/zebrafish/
Electrophoretic mobility shift assay
Nuclear extracts were prepared as follows. Embryos were added to a Dounce homogenizer and washed with 5 mL of ice-cold PBS. After removing the PBS, 5 mL of buffer A [10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF] that contained 0.6% NP-40 was added. Embryos were homogenized with ten strokes of pestle A. Homogenates were spun at 500 g in for 30 sec to remove unbroken tissue. Nuclei were pelleted by centrifuging for 5 min at 3,000 g, washed in buffer A, lysed by incubating in buffer C [20 mM HEPES pH 7.9, 25%(v/v) glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF] for 20 min on ice, and centrifuged for 5 min at 14,000 g. Supernatants were collected and stored at −80°C. Extracts from untreated WEHI-231 cells were prepared as described previously.21
EMSA was carried out using a digoxygenin (DIG) gel shift kit (Roche Diagnostic Corporation, Indianapolis, IN) according to the manufacturer's instructions. Briefly, oligonucleotides containing the mammalian consensus binding sequence for NF-κB (κB: 5′ -AGTTGAGGGGACTTTCCCAGGC-3′) and the EZR1-κB site (EZR1: 5′-CACGCTGGAAAATTCCAC-3′) (κB sites are underlined) were DIG-labeled. The DNA probe was incubated with 15–20 μg (zebrafish) or 0.5 μg (WEHI-231) of nuclear extract for 20 min at RT. Protein–DNA complexes were separated on a 5% native polyacrylamide gel run in 0.5X TBE buffer and electrophoretically transferred to a nylon membrane (Roche) for chemiluminescent band detection. In competition assays, nuclear extracts were incubated with unlabeled oligonucleotide pairs at 100–500 times molar excess for 10 min at RT prior to addition of the DNA probe (see figure legends for sequences).
Results
Characterization of TR004 transcripts
Sequencing of the ESTs showed the TR004 cluster to consist of 19 nonidentical clones, ranging in length from <250 bp to nearly 2 kb. Aligning these transcript fragments yields a single assembly of 1915 bp in length, terminating at a common poly-A tail. Although having obvious sequence similarity, this alignment revealed a large number of single nucleotide mismatches and ambiguities, as well as a region of ∼500 bp near the 3′ end that contained several sites of distinct sequence motifs and significant insertions/deletions. A combination of RT-PCR and 5′RACE was used to obtain at least two-fold coverage of a putative full-length transcript. All sequences were assembled into a single contig of 4312 bp (Fig. 1).
FIG. 1.
Schematic illustration of EZR1 sequence assembly (top), including TR004 ESTs (black), PCR (light gray), and 5′ RACE (dark gray) products, and the resultant consensus sequence (bottom). Gaps in sequence coverage are indicated by dotted lines. Regions of concentrated sequence variability (pale gray), the integrase ORF (Int), and a noncoding region of homology to retroviral env genes (stippled) are depicted on the consensus sequence schematic. Flanking direct repeats are indicated by black triangles, with sequences inset. Color images available online at www.liebertpub.com/zeb
No two clones in this assembly were identical, and 9.8% of the consensus sequence consisted of ambiguities. The primary sources of variation were two regions of 80 bp, with >50% polymorphism rate and significant insertions/deletions in some subset of clones. Outside these variable regions, the mismatch rate was approximately 1/39 nucleotides, more than twice the polymorphism rate expected in zebrafish coding sequence based on allelic variation.22,23 The 5′ half of the assembly appeared to be more highly conserved than the 3′ half, likely due to lesser sequence coverage (i.e., 2 clones versus 23).
The consensus sequence of the complete assembly was found to contain a single open reading frame spanning positions 1136 bp to 2518 bp, with a conserved poly-adenylation signal (AATAAA) at position 2521 bp (Fig. 1). The 462 aa protein sequence putatively encoded by this open reading frame was highly similar to retroviral pol polyproteins. Domain searches revealed a complete integrase core catalytic domain but no significant similarity to reverse transcriptase or other typical pol constituents. To reflect this fact, this open reading frame was designated int, rather than pol. Due to a lack of an identifiable RT, EZR1 was not detected in a previous survey of zebrafish (and other fish) retroid agents.24
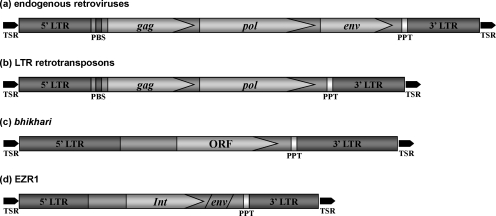
The region downstream of the int ORF had moderate (e-values≤4×10−4) amino acid similarity to retroviral env sequences. However, no open reading frame was detected in this region when either the TR004 consensus sequence or individual clones were interrogated. No similarity to known gag genes was detected. Based on the similarity between the sequence and gene order of TR004 and those of endogenous retroviruses (Fig. 2), the described transcripts were collectively renamed EZR1, for Expressed Zebrafish Retroelement group 1.
FIG. 2.
Schematic depiction of canonical domain structures for vertebrate endogenous retroviruses (a), LTR retrotransposons (b), zebrafish bhikhari (c), and EZR1 (d). Arrows indicate protein coding regions. LTR, long terminal repeat; ORF, open reading frame; PBS, primer binding site; PPT, poly-purine tract; TSR, target site repeat (duplication); and env, envelope protein gene; gag, group antigen gene; int, integrase gene; pol, poly-protein gene.
