Background: Thermobifida fusca β-xylosidase Xyl43A contains a catalytic and an ancillary module.
Results: Enzymatic activity is lost when the modules are expressed independently but is restored upon specific noncovalent re-association.
Conclusion: The catalytic and ancillary modules of Xyl43A behave as a single functional entity.
Significance: Two phylogenetically distinct modular components of an enzyme evolved together to form an enzymatically active unit.
Keywords: Enzyme Mechanisms, Glycoside Hydrolases, Protein Domains, Protein Evolution, Protein-Protein Interactions, GH43, Thermobifida fusca, Hemicellulase
Abstract
β-Xylosidases are hemicellulases that hydrolyze short xylo-oligosaccharides into xylose units, thus complementing endoxylanase degradation of the hemicellulose component of lignocellulosic substrates. Here, we describe the cloning, characterization, and kinetic analysis of a glycoside hydrolase family 43 β-xylosidase (Xyl43A) from the aerobic cellulolytic bacterium, Thermobifida fusca. Temperature and pH optima of 55–60 °C and 5.5–6, respectively, were determined. The apparent Km value was 0.55 mm, using p-nitrophenyl xylopyranoside as substrate, and the catalytic constant (kcat) was 6.72 s−1. T. fusca Xyl43A contains a catalytic module at the N terminus and an ancillary module (termed herein as Module-A) of undefined function at the C terminus. We expressed the two recombinant modules independently in Escherichia coli and examined their remaining catalytic activity and binding properties. The separation of the two Xyl43A modules caused the complete loss of enzymatic activity, whereas potent binding to xylan was fully maintained in the catalytic module and partially in the ancillary Module-A. Nondenaturing gel electrophoresis revealed a specific noncovalent coupling of the two modules, thereby restoring enzymatic activity to 66.7% (relative to the wild-type enzyme). Module-A contributes a phenylalanine residue that functions as an essential part of the active site, and the two juxtaposed modules function as a single functional entity.
Introduction
Xylan is the major constituent of hemicellulose, which represents 20–30% of lignocellulosic plant biomass (1) and represents an important industrial target for degradation (e.g. bleaching in the pulp and paper industry (2)). Xylan is a linear polysaccharide, consisting of β-1,4 linked d-xylose units with a large variety of side-chain substituents. Consequently, the contribution of multiple hemicellulases is required for complete hydrolysis of xylan by synergistically complementary enzymes.
The well studied aerobic thermophilic soil bacterium, Thermobifida fusca, can use cellulose and xylan as sole carbon sources (3, 4). For this purpose, the bacterium produces a set of six cellulases and several hemicellulases such as xylanases, a β-xylosidase, an α-l-arabinofuranosidase, and an acetyl xylan esterase (5, 6). Two T. fusca endoxylanases (Xyn11A and Xyn10B) have been expressed and fully characterized (7, 8). A third endoxylanase, Xyn10A, was characterized from a related species Thermomonospora alba (9), and a nearly identical gene coding for this enzyme was found in T. fusca. Because of their potential industrial applications, the cloning and characterization of other thermostable xylanases deserve particular attention. We have therefore investigated in this communication the putative hemicellulase encoded by the T. fusca gene tfu1616, which was related in sequence to the family GH43 glycoside hydrolases (GH43), the members of which are known to possess β-xylosidase (EC 3.2.1.37), β-1,3-xylosidase (EC 3.2.1.72), α-l-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), or galactan 1,3-β-galactosidase (EC 3.2.1.145) activities (10). To date, over 1500 bacterial genes have been associated with this family. Over 50 bacterial enzymes from this family have been examined for their enzymatic activities, but only a part of them have been fully characterized, the results of which were published in the primary literature. Among them, β-xylosidases (11–16), endoxylanases (17, 18), arabinanases (19–22), arabinofuranosidases (23–25), and galactanases (26) were described.
This study reports the cloning of the tfu1616 gene of T. fusca and the characterization of the protein. The gene was expressed in Escherichia coli, and the protein was purified to near homogeneity. Data on physicochemical parameters and enzyme activity of the intact Xyl43A are provided. The two modules of the protein (a GH43 catalytic module and an associated ancillary module, Module-A) were produced as separate protein entities; the catalytic and binding activities were studied for both the separated modules as well as their in vitro combined form.
EXPERIMENTAL PROCEDURES
Cloning
Xyl43A (tfu1616, GenBankTM accession number AAZ555651) was cloned from T. fusca YX genomic DNA. Primers were designed with the program Oligo Primer Analysis Software version 5.1 and ordered at the Weizmann Institute of Sciences facility (N-terminal primer 5′-TCATGACATATGCACCATCACCATCACCATACTTCTCCCCAAGTCACGTCCT-3′ and C-terminal primer 5′-TGATTGCTCGAGTTAGGAGGGGGACTGAGGCCGGTA-3′ (NdeI and XhoI sites in boldface). The PCR product was inserted and ligated into linearized pET21a to form pXyl43A.