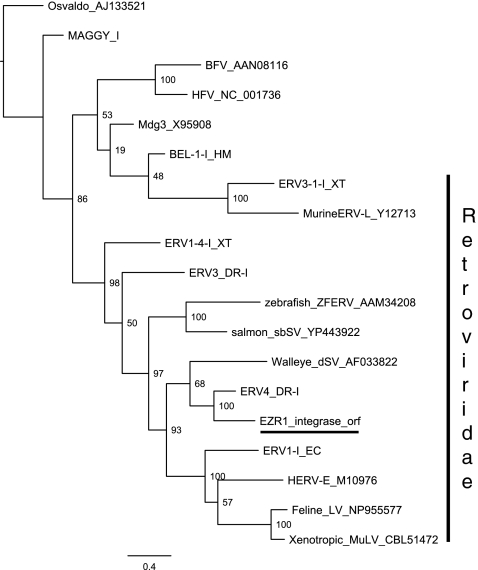
Phylogenetic analysis of the amino acid sequence of the putative EZR1 int sequence with other retroviral int proteins (Fig. 3) shows that EZR1 clusters with ERV4-DR1 (from RepBase), and appears to be related to the walleye (Stizostedion vitreum) dermal sarcoma virus described by Martineau et al.25–28 EZR1 integrase is less similar to the zebrafish endogenous retrovirus (ZFERV, ZFERV-2-I_DR) integrase described by Shen and Steiner29,30 (23% amino acid identity), but in general appears to be related to the murine leukemia virus (MVL) and related viruses (Class I Retroviridae).29
FIG. 3.
Phylogenetic analysis of the putative amino acid sequence of the EZR1 integrase sequence with other retroviral int proteins. The putative EZR1 integrase clusters with ERV4-DR1 and appears to be related to the walleye (Stizostedion vitreum) dermal sarcoma virus and other Retroviridae.
LTR structure of EZR1
The 5′ and 3′ termini of the EZR1 sequence assembly were found to constitute a 27 bp identical direct repeat, suggesting the presence of long terminal repeats (Fig. 1). Searching the zebrafish genome revealed several genomic sequences with regions of ≥98% identity to the final 185 bp of the EZR1 transcripts, presumed to be an LTR sequence. Dot-plot comparison of the complete EZR1 consensus sequence to a single genomic sequence contig revealed two direct repeats of a sequence composed of ∼500 bp from the 3′ end of the EZR1 transcripts, followed by a region of similarity to the first ∼50 bp of EZR1 transcripts (Supplementary Fig. S1; Supplementary Material is available online at www.liebertonline.com/zeb).
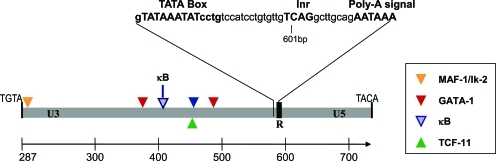
The putative LTRs are marked by the canonical tetranucleotide sequence TACA at the 3′ end, and the 5′-most tetranucleotides TGAG (inverted repeat of the final tetranucleotide) within the region conserved among all cDNA clones. This designation is supported by the presence of an 11 bp poly-purine tract immediately upstream of this location. We observed a diversity of 5′-most tetranucleotides in the genomic copies of EZR1. The 5′-most tetranucleotide was TGTA in 18 of 48 cases (37%) and TGAG in 20 cases (42%); other major observed patterns include TGAC (12%). Notably, the 5′-most tetranucleotide observed in all sequences in the TCDD-inducible clade was TGAG. Assuming these boundaries yields a putative LTR of ∼437 bp in length (alignment length of 724 bp; see Supplementary Fig. S2), containing U3, R, and U5 domains. Genomic copies of the EZR1 LTR ranged in length from 104 to 999, but averaged 437 bp. The 5′ boundary of the R domain is defined by a consensus transcription initiation sequence (TCAG) at LTR consensus position 614 bp, directly abutting the observed 5′ mRNA terminus at 624 bp (Fig. 4). Analysis of the genomic copies of EZR1 with LTR-Finder revealed that the 3′ end of the LTR containing the U5 region is a further 120 bp downstream, at consensus position 726. A canonical TATA box (overall 96% identity to 15 bp motif matrix) is 28 bp upstream of the putative initiation site, and a consensus poly-adenylation signal is at consensus positions 626–631, 15 bp upstream of cloned poly-A tails (Fig. 4). In addition, a strong GT-rich retroviral polyadenylation downstream sequence (PADS) element (matrix similarity score of 91.2%) is located in the putative U3 region at positions 343–357. Thus, all data supported the existence of a typical (32 bp) central R domain, flanked by U3 and U5 regions of ∼300 bp and 79 bp, respectively (Fig. 4; see also Supplementary Fig. S2).
FIG. 4.
Schematic illustration of the putative EZR1 LTR structure, showing U3, R, and U5 domain boundaries, and part of the R domain sequence (inset). Triangles depict locations of predicted binding sites for GATA-1 (black), MAF-1/Ik-2 (light gray), TCF-11 (gray), and the EZR1-κB site (light gray outlined). Triangles above the gray bar indicate motifs on the plus strand, those below represent motifs on the minus strand. The alignment positions refer to the complete alignment of sequenced clones and genomic copies, in the Supplementary Data. Color images available online at www.liebertpub.com/zeb
Tissue expression of EZR1
EZR1 transcripts were found to be highly abundant in cDNA libraries from wild-type adult heart tissue, regardless of genetic strain or originating facility. EZR1 clones comprised approximately 0.4% (19 of 4896) of adult heart cDNAs randomly selected for use in constructing the cDNA microarrays with which EZR1 was identified.4 Similarly, a single UniGene cluster composed of ESTs that were greater than 90% identical to the EZR1 int region accounted for 1.6% and 2.2% of sequences in two independent adult heart cDNA libraries (UniGene Libraries Lib.19739 and Lib.19740).
EZR1-like ESTs also were detected in cDNA libraries from a variety of tissues and developmental stages. BLAST searching the zebrafish EST database at GenBank revealed over 750 matches with e-value=0.0 (August 2011). These ESTs were drawn from tissues including heart, brain, liver, kidney, ovary, and testis, eye, olfactory rosettes, fin, and whole embryos ranging from shield stage (6 hpf) to 5 days post fertilization.