GH43 was cloned using Xyl43A WT forward primer and 5′-TACGCTCTCGAGCTACGGCCACGGGTGCGGGG-3′ as a reverse primer (XhoI site in boldface) and Module-A using 5′-TTAAGCCATATGCAGCCGTCAGAGACCGACCACTTCGACGA-3′ and 5′-TTATGTCTCGAGGGAGGGGGACTGAGGCCGGT-3′ (NdeI and XhoI sites in boldface).
PCRs were performed using ABgene Reddymix x2 (Advanced Biotechnologies Ltd., Epsom, UK). DNA samples were purified using a HI YieldTM Gel/PCR fragments extraction kit (Real Biotech Corp., Banqiao City, Taiwan).
PCR mutagenesis was performed for the preparation of Xyl43A(F518A) and Module-A(F518A) plasmids using phosphorylated primers 5′-GGTGCCACGGGAGCGTTCCTCGGCCTGTGGG-3′ and 5′-CCACATGATGGGGTCGTTGC-3′ (ordered from Syntezza Bioscience Ltd., Jerusalem), and PCRs were performed using Pfu UltraII DNA polymerase (Agilent Technologies, Santa Clara, CA).
Protein Expression and Purification
Plasmids containing genes coding for the full-length Xyl43A enzyme, the GH43 module, ancillary Module-A, and mutants Xyl43A(F518A) and Module-A(F518A) were expressed in E. coli BL21 (lDE3) pLysS cells, and the expressed His-tagged proteins were purified on a nickel-nitrilotriacetic acid column (Qiagen), as reported earlier (27). A gel filtration purification step was conducted for association of the GH43 module and the Module-A, using an AKTA-prime system and a Hiload 16/60 Superdex 75 column (GE Healthcare). SDS-PAGE was employed to test the purity of the recombinant proteins (12% acrylamide gels). The fractions with the highest degree of purity were pooled, and the concentrations of the recombinant proteins were estimated by absorbance at 280 nm based on their amino acid composition using the ProtParam program. Proteins were stored in 50% (v/v) glycerol at −20 °C.
Substrates
Microcrystalline cellulose (Avicel) was purchased from FMC Biopolymer (Philadelphia, PA) and was used for the preparation of phosphoric acid swollen cellulose (PASC 7.5 mg ml−1, pH 7). Insoluble xylan was prepared by boiling oat-spelt xylan (Sigma) for 30 min in distilled water and recovering the pellet by centrifugation; this was followed by three cycles of washes with distilled water to remove soluble sugars and determination of dry weight (28). Birchwood xylan, beechwood xylan, chitin, p-nitrophenyl-β-d-glucopyranoside, p-nitrophenyl-α-l-arabinofuranoside, p-nitrophenyl-β-d-cellobioside, p-nitrophenyl-β-d-xylopyranoside (pNPX)4 and xylobiose were purchased from Sigma. Debranched linear arabinan was purchased from Megazyme International, Ltd. (Wicklow, Ireland). Hatched wheat straw (0.2–0.8 mm) was provided by Valagro (Poitiers, France) and treated as described previously (29, 30).
pH Studies
The β-xylosidase activity over a pH range of 3.5–10 was investigated using citrate buffer from 3.5 to 8, Tris buffer from 7 to 9, and glycine/NaOH buffer from 9 to 10. Xyl43A was assayed at 50 °C for 7 min in a 700-μl reaction mixture containing 0.05 μm enzyme, 100 mm buffer, 1 mg/ml bovine serum albumin (BSA), and 5 mm pNPX. After 7 min, the tubes were placed on ice; the reaction was terminated by adding 200 μl of 1 m NaOH, and the absorbance was measured at 420 nm.
Temperature Dependence and Stability
The influence of temperature upon activity was assayed as follows. The release of p-nitrophenol by 0.05 μm Xyl43A at temperatures ranging from 30 to 80 °C was measured at 420 nm after a 7-min reaction and 15 min cooling on ice-water in a 700-μl final volume (100 mm buffer citrate, pH 6, 1 mg/ml BSA, 5 mm pNPX). The effect of temperature on the stability of Xyl43A was determined by incubating the enzyme without substrate over a 15-min period for 50 h at 50, 60, and 70 °C at pH 6. Similarly, the purified GH43 catalytic module, and Module-A were incubated at 50 °C over the same time period, either before or after combining the GH43/Module-A at 1:0.8 molar ratio. After adding substrates, 7-min reactions (as specified above) were carried out at 50 °C. The absorbance at 420 nm was then measured after a 15-min incubation on ice.
Kinetic Studies
Temperature-equilibrated reactions of a 180-μl final volume (100 mm buffer citrate, pH 6, 1 mg/ml BSA, 0–15 mm pNPX at 50 °C) were initiated by adding 10 μl of the enzyme (final concentration of 0.05 μm). The absorbance was monitored at 420 nm at 14-s intervals for 10 min, using an ELISA plate reader (BioTek Synergy HT, Winooski, VT). Absorbances (420 nm) were then converted to molarities using an extinction coefficient of 3365 m−1·cm−1 for the absorbance of p-nitrophenol in citrate buffer, pH 6, at 50 °C.
Effect of Salts and Chemicals
Various salts and chemicals were added at a final concentration of 1 mm to the standard enzymatic reaction mixtures to check potential inhibitory effects on enzymatic activity.
Binding to Insoluble Polysaccharides
The binding of the proteins to insoluble xylan and microcrystalline cellulose was determined qualitatively using SDS-PAGE. In a final volume of 100 μl, 10 μg (5 μg for cellulose binding experiments) of pure protein in 50 mm citrate buffer, pH 6.0, containing 12 mm CaCl2 and 2 mm EDTA was mixed with 0.5 mg of insoluble xylan (or 10 mg of microcrystalline cellulose). Tubes were mixed gently at 4 °C for 1 h and centrifuged at 14,000 rpm for 1 min. The supernatant fluids (containing the unbound fraction of the proteins) were transferred to new tubes and supplemented with SDS-containing buffer. The pellets (containing the bound fraction of the proteins) were washed once by resuspension in 100 μl of the above-described buffer, centrifuged, and resuspended in 60 μl of SDS-containing buffer. Boiled samples were subjected to SDS-PAGE (12% gels) for analysis. BSA served as a negative control for binding specificity.
Thin Layer Chromatography (TLC)
The hydrolysis of xylobiose by Xyl43A was monitored by TLC analysis. The xylobiose hydrolysis reaction included 5 mg of xylobiose, 0.2 mm Xyl43A, and 100 mm citrate buffer, pH 6. The reaction was performed at 50 °C, and at different time points (5, 15, and 30 min and 2 h), samples were taken, and the reaction was terminated by adding Hg2+ to a final concentration of 1 mm (which completely inhibits the enzyme). Chromatography was carried out on Silica Gel 60 plates (Merck), using butanol/ethanol/water (3:2:2) as a developing solvent. Spots were visualized by charring with 0.2% orcinol (in methanol and 20% phosphoric acid in methanol, 1:1 v/v) and heating for 2 min at 150 °C.
Multiple Amino Acid Sequence Alignment
The Clustal W2 program was employed to analyze the amino acid sequence-based alignment between the characterized β-xylosidases in glycoside hydrolase family 43 (11–16) and T. fusca Xyl43A. This alignment served to identify the homologous conserved residues between Geobacillus stearothermophilus and T. fusca β-xylosidases that appear to be involved in the interface between the catalytic and the C-terminal Module-A of Xyl43A.
RESULTS
Sequence Analysis and Production of Xyl43A (tfu1616)
The gene encoding for Xyl43A is part of the hemicellulolytic system of T. fusca. It encodes a 550-amino acid protein with a calculated molecular mass of 61,686 Da. The amino acid sequence of the Xyl43A protein was subjected to BLAST analysis (blastp, nr data base, blast.ncbi.nlm.nih.gov) and showed significant homology (from 44 to 59% sequence identity) to β-xylosidases and α-l-arabinofuranosidases that belong to glycoside hydrolase family 43. The ORF has a G + C content of 68.4%, not significantly different from that (67%) of the whole genome of T. fusca, which is typical of many thermophilic organisms (31). The gene encoding for Xyl43A does not include any recognizable signal peptide as determined by the SignalP server. Thus, the protein is assumed to remain intracellularly in T. fusca. SDS-PAGE analysis of the purified Xyl43A protein revealed a single protein band of about 60 kDa, which is in good agreement with the calculated molecular mass. A typical Xyl43A preparation yielded 31.2 mg/liter purified protein.
Bioinformatic analysis of the protein revealed the presence of a GH43 catalytic module located between residues 16 and 319 and a second module of undefined function (ancillary module, Module-A) located between residues 338 and 550 in the C-terminal part of the protein. To evaluate the binding properties and the enzymatic activity of both N-terminal (catalytic core) and C-terminal (ancillary) modules, we produced two distinct chimeras, GH43 and Module-A. Because there is 45% identity and 60% similarity between the previously characterized GH43 from G. stearothermophilus (15, 32), its structure was used and compared with the predicted structure of T. fusca Xyl43A, obtained using Swiss Model (Fig. 1). The quality of the model was estimated using the ProSA-web site. The Z-score of the model was −6.57, which is well within the range of scores typically found for native proteins of similar size.
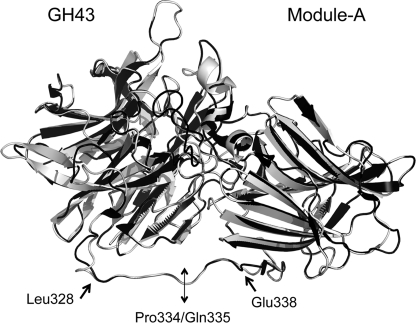
FIGURE 1.