Genomic distribution of EZR1
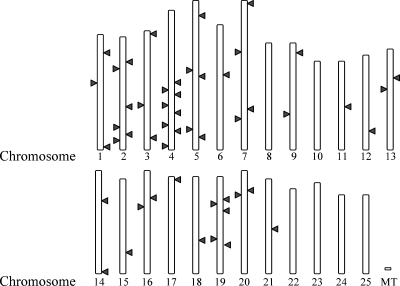
The excessive polymorphism rate and putative identification as a retroelement strongly suggested the possibility of multiple genomic copies of EZR1. Radiation hybrid mapping (T51 panel; data not shown) using PCR primers targeted to the int ORF region suggests that most linkage groups carry at least one EZR1 copy. Likewise, searching the zebrafish genome (Zv9, Ensembl release 62) with a profile hidden Markov model (HMM) derived from the alignment of putative LTR sequences revealed nearly exact copies of the complete EZR1 LTR on every chromosome (e-values<5×10−74). More than 150 LTR copies, often incomplete, were found, (e-value range 3.7×10−308–1.2×10−7).
Blast searches with the full-length (4.35 kb) EZR1 consensus sequence yielded a more conservative estimate of genomic copy number. These searches identified at least 50 unique insertions sites 4.1 kb in length or longer distributed over 19 chromosomes and 8 other unassembled regions (Fig. 5). Four copies had additional insertions ranging from 395 bp to 5.5 kb, giving one copy a total length of 11.5 kb. This long copy (ENSDARG00000086238; CR847511.1-201) located on Chromosome 4 is unique in that it contains an open reading frame encoding a putative reverse transcriptase and RNaseH domain (pol protein) but no endonuclease. The formation of this long unique copy is possibly due to the insertion of a non-LTR transposon (I-2_DR-like) into a copy of EZR1. The putative I-2_DR-like pol protein exhibits high identity to pol proteins in insects, bloodfluke, and trypanosomes, and is also present in hundreds of copies in the zebrafish genome. A second long EZR1-like copy on Chromosome 5 (3,724,204–3,734,203) appears to contain full protease/reverse transcriptase/ RNaseH /integrase (PR/RT/RH/IN) domains, with near identity between a portion of the integrase orf and the EZR1 int. However, this copy has a low similarity to EZR1 LTR or other regions, suggesting either rapid LTR evolution or recombination to product EZR1. Thus, EZR1 may be transcriptionally active but currently immobile, and the copy which was responsible for its transposition may have been recently deleted from the genome.
FIG. 5.
Distribution of 51 EZR1 BLAST hits over the zebrafish genome (Zv9; Ensembl 62). Only BLAST hits with e-values <1e-75 were retained. Note that some EZR1 sites are not currently assembled to a chromosome. The longest EZR1 genomic copy is located on Zv9 Chromosome 4 (ENSDARG00000086238).
Primer binding sites adjacent to the LTR were identified in the 48 genomic copies of EZR1 that were analyzed. Thirty-six of these PBS regions had tRNA-Pro, nine had tRNA-Asn and others had tRNA-Arg and -Tyr (2 and 1, respectively.) All TCDD-inducible copies of EZR1 had tRNA-Pro. In addition, 5 bp Target Site Repeats (also known as target site duplications) occurred in 70% of the genomic copies of EZR1.
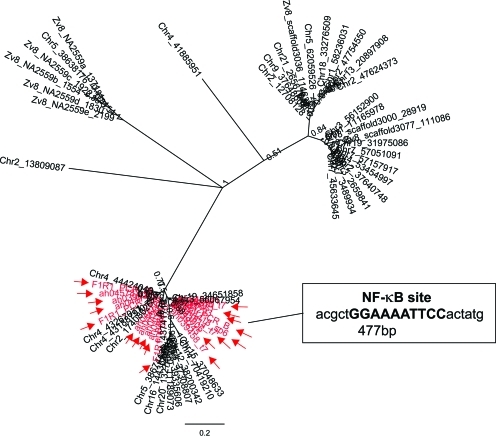
EZR1 copies can be divided into two distinct classes based on their LTR sequences. LTR positions 316 bp to 606 bp were used for phylogenetic analyses, as sequence information for this region was available for all cDNA clones (excepting RACE products). Both the Bayesian most probable tree and the maximum likelihood 50% consensus tree placed the majority of EZR1 transcripts into two primary clades (Fig. 6 and Supplementary Fig. S3). Resolution within clades is extremely poor and there were several minor discrepancies between results generated by the two phylogenetic methods. Notably, however, all of the ESTs induced by TCDD consistently fall into a single clade, along with the 11.5kb I-2_DR-containing genomic copy and 15 of the 34 deletion derivatives (Fig. 6) This clade is distinguished by a 125 bp insertion in the U3 portion of the LTR; this region is completely absent from all members of the other clade.
FIG. 6.
Consensus tree morphology for EZR1 LTR sequences, as determined by Bayesian inference of phylogeny. Nodes with >50% support are labeled with Bayesian posterior probabilities. Sequences in gray indicated with arrows are cloned TCDD-inducible sequences. Inset: EZR1-κB binding site. Color images available online at www.liebertpub.com/zeb
Putative regulatory elements in the LTR
We searched the TransFac 4.0 database of known binding motifs19 to identify potential transcription factor binding sites in the EZR1 LTR sequences. Matches were accepted only if all copies of the LTR sequence contained an absolutely conserved core sequence within a motif ≥85% identical to the complete corresponding weight matrix. Most predicted transcription factor binding sites were located in the putative U3 region; 6 matches were discarded based on coincidence with either the TATA box or transcription initiation site, and no predicted sites fell within the U5 region. In all, 18 binding sites for 14 individual transcription factors were predicted (Fig. 4 and Supplementary Table S1).
Despite being TCDD-inducible, no AHR/ARNT binding sites, nor dioxin response elements (DREs), were found in the putative LTR sequences. Vogel et al.10,31 recently identified a RelB-AHRE, by which gene expression was altered independent of ARNT and the traditional AHRE. We did not find RelB-AHRE binding sequences in the LTRs. Binding sites for hematopoietic transcription factors, particularly GATA-1, accounted for a significant fraction of all matches. Of particular note, two strong (>94% matrix similarity) GATA-1 binding sites were predicted. These sites also were identified as matching recognition sites for LMO-2/GATA-1 complexes. However, similarity was restricted to the GATA-1 half of the motif, with no corresponding similarity to LMO-2 specific sequences. A predicted BRN-2 binding site overlapped the GATA-1 site at position 483 bp over two-thirds of its length; the significance (if any) of this finding is unknown. Similarly, the region beginning at 278 bp might be predicted to interact with either Ikaros-2 or MZF-1, as the predicted Ik-2 binding sequence is entirely encompassed by the MZF-1 motif. Finally, a well-conserved (92.5% matrix similarity) serum response factor recognition site was identified at position 534 bp.
Binding motifs for activator proteins AP-4 also were conspicuous. Most notably, though, the LTR region unique to members of the TCDD-inducible clade was found to contain a single potential NF-κB p65/cRel binding motif, GGAAAaTtCC. Capital letters indicate those nucleotides that match a motif known to bind zebrafish NF-κB.
TCDD-stimulated nuclear protein binding to EZR1 promoter element
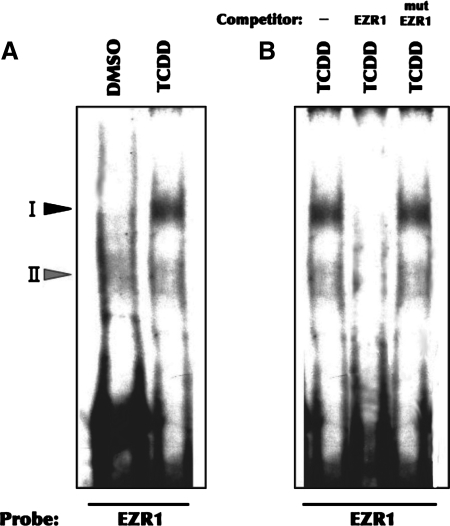
EMSA analysis was used to examine the potential contribution of NF-κB-DNA binding to regulation of EZR1 expression by TCDD. Nuclear proteins isolated from vehicle (DMSO)-treated zebrafish embryos exhibited minimal binding to the putative NF-κB binding site in the EZR1 promoter (EZR1-κB), forming two weak bands: a slower and a faster migrating band, designated complexes I and II, respectively (Fig. 7A). Nuclear proteins from TCDD-treated zebrafish embryos also bound to the EZR1-κB site, with a significant increase in binding occurring specifically in complex I (Fig. 7A). The specificity of the observed bands was demonstrated by complete competition by a 100-fold excess of cold probe and lack of competition by cold probe that had been mutated at a single site (Fig. 7B). These data are consistent with TCDD upregulating EZR1 transcription by stimulating NF-κB binding.
FIG. 7.
TCDD activates the binding of nuclear protein to EZR1-κB site. Nuclear extracts were prepared from 74-hpf zebrafish embryos exposed to DMSO (vehicle) or 5 nM TCDD for 2 h starting at 2-hpf. (A) EMSA analysis of EZR1-κB (5′-CACGCTGGAAAATTCCAC-3) binding; (B) Competition analyses were performed by including a 100x excess of unlabeled EZR1-κB or mutant EZR1-κB (5′-CACGCTGGAAAATTGCACTATG-3′) to verify the specificity of the complexes. The positions of protein–DNA complexes are indicated by arrowheads. Data are representative of three experiments.
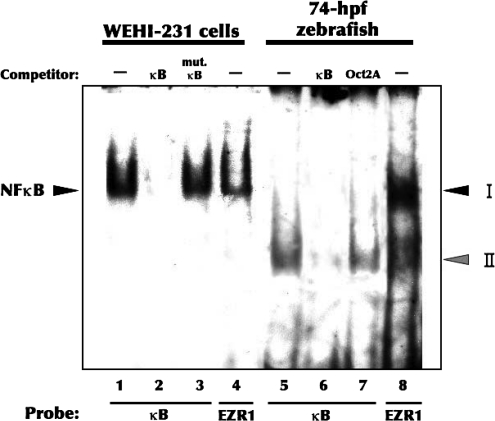
Binding of nuclear proteins from zebrafish embryos to a known κB consensus sequence and to the EZR1-κB-like site was compared to binding of proteins of nuclear extracts from WEHI-231 cells. WEHI-231 is a mouse lymphoma cell line with high levels of constitutively active NF-κB. Nuclear proteins from untreated WEHI-231 cells formed one complex with the EZR1-κB site, which migrated in a similar fashion to the complex formed with the mammalian consensus κB site (Fig. 7A). Interestingly, the zebrafish EZR1 complex I that was upregulated by TCDD had a mobility different from that of the complex resulting from binding to the consensus κB site (Fig. 8B). Unlabeled EZR1-κB site sequence competed with the labeled consensus κB for binding of nuclear proteins. Likewise, increasing amounts (250- and 500-fold molar excess) of unlabeled consensus κB site also partially competed with the labeled EZR1-κB for nuclear proteins, although to a lesser extent than competition by the unlabeled EZR1-κB-like sequence. These studies suggest that the EZR1 is regulated by interaction with NF-κB (or NF-κB-like) proteins that are different from the NF-κB proteins that bind to the consensus κB site.
FIG. 8.
Binding of murine nuclear proteins to the EZR1-κB site. Nuclear protein extracts were prepared from untreated WEHI-231 cells, and from zebrafish embryos treated with 5 nM TCDD for 2 h starting at 2 hpf. EMSA analysis was performed using a consensus NF-κB site (5′ -AGTTGAGGGGACTTTCCCAGGC-3′) and the EZR1-κB site. In competition analyses, unlabeled double-stranded κB (Lanes 2 and 6), single-mutated κB (mutant NF-κB; 5 -AGTTGAGGGCACTTTCCCAGGC-3) (Lane 3), or Oct2A (Oct2A; 5'-GTACGGAGTATCCAGCTCCGTAGCATGCAAATCCTGG-3') (Lane 7) oligonucleotides were included at 200-fold molar excess to verify the identity of shift complexes. The positions of protein–DNA complexes are indicated by arrowheads.