Superposition of the solved structure of G. stearothermophilus XynB3 and the predicted model of T. fusca Xyl43A. The known structure of G. stearothermophilus XynB3 (2exh) is shown in black and the predicted structure of T. fusca Xyl43A, determined using Swiss model, in gray. Amino acid residues delimiting the linker portion (Leu-328 and Glu-338) are given as well as the residues (Pro-334/Gln-335) where the N-terminal catalytic module and C-terminal module were separated.
The model was especially helpful for determining the linker region of T. fusca Xyl43A. In G. stearothermophilus, the linker extends from Val-319 to Asp-329; by analogy, we deduced that the linker region in T. fusca Xyl43A would extend similarly from Leu-328 to Glu-338. The most appropriate juncture between the two modules was determined to be the bond between the adjacent Pro-334 and Gln-335, where there seemed to be little or no interactions between the linker and the modules in the predicted model. The two modules were therefore cloned separately (each with an N-terminal His tag), expressed, and purified by metal ion affinity chromatography. The resultant GH43 module and ancillary Module-A exhibited their expected molecular masses (35.8 and 25.9 kDa, respectively) in the SDS-polyacrylamide gel, and purity above 95% was estimated (data not shown). A schematic representation of the recombinant proteins expressed in this study is shown in Fig. 2.
FIGURE 2.
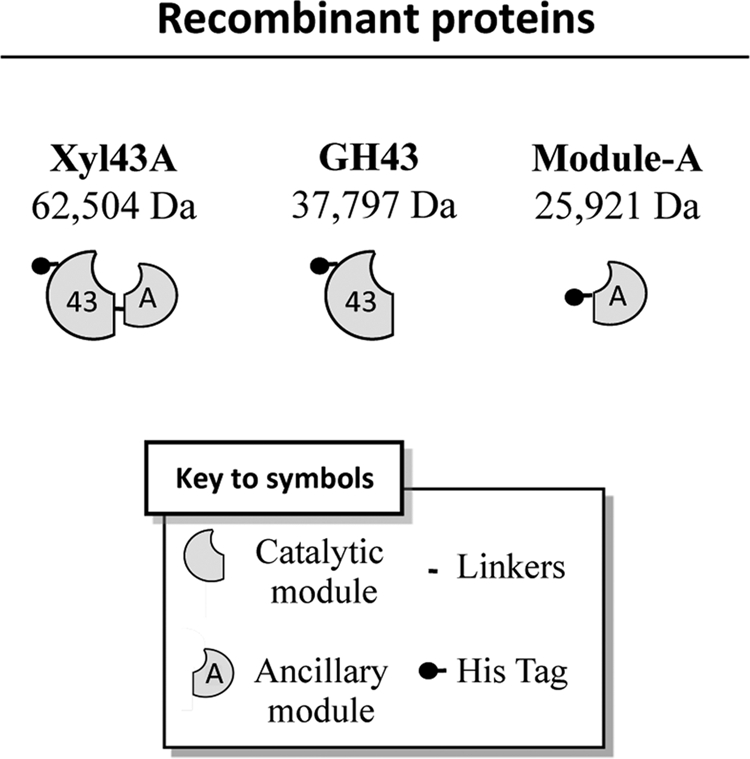
Schematic representation of the recombinant proteins used in this study and their calculated molecular masses.
Substrate Specificity
The activity of the recombinant enzyme was tested on a variety of substrates. No enzymatic activity was detected on xylans (from birch wood, beech wood, and oat-spelt, both soluble and insoluble), cellulosic substrates (Avicel and PASC), chitin, wheat straw, arabinan p-nitrophenyl-β-d-glucopyranoside, p-nitrophenyl-β-d-cellobioside, and p-nitrophenyl-α-l-arabinofuranoside. Detectable levels of hydrolysis were only observed for pNPX and xylobiose, confirming that the enzyme belongs to the β-xylosidase class. Residual activity (kcat/Km of 0.75 s−1·mm−1 versus 12.25 for pNPX) was observed with p-nitrophenyl-α-l-arabinofuranoside, due to the structural similarity between β-d-xylopyranoside and α-l-arabinofuranoside. These results support the premise that the conserved arginine (position 288 in T. fusca Xyl43A) allows the distinction between β-xylosidase and arabinofuranosidase classes (32). This arginine is believed to be conserved only in β-xylosidases. Neither the GH43 nor Module-A alone had measurable activities on any substrates (including xylans and arabinoxylan).