Discussion
A novel class of nonautonomous LTR retroelements
In these studies, we analyzed a family of expressed sequences originally observed among transcripts upregulated in zebrafish embryos by TCDD, and determined that these sequences collectively constitute a novel LTR-class retroelement, which we term EZR1. EZR1 transcribed sequences begin and end with direct repeat sequences, which can be recombined to generate a putative LTR sequence with all features of canonical LTRs—flanking inverted tetranucleotide repeats, a central R domain with strong initiation and poly-adenylation signals, and a U3 region rich in putative regulatory elements. EZR1 also contains other retroelement-specific sequence motifs, including a retroviral PADS element in the U3 region and a poly-purine tract immediately upstream of the 3′ LTR.
The internal sequence and structure of EZR1 also bears a resemblance to retroviruses and derived retroelements. The single EZR1 open reading frame appears to encode a retroviral-type integrase with a conserved catalytic domain that suggests the possibility of an active enzyme, but this requires confirmation. The region downstream of the int ORF lacks open reading frames, yet the translated sequence is highly similar to retroviral env proteins. This suggests the presence, at some point in the past, of an env gene that has been degraded by subsequent mutation. In vertebrate retroviruses, as well as Ty3/gypsy and BEL retrotransposons, integrase is the final domain of the pol gene. Env genes are generally absent from LTR retroelements, but are found downstream of the pol gene in retroviruses.32,33 Thus, the apparent int-env organization of EZR1 is strongly reminiscent of the 3′ half of retroviral genomes (Fig. 2).
In contrast to the above, EZR1 lacks elements common to the 5′ half of retroviral genomes. There is no evidence in EZR1 sequences of a gag gene, or of the reverse transcriptase, RNase H, or protease domains of pol. As the current scheme of LTR retroelement classification is based primarily on aspects of the pol gene, EZR1 cannot be fit into any existing LTR retroelement class.32,33 Furthermore, without gag and pol, EZR1 lacks the means to generate reverse transcription machinery necessary for autonomous replication and retrotransposition. In this regard, EZR1 is similar to another zebrafish retroelement, bhikhari.34 However, bik appears to be even more remotely related to other retroelements, as it contains a single ORF encoding a protein with no significant homology to any known proteins.
EZR1 and bik are distinct from retroelement pseudogenes, the only other known nonautonomous LTR retroelements. Most pseudogenes differ from active relatives by single nucleotide mutations or modest deletions or rearrangements. Furthermore, retroelement pseudogenes generally are not replicated, and thus, are found at very low copy numbers.7 In contrast, EZR1 and bik appear to represent independent, replicating lineages. Their internal gene content is vastly different from any autonomous retroelements, and both are represented by 25–100 copies per genome,34 suggesting amplification in the absence of autonomous retrotransposition capability. The paired (5′/3′) LTR identity of the EZR1 genomic copies averages 97% (range of 85%–100% identity). Assuming the paired LTR regions experience independent neutral evolution, this high identity suggests that the separate LTR ends of the identified copies have not had time to diverge from each other, and therefore that EZR1 is not ancient. Thus, EZR1 and bik seem to constitute a novel class of nonautonomous LTR retroelements. The broad distribution within the genome implies that retrotransposition has been common. Whether these elements are currently active (i.e., transposing) is unknown.
Regulation of EZR1 expression
The evidence indicates that complete 4.35 kb retroelement transcripts are expressed under control of the EZR1 LTR. Experimental data and in silico predictions were in absolute agreement regarding the site of transcriptional initiation. Similarly, all EZR1 ESTs terminated at a single poly-adenylation site that was strongly supported by the presence of a retroviral PADS element upstream of the conserved poly-adenylation signal. Thus, while EZR1 transcripts likely originate from numerous loci in the zebrafish genome, all appear to conform to the canonical domain structure R-U5-internal-U3-R. Furthermore, numerous nonidentical transcripts, presumably originating from multiple loci, exhibited similar patterns of expression. These similarities argue that EZR1 expression is driven by common LTR sequences, rather than regulatory elements in a specific genomic context.
Putative regulatory elements might account for many aspects of the observed expression patterns. In situ hybridization indicates that EZR1 is expressed at low to moderate levels throughout zebrafish embryos. While the transcription initiation site and upstream TATA box both conform to expectations for a strong initiation site, no CCAAT box was found. Still, moderate basal expression might be accomplished, even without additional enhancers. TCF-11 is a ubiquitous transcriptional enhancer that might also contribute to general expression via the binding site predicted in the EZR1 U3 region.
The high levels of cardiac-specific expression inferred from EZR1-like EST abundance in adult heart cDNA libraries may be attributed to a combination of cardiac- and blood-specific transcription factors. We identified a moderately conserved Nkx-2.5 binding site. While Nkx-2.5 is a cardiac-specific homeobox transcription factor of known importance in heart development and function in zebrafish,35,36 it seems unlikely that Nkx-2.5 activity at a single site could drive EZR1 expression to levels of 0.5%–2.5% of all cardiac transcripts. Based on the number of hematopoietic regulatory elements detected (i.e., GATA-1, MZF-1, Ikaros-2), it seems likely that EZR1 is also expressed in hematopoietic lineages. Whether EZR1 expression would be restricted to certain blood cell-types is uncertain, as GATA-1, MZF-1 and Ikaros-2 are active primarily in erythroid, myeloid, and lymphoid lineages, respectively. Blood trapped in dissected hearts likely contributed to cardiac cDNA libraries, but blood cell-specific expression alone is not likely to account for observed EZR1 EST quantities. Certainly, no other blood cell-specific genes were found at remotely comparable levels in these libraries. The most likely explanation for the extreme abundance of EZR1 transcripts in cardiac cDNA libraries is a combination of multiple transcription factors driving expression from numerous genomic copies of EZR1.