pH Studies
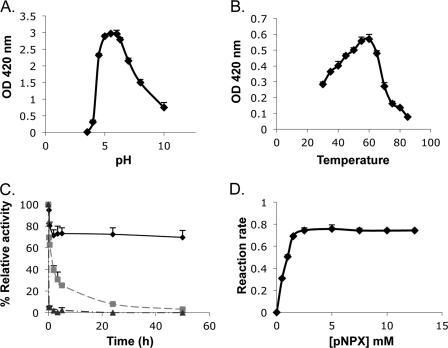
The optimal pH range of Xyl43A was 5.5–6 for β-xylosidase activity (Fig. 3A) with more than 70% activity retained at pH 4.5 and 7. Xyl43A is more stable at alkaline pH than at acidic pH and is drastically inactivated at pH 4. It is indeed known that changes in pH affect the ionic or electric charge of active site amino acids (that participate in substrate binding and catalysis) and either enhance and stabilize interactions with the substrate or break intra- and intermolecular bonds, changing the shape of the enzyme, therefore affecting its efficacy (33). Activity profiles of glycosidases versus pH are typically bell-shaped, reflecting the ionization state of, at least, two essential amino acids residues. The expected pKa values of the general base and general acid ionizable groups in the enzyme (derived from the pH-dependence graph) are 4.5 and 8, respectively, close to the values obtained for G. stearothermophilus XynB3 (15).
FIGURE 3.
Characterization of recombinant T. fusca Xyl43A. A, effect of pH on β-xylosidase activity. B, effect of temperature on β-xylosidase activity. C, thermostability of Xyl43A β-xylosidase activity at different incubation temperatures, 50 (♦), 60 (■), and 70 °C (▴) at pH 6. D, effect of substrate concentration on β-xylosidase activity. Unless otherwise stated, the reactions were monitored for 10 min at 50 °C, pH 6. Reaction rates are given in μmol of p-nitrophenol·s−1 × 106. Reactions were performed in triplicate, and standard deviations are indicated.
Thermostability
The hydrolysis rate increased from 30 to 60 °C, and the temperature coefficient (Q10) was calculated to be 1.3. An Arrhenius plot exhibited a straight line from 30 to 60 °C, typical of a single rate-limited thermally activated process, indicating an increase in enzyme activity with increasing temperature. The activation energy and the pre-exponential factor were determined to be 18.9 kJ·mol−1 and 390, respectively. Under the reaction conditions, the temperature optimum was found to be 55–60 °C for β-xylosidase activity (Fig. 3B); however, the enzyme underwent rapid inactivation at 60 °C (Fig. 3C). Purified Xyl43A demonstrated moderate thermostability, and even after 50 h at 50 °C, 70% of the β-xylosidase activity remained. Incubation at 60 °C resulted in about 50% loss of activities after only 2 h and almost no activity after 24 h. Treatment at 70 °C resulted in complete inactivation of the enzyme in less than 15 min (Fig. 3C).
Kinetic Constants
The kinetic constants of Xyl43A on pNPX were determined by running the reaction at different substrate concentrations (Fig. 3D). At 50 °C and at pH 6, the release of p-nitrophenol was linear with time and proportional to enzyme concentration. The apparent Km value was 0.55 mm pNPX, and the kcat was 6.72 s−1.
Effect of Various Chemicals
The effects of different chemicals on Xyl43A activity are shown in Table 1. The enzyme retained almost full activity in the presence of 1 mm Li+, Na2+, K+, Mg2+, Ca2+, Fe3+, Mn2+, Co2+, Cu2+, and the reagent SDS. The addition of EDTA had no effect on the activity, suggesting that no metals are needed for the enzyme reactions. Group IIb metals such as Hg2+ and Ag2+ gave stronger inhibition (mercury inhibition is indeed well known for most enzymes, including cellulases and xylanases, indicating the existence of essential thiol groups (7)).
TABLE 1.
Effect of various chemicals at a concentration of 1 mm on Xyl43A activity
Relative activity is shown in % ± S.D.
| Chemical | Relative activity | Chemical | Relative activity |
|---|---|---|---|
| % | % | ||
| None | 100 | CuSO4 | 81 ± 3 |
| LiCl | 98 ± 3 | AgNO3 | 10 ± 1 |
| NaCl | 91 ± 2 | ZnCl2 | 100 |
| KCl | 92 ± 5 | HgCl2 | 8 ± 4 |
| MgCl2 | 89 ± 5 | EDTA | 100 |
| CaSO4 | 86 ± 1 | SDS | 80 ± 1 |
| FeSO4 | 91 ± 4 | Urea | 78 ± 6 |
| MnCl2 | 85 ± 6 | Citric acid | 73 ± 2 |
| CoCl2 | 96 ± 1 | Boric acid | 61 ± 4 |
Binding to Insoluble Polysaccharides
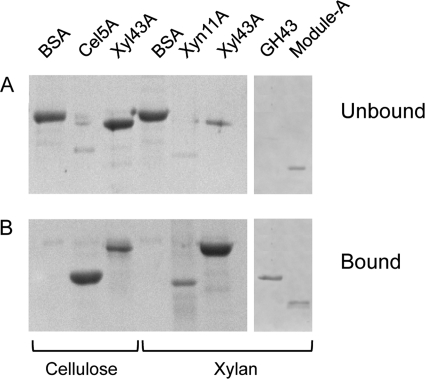
The binding capacity of Xyl43A to insoluble xylan and microcrystalline cellulose was investigated. Full-length Xyl43A exhibited strong binding capacity for insoluble xylan, whereas weak binding to microcrystalline cellulose was observed (Fig. 4). As expected, the bulk of the BSA negative control was found in the unbound fraction, and the respective insoluble xylan and microcrystalline cellulose-positive controls were located in the bound fractions. The GH43 catalytic module alone demonstrated full binding capacity for insoluble xylan, whereas Module-A was found in bound and unbound fractions in almost equal proportions. Thus, it seems that the xylan binding capacity of Xyl43A is due mainly to the residues present in the catalytic module, and the ancillary Module-A plays a minor role in the binding interaction (Fig. 4).
FIGURE 4.
Binding of the proteins to insoluble polysaccharides as assessed by SDS-PAGE. A, unbound fractions; B, bound fractions. Each gel was loaded in the following order: BSA, Cel5A, Xyl43A, BSA, Xyn11A, Xyl43A, GH43, and Module-A. 1st to 3rd lanes, binding experiment with microcrystalline cellulose; 4th to 8th lanes, interactions with insoluble xylan.
Re-association of GH43 and Module-A
Noncovalent association of the separately expressed and purified GH43 and Module-A components at room temperature was analyzed by nondenaturing PAGE. The catalytic module was mixed with increasing amounts of the helper module in molar ratios ranging from 1:0.2 to 1:1.8, and the mobility pattern of each mixture was compared with that of each module alone and with that of the full-length Xyl43A enzyme. Mixtures of GH43 and Module-A resulted in complex formation (Fig. 5A). The mobility pattern of the complex was nearly identical to that of wild-type Xyl43A, revealing a specific interaction between the catalytic and ancillary modules. The intensity of the band, corresponding to the interaction of GH43 and Module-A, increased until a 1:0.8 ratio (GH43/Module-A), whereas the intensity of the bands of each of the individual modules decreased. At this ratio, the Module-A band interacted completely with GH43. A relatively small part of the latter fails to form the complex, probably because of incorrect folding of a portion of the GH43 fraction. Up to 25% of the wild-type β-xylosidase activity was thus recovered upon association of the two modules (Fig. 5B).
FIGURE 5.
Physical association of GH43 and Module-A. A, specific association of GH43 and Module-A revealed by nondenaturing gel electrophoresis (7.5%). Mixtures of GH43 and Module-A (at the indicated molar ratio relative to GH43) were subjected to nondenaturing PAGE, and the mobility pattern of each mixture was compared with that of the individual modules and to wild-type Xyl43A. Mixtures of the two modules resulted in formation of a complex (thick arrow). 1st lane, full-length Xyl43A enzyme (150 pmol); 2nd lane, GH43 (150 pmol); 3rd lane, Module-A (150 pmol); 4th to 10th lanes, GH43 (150 pmol) mixed with increasing amounts of Module-A (30–290 pmol), at the indicated molar ratios. B, recovery of β-xylosidase activity upon association of GH43 and Module-A. A fixed amount (150 pmol) of the GH43 was mixed with increasing amounts of Module-A, and their respective activities were compared with that of the intact Xyl43A (set as 100%). C, thermostability of GH43, Module-A, and the re-associated form of Xyl43A over time at 50 °C and pH 6. Curves are labeled as follows: heat treatment of GH43 and Module-A prior to their interaction (gray triangles) and following re-association at a 1:0.8 molar ratio of GH43:Module-A (black square). Reactions were performed in duplicate, and standard deviations are indicated.
Gel filtration was used to purify the reassociated enzyme from the two modules at a 1:0.8 ratio, and the kinetics parameters were determined for the purest fraction obtained. The apparent Km value was 0.6 mm pNPX (similar to that of the wild-type enzyme), whereas the kcat was 4.83 s−1. Up to 71.8% of the wild-type β-xylosidase activity was thus recovered upon association of the two modules.
The moderate thermostability characteristics observed for the wild-type enzyme (Fig. 3C) raised the question as to whether the re-associated Xyl43A would also be stable to heat treatment under the same conditions. Moreover, it was of interest to determine whether exposure of the two constitutive modules to heat would result in reduced stability. We therefore subjected the two isolated modules to heat treatment at 50 °C both prior to and after re-association. Heat treatment of the two modules separately before re-association led to a marked loss of stability (Fig. 5C). About 60% of the residual activity was obtained after 2 h of heat treatment, and only 15% remained after 5 h. In contrast, re-association of the two modules before heat treatment resulted in increased thermostability; about 80% relative activity remained after 5 h of heat treatment and 50% after 48 h (Fig. 5C). The results indicated that the individual modules exhibit decreased stability in solution but upon re-association form a stable complex.