EZR1 induction by TCDD
The initial discovery of EZR1 was based on its transcriptional induction by TCDD, and finding a possible mechanism for this induction was an impetus for the current work. Most TCDD-induced toxic effects and effects on gene expression are mediated by the AHR.1,37 However, the absence of DREs in the LTR suggests that TCDD is not acting directly by activating AHR/ARNT binding to the EZR1 promoter. Accordingly, the EZR1 promoter sequence was examined further for binding sites of other transcription factors.
A putative NF-κB binding site was identified in the EZR1 promoter region, and EMSA analyses support the conclusion that zebrafish Rel family protein homologs bind to this site. Zebrafish express at least three NF-κB proteins (p65, c-Rel, and p100).38 However, the lack of competition of EZR1 binding by a mammalian consensus κB sequence suggests that the EZR1 site is not a canonical NF-κB site. This is not necessarily surprising, as small changes in binding site sequence can dramatically affect NF-κB binding.39
We hypothesized that the EZR1-κB site was a target for TCDD-induced activation, as TCDD and structurally similar polychlorinated biphenyls have been shown to stimulate NF-κB activation.11,12,14,40 Indeed, nuclear protein binding to the EZR1-κB site was enhanced significantly in TCDD-treated embryos. Even though the AHR does not bind directly to the EZR1 promoter, it could be involved indirectly in the induction of EZR1 expression by TCDD. CYP1A is highly induced in TCDD-treated zebrafish embryos.4 Access of planar halogenated aromatic hydrocarbons to the active site of CYP1As is restricted,41 resulting in uncoupling of oxidative metabolism and production of reactive oxygen species (ROS).42,43 Interaction of TCDD with the highly induced CYP1A in zebrafish embryos likely results in production of ROS, important effector molecules in the activation of NF-κB.44 TCDD-induced embryotoxicity has been shown to be suppressed by co-treatment with an antioxidant.45,46 Moreover, TCDD-induced activation of binding of NF-κB in the LTR of HIV occurs through an AHR- and CYP1A1-dependent oxidative signal.15 Thus, TCDD-induced, AHR-dependent ROS production, whether via a CYP1, COX2, or some other path, is a potential mechanism for both EZR1 activation and cardiovascular toxicity, although not directly investigated here.47 This is particularly intriguing in light of findings that induction of a novel LINE-1retrotransposon (L1Md-A2) in mouse vascular smooth muscle cells by benzo[a]pyrene also involves a redox-sensitive mechanism.48
It is conceivable that glucocorticoid receptor (GR) and/or the AP-1 complex might contribute to TCDD induction of EZR1 expression. GR has been implicated in TCDD-responsiveness of sequences under the control of the murine mammary tumor virus LTR.49 Cross-talk between AHR and GR signaling also has been observed in other systems.50 Thus, the glucocorticoid response element in the EZR1 LTR could be a target for indirect effects of TCDD. Likewise, TCDD has been shown to enhance both expression of constituent proteins and AP-1 DNA binding activity under some conditions.40,51 Thus, the potential AP-1 binding sites in the EZR1 LTR might be involved in the observed increase in EZR1 transcript levels
Biological implications of EZR1 expression
Retroelements make up nearly 40% of some vertebrate genomes and can influence gene expression and genome rearrangement. EZR1 is abundantly expressed in cardiac tissue, possibly in response to specific LTR elements, but the implications of this specific retroelement expression independent of transposition are matters for speculation. The repercussions of EZR1 induction by TCDD are, likewise, difficult to predict. There is evidence to indicate that diverse mobile elements are activated by a variety of stimuli, including chemicals similar to TCDD.48,52 Particularly interesting is the correlation between elevated levels of certain endogenous retroviral transcripts in myocardium and cardiovascular disease in rats,53,54 and the suggestion that L1Md may be involved in atherogenesis.55 EZR1 transcript abundances were dose-dependently increased by doses of TCDD that cause cardiovascular toxicity and disrupted cardiomyocyte gene expression.4 Regardless, the evidence indicates that TCDD induction of EZR1 involves activation of a zebrafish NF-κB-like factor. The direct stimulus for this activation, whether a change in redox status involving uncoupling of CYP1A or some other AHR-dependant path to oxidative stress, or a direct involvement of AHR with a κB component is not clear. This mechanism and the connection between EZR1 expression and cardiac malfunction in zebrafish warrant further investigation.
Supplementary Material
Acknowledgments
This work was supported in by the National Institutes of Health [Grant Number R01ES015912 to JJStegeman, P42ES007381 [Superfund Research Program at Boston University, JJStegeman and JJSchlezinger]. We would also like to acknowledge the detailed comments of an anonymous reviewer. The sponsors had no involvement in performing or in the decision to publish this study. The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon.
Disclosure Statement
No competing financial interests exist.
References
- 1.Fernandez-Salguero PM. Hilbert DM. Rudikoff S. Ward JM. Gonzalez FJ. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol App Pharmacol. 1996;140:173–179. doi: 10.1006/taap.1996.0210. [DOI] [PubMed] [Google Scholar]
- 2.Carney SA. Chen J. Burns CG. Xiong KM. Peterson RE. Heideman W. Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol Pharmacol. 2006;70:549–561. doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- 3.Henry TR. Spitsbergen JM. Hornung MW. Abnet CC. Peterson RE. Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio) Toxicol App Pharmacol. 1997;142:56–68. doi: 10.1006/taap.1996.8024. [DOI] [PubMed] [Google Scholar]
- 4.Handley-Goldstone HM. Grow MW. Stegeman JJ. Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol Sci. 2005;85:683–693. doi: 10.1093/toxsci/kfi116. [DOI] [PubMed] [Google Scholar]
- 5.Lander ES. Linton LM. Birren B. Nusbaum C. Zody MC. Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.Waterston RH. Lindblad-Toh K. Birney E. Rogers J. Abril JF. Agarwal P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 7.Deininger PL. Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 8.Houwing S. Kamminga LM. Berezikov E. Cronembold D. Girard A. van den Elst H, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y. Ah receptor and NF-kappaB interplay on the stage of epigenome. Biochem Pharmacol. 2009;77:670–680. doi: 10.1016/j.bcp.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 10.Vogel CF. Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho IA. Singh N. Hegde VL. Nagarkatti M. Nagarkatti PS. Treatment of mice with 2,3,7,8-tetrachlorodibenzo-p-dioxin leads to aryl hydrocarbon receptor-dependent nuclear translocation of NF-kappaB and expression of Fas ligand in thymic stromal cells and consequent apoptosis in T cells. J Immunol. 2005;175:90–103. doi: 10.4049/jimmunol.175.1.90. [DOI] [PubMed] [Google Scholar]
- 12.Hennig B. Meerarani P. Slim R. Toborek M. Daugherty A. Silverstone AE, et al. Proinflammatory properties of coplanar PCBs: In vitro and in vivo evidence. Toxicol App Pharmacol. 2002;181:174–183. doi: 10.1006/taap.2002.9408. [DOI] [PubMed] [Google Scholar]
- 13.Puga A. Barnes SJ. Dalton TP. Chang C. Knudsen ES. Maier MA. Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J Biol Chem. 2000;275:2943–2950. doi: 10.1074/jbc.275.4.2943. [DOI] [PubMed] [Google Scholar]
- 14.Schlezinger JJ. Blickarz CE. Mann KK. Doerre S. Stegeman JJ. Identification of NF-kappaB in the marine fish Stenotomus chrysops and examination of its activation by aryl hydrocarbon receptor agonists. Chem Biol Interact. 2000;126:137–157. doi: 10.1016/s0009-2797(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 15.Yao Y. Hoffer A. Chang CY. Puga A. Dioxin activates HIV-1 gene expression by an oxidative stress pathway requiring a functional cytochrome P450 CYP1A1 enzyme. Environ Health Perspect. 1995;103:366–71. doi: 10.1289/ehp.95103366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisler R. Rauch GJ. Baier H. van Bebber F. Bross L. Dekens MP, et al. A radiation hybrid map of the zebrafish genome. Nature Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- 17.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 19.Quandt K. Frech K. Karas H. Wingender E. Werner T. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z. Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlezinger JJ. Jensen BA. Mann KK. Ryu HY. Sherr DH. Peroxisome proliferator-activated receptor gamma-mediated NF-kappa B activation and apoptosis in pre-B cells. J Immunol. 2002;169:6831–6841. doi: 10.4049/jimmunol.169.12.6831. [DOI] [PubMed] [Google Scholar]
- 22.Coe TS. Hamilton PB. Griffiths AM. Hodgson DJ. Wahab MA. Tyler CR. Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology (London, England) 2009;18:144–150. doi: 10.1007/s10646-008-0267-0. [DOI] [PubMed] [Google Scholar]
- 23.Guryev V. Koudijs MJ. Berezikov E. Johnson SL. Plasterk RH. van Eeden FJ, et al. Genetic variation in the zebrafish. Genome Res. 2006;16:491–497. doi: 10.1101/gr.4791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basta HA. Buzak AJ. McClure MA. Identification of novel retroid agents in Danio rerio, Oryzias latipes, Gasterosteus aculeatus and Tetraodon nigroviridis. Evol Bioinform Online. 2007;3:179–195. [PMC free article] [PubMed] [Google Scholar]
- 25.Holzschu DL. Martineau D. Fodor SK. Vogt VM. Bowser PR. Casey JW. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martineau D. Bowser PR. Renshaw RR. Casey JW. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martineau D. Bowser PR. Wooster G. Forney JL. Histologic and ultrastructural studies of dermal sarcoma of walleye (Pisces: Stizostedion vitreum) Vet Pathol. 1990;27:340–346. doi: 10.1177/030098589002700506. [DOI] [PubMed] [Google Scholar]
- 28.Martineau D. Bowser PR. Wooster GA. Armstrong LD. Experimental transmission of a dermal sarcoma in fingerling walleyes (Stizostedion vitreum vitreum) Vet Pathol. 1990;27:230–234. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- 29.Shen CH. Steiner LA. Genome structure and thymic expression of an endogenous retrovirus in zebrafish. J Virol. 2004;78:899–911. doi: 10.1128/JVI.78.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurka J. Kohany O. LTR retrotransposons from zebrafish. Repbase Rep. 2008;8:1608. [Google Scholar]
- 31.Vogel CF. Sciullo E. Li W. Wong P. Lazennec G. Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flavell AJ. Retroelements, reverse transcriptase and evolution. Comp Biochem Physiol. 1995;110:3–15. doi: 10.1016/0305-0491(94)00122-b. [DOI] [PubMed] [Google Scholar]