To gain insight into the molecular basis of re-association of the GH43 and Module-A, we employed the Protein Interaction Calculator server (34) to analyze the known three-dimensional structure of the family GH43 XynB3 β-xylosidase from G. stearothermophilus. Using this server, the various interactions involved in the interface between the catalytic and the C-terminal Module-A were determined, and the homologous conserved residues in T. fusca Xyl43A were then identified. Table 2 shows the major residues, predicted to be involved in the intramodular interface. The data suggest strong reciprocal binding interaction between the two modules of T. fusca Xyl43A and provide an explanation for their observed re-association.
TABLE 2.
Major conserved residues likely to be involved in the strong binding interaction between the two modules of the T. fusca Xyl43A, based on the solved structure of G. stearothermophilus
| Residues | Intermodular interactions |
||||
|---|---|---|---|---|---|
| Hydrophobic | Hydrogen bonding | Ionic | Aromatic | Cation Pi | |
| Phe-28 | Phe-522 | Arg-392 | Phe-522 | Arg-392 | |
| Trp-50 | Tyr-420 | ||||
| Tyr-51 | Pro-357 | Thr-353 | |||
| Lys-262 | Asp-502 | ||||
| Arg-296 | Phe-518 | Asp-502 | Phe-518 | ||
| Ser-352 | Trp-50 (2)a | ||||
| Pro-52 (2) | |||||
| Thr-353 | Tyr-51 | ||||
| Arg-355 | Arg-83 (3) | ||||
| Thr-423 | Asp-85 (2) | ||||
| Glu-503 | Trp-225 | Lys-262 | |||
| Lys-262 | |||||
| Cys-292 | |||||
| Phe-518 | Phe-48 | Phe-48 | |||
| Trp-90 | Trp-90 | ||||
a Number of interactions (greater than 1) of a given residue is shown parenthetically.
Production of Mutated Xyl43A(F518A)
According to the crystal structure of the family GH43 β-xylosidase (XynB3) from G. stearothermophilus T-6 (32), the active site of the enzyme possesses a pocket topology, mainly constructed from β-propeller domain residues. In addition, a conserved phenylalanine residue (Phe-518 of T. fusca Xyl43A, homologous to Phe-506 of G. stearothermophilus XynB3) is located on a loop of Module-A in the latter structure. The loop serves to close one side of a cleft, formed by the catalytic module, and inserts Phe-506 therein. This phenylalanine is the only residue in Module-A, which participates in the active site and presumably plays a role in substrate binding and specificity. To further investigate the role of this amino acid in the catalytic activity of T. fusca Xyl43A, the conserved Phe-518 was replaced with Ala.
The kinetic constants of Xyl43A(F518A) for pNPX hydrolysis were determined by interaction of the mutant enzyme at different substrate concentrations as performed for the wild-type enzyme (data not shown). At 50 °C and pH 6, the release of p-nitrophenol was linear with time and proportional to enzyme concentration. The apparent Km value was 0.98 mm pNPX, and the kcat was 0.059 s−1. The replacement of the phenylalanine by alanine affected the activity of the enzyme dramatically, the activity of the mutant representing less than 1% of the wild-type activity on pNPX (0.89%).
Re-association of GH43 and Mutated Module-A(F518A)
To further examine the role of the Phe-518 residue in the catalysis, Module-A was replaced with alanine in a manner similar to that of the wild-type enzyme. As described for the wild-type modules, noncovalent association of GH43 and the mutated Module-A(F518A) was performed. A strong interaction between the two modules was observed (data not shown), suggesting that Phe-518 does not play a dominant role in the strong association between the two modules. Nevertheless, upon association of the modules, β-xylosidase activity was not recovered, suggesting that the role of this particular amino acid is directly related to catalysis per se.
DISCUSSION
A number of GH43 family members (as well as GH3, GH51, and GH54 families) have been reported to exhibit simultaneously both β-xylosidase and α-l-arabinofuranosidase activities, probably because of the active site geometry that does not allow the enzyme to distinguish between the two saccharides. Indeed, both activities seem to occur in the same active site (35, 36). The T. fusca Xyl43A, characterized here, exhibited primarily β-xylosidase activity and only nominal levels of α-l-arabinofuranosidase activity.