- 33.Eickbush TH. Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel AM. Gerster T. Promoter activity of the zebrafish bhikhari retroelement requires an intact activin signaling pathway. Mech Develop. 1999;85:133–146. doi: 10.1016/s0925-4773(99)00104-5. [DOI] [PubMed] [Google Scholar]
- 35.Chen CY. Croissant J. Majesky M. Topouzis S. McQuinn T. Frankovsky MJ, et al. Activation of the cardiac alpha-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Devel Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Skerjanc IS. Petropoulos H. Ridgeway AG. Wilton S. Myocyte enhancer factor 2C and Nkx2-5 upregulate each other's expression and initiate cardiomyogenesis in P19 cells. J Biol Chem. 1998;273:34904–34910. doi: 10.1074/jbc.273.52.34904. [DOI] [PubMed] [Google Scholar]
- 37.Prasch AL. Teraoka H. Carney SA. Dong W. Hiraga T. Stegeman JJ, et al. Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol Sci. 2003;76:138–150. doi: 10.1093/toxsci/kfg202. [DOI] [PubMed] [Google Scholar]
- 38.Correa RG. Tergaonkar V. Ng JK. Dubova I. Izpisua-Belmonte JC. Verma IM. Characterization of NF-kappa B/I kappa B proteins in zebra fish and their involvement in notochord development. Mol Cell Biol. 2004;24:5257–5268. doi: 10.1128/MCB.24.12.5257-5268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins ND. Schmid RM. Duckett CS. Leung K. Rice NR. Nabel GJ. Distinct combinations of NF-kappa B subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puga A. Barnes SJ. Chang C. Zhu H. Nephew KP. Khan SA, et al. Activation of transcription factors activator protein-1 and nuclear factor-kappaB by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochem Pharmacol. 2000;59:997–1005. doi: 10.1016/s0006-2952(99)00406-2. [DOI] [PubMed] [Google Scholar]
- 41.Prasad JC. Goldstone JV. Camacho CJ. Vajda S. Stegeman JJ. Ensemble modeling of substrate binding to cytochromes P450: Analysis of catalytic differences between CYP1A orthologs. Biochemistry. 2007;46:2640–2654. doi: 10.1021/bi062320m. [DOI] [PubMed] [Google Scholar]
- 42.Schlezinger JJ. Struntz WD. Goldstone JV. Stegeman JJ. Uncoupling of cytochrome P450 1A and stimulation of reactive oxygen species production by co-planar polychlorinated biphenyl congeners. Aquat Toxicol (Amsterdam, Netherlands) 2006;77:422–432. doi: 10.1016/j.aquatox.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 43.Schlezinger JJ. White RD. Stegeman JJ. Oxidative inactivation of cytochrome P-450 1A (CYP1A) stimulated by 3,3',4,4'-tetrachlorobiphenyl: Production of reactive oxygen by vertebrate CYP1As. Mol Pharmacol. 1999;56:588–597. doi: 10.1124/mol.56.3.588. [DOI] [PubMed] [Google Scholar]
- 44.Gloire G. Legrand-Poels S. Piette J. NF-kappaB activation by reactive oxygen species: Fifteen years later. Biochem Pharmacol. 2006;72:1493–1505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Dong W. Teraoka H. Yamazaki K. Tsukiyama S. Imani S. Imagawa T, et al. 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in the zebrafish embryo: Local circulation failure in the dorsal midbrain is associated with increased apoptosis. Toxicol Sci. 2002;69:191–201. doi: 10.1093/toxsci/69.1.191. [DOI] [PubMed] [Google Scholar]
- 46.Cantrell SM. Lutz LH. Tillitt DE. Hannink M. Embryotoxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): The embryonic vasculature is a physiological target for TCDD-induced DNA damage and apoptotic cell death in Medaka (Orizias latipes) Toxicol App Pharmacol. 1996;141:23–34. doi: 10.1006/taap.1996.0256. [DOI] [PubMed] [Google Scholar]
- 47.Goldstone HM. Stegeman JJ. Molecular mechanisms of 2,3,7,8-tetrachlorodibenzo-p-dioxin cardiovascular embryotoxicity. Drug Metab Rev. 2006;38:261–289. doi: 10.1080/03602530600570099. [DOI] [PubMed] [Google Scholar]
- 48.Lu KP. Hallberg LM. TomLinson J. Ramos KS. Benzo(a)pyrene activates L1Md retrotransposon and inhibits DNA repair in vascular smooth muscle cells. Mutation Res. 2000;454:35–44. doi: 10.1016/s0027-5107(00)00095-6. [DOI] [PubMed] [Google Scholar]
- 49.Jankun J. Matsumura F. Kaneko H. Trosko JE. Pellicer A. Greenberg AH. Plasmid-aided insertion of MMTV-LTR and ras DNAs to NIH 3T3 fibroblast cells makes them responsive to 2,3,7,8-TCDD causing overexpression of p21ras and down-regulation of EGF receptor. Mol Toxicol. 1989;2:177–186. [PubMed] [Google Scholar]
- 50.Abbott BD. Perdew GH. Buckalew AR. Birnbaum LS. Interactive regulation of Ah and glucocorticoid receptors in the synergistic induction of cleft palate by 2,3,7,8-tetrachlorodibenzo-p-dioxin and hydrocortisone. Toxicol Appl Pharmacol. 1994;128:138–150. doi: 10.1006/taap.1994.1191. [DOI] [PubMed] [Google Scholar]
- 51.Enan E. Matsumura F. Regulation by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) of the DNA binding activity of transcriptional factors via nuclear protein phosphorylation in guinea pig adipose tissue. Biochem Pharmacol. 1995;50:1199–1206. doi: 10.1016/0006-2952(95)00258-2. [DOI] [PubMed] [Google Scholar]
- 52.Perkins E. Lotufo G. Farrar D. Transposition of retrotransposons in the amphipod Leptochierus plumulosus in response to pollutant stress. Marine Environ Res. 2004;58:579. [Google Scholar]
- 53.Bing OH. Sirokman G. Humphries DE. Hypothesis: Link between endogenous retroviruses and cardiovascular disease. J Mol Cell Cardiol. 1998;30:1257–1262. doi: 10.1006/jmcc.1998.0723. [DOI] [PubMed] [Google Scholar]
- 54.Sirokman G. Humphries DE. Bing OH. Endogenous retroviral transcripts in myocytes from spontaneously hypertensive rats. Hypertension. 1997;30:88–93. doi: 10.1161/01.hyp.30.1.88. [DOI] [PubMed] [Google Scholar]
- 55.Lu KP. Ramos KS. Redox regulation of a novel L1Md-A2 retrotransposon in vascular smooth muscle cells. J Biol Chem. 2003;278:28201–28209. doi: 10.1074/jbc.M303888200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.