In previous research (37), Asp and Glu residues of family GH43 enzymes have been directly implicated in substrate hydrolysis, in line with the classical general acid (proton donor) and nucleophile/base that characterize enzymatic hydrolysis of the glycosidic bond (38). Multiple sequence alignment of glycoside hydrolase families 32, 43, 62, and 68 revealed three conserved blocks, each containing an acidic residue at an equivalent position in all of the enzymes; site-directed mutagenesis studies have shown that Asp/Glu (block I), Asp (block II), and Glu (block III) are essential for catalytic activity of GH43. This indicates direct involvement of the conserved residues in substrate binding and hydrolysis, and substitutions of these specific residues inactivate the enzyme or considerably reduce its activity. These three strictly conserved key acidic residues are located in a funnel-shaped active site that includes two subsites with a single route for access by ligands in a β-propeller architecture, and the three residues operate with the canonical reaction mechanism of inversion of anomeric configuration (32, 39). More recently, the crystal structures of Cellvibrio japonicus arabinanase 43A and G. stearothermophilus β-xylosidase further demonstrated the role of these three carboxylates in catalytic activity (19, 31). Similar blocks and essential residues were found in T. fusca Xyl43A by sequence comparison as follows: block I 29YPDPS33, block II 142GFDPS146, and block III 201VTEAPH206.
In the GH43 family, several modular architectures are observed (40). One group of GH43 enzymes is composed only of a single GH43 module and lacks the C-terminal module (Module-A). These enzymes belong to the arabinanase class (20–22, 41). Another group of family GH43 enzymes comprising both modules is defined as either β-xylosidases or α-l-arabinofuranosidases (32, 42). One possibility is that the C-terminal module in family GH43 would be responsible for switching substrate specificity from arabinan to xylobiose. However, in this study, the T. fusca GH43 module alone was unable to degrade arabinan, and no binding ability of either of the two Xyl43A modules toward arabinan was observed, thus suggesting that the difference in specificities is a function of the topography and composition of the active site. According to Alhassid et al. (22), the topology of the active site in G. stearothermophilus T-6 arabinanase (AbnB) resembles a groove with a narrow bridge formed by Phe-104 (conserved in family GH43 arabinanases) and Cys-222. A neighboring cysteine (Cys-221) bridges with the Cys-222 and covers the cleft formed in the active site. The Phe and both Cys residues are absent from β-xylosidases in general, and T. fusca β-xylosidase in particular. Moreover, according to Brux et al. (32), in the β-xylosidases, an extended arabinopyranose ligand would clash with the hydrophobic area generated by conserved xylosidase residues, i.e. Trp-74, Phe-127, Phe-32, and Phe-506 of G. stearothermophilus XynB3. In the case of galactopyranose, there is also a steric clash caused by the additional hydroxymethyl group, which encounters Phe-506 (1.4 Å). These criteria explain the specificity of the T. fusca catalytic GH43 module and underscore the sophisticated divergence of the conserved GH43 arabinanases and β-xylosidases.
Physical separation and reassociation of the two modular components of Xyl43A (GH43 and Module-A) enabled us to further analyze the enzymatic and binding properties of this enzyme. Separating the two modules led to a complete loss in enzymatic activity of the N-terminal module GH43 but did not affect its ability to bind xylan, suggesting that the active site would thus need the combined conformation of the complete protein to degrade its substrate correctly. Indeed, a strong noncovalent interaction occurs between the two modules of T. fusca Xyl43A. This physical interaction led to substantial recovery of β-xylosidase activity (71.8%), demonstrating the role of the Module-A in enzymatic degradation of its substrate. Based on the solved structure of G. stearothermophilus (32, 43), several residues play an essential role in formation of the strong interface between the two modules of the enzyme (Table 2).
The re-association of GH43 and Module-A in T. fusca Xyl43A provided a key functional residue for β-xylosidase activity. The phenylalanine located at position 518 of Module-A was thus demonstrated to be inserted in a critical position and essential for Xyl43A enzymatic activity. The F518A replacement, either in the intact enzyme Xyl43A or in Module-A, resulted in the near-complete loss of enzymatic activity. According to Brux et al. (32), this conserved phenylalanine emerges from a loop originated from the β-sandwich domain and contributes to substrate binding by a stacking interaction with the xylose unit at the −1 subsite. Interestingly, the amino acid replacement does not appear to affect the overall folding of the enzyme according to the re-association of the separated GH43 and mutated Module-A.
In a recent article (44), physical re-association of a family GH9 endoglucanase catalytic module and its ancillary family 3c CBM was also demonstrated with attendant regain of enzyme activity. The latter study, together with this study, promotes the original description of the combined S-protein and S-peptide to generate an active form of ribonuclease A (45). In the current studies on the glycoside hydrolases, however, the two interacting modules are clearly of distinct evolutionary origin.
Indeed, the two separate modules of Xyl43A fold on their own, suggesting that originally they may have had distinct functions. The evolutionary source of the C-terminal Module-A, however, is unclear, because it is phylogenetically unconnected with any known protein in the databases other than those associated with the family GH43 enzymes. Perhaps an ancestral form of Module-A was originally a CBM, because it possesses reminiscent characteristics of a CBM (e.g. β-sandwich structure); its binding role would presumably have been lost during evolvement of its singular catalytic contribution to the GH43 xylosidases. Moreover, because Xyl43A does not contain any signal peptide, the enzyme appears to remain in the intracellular milieu in T. fusca and would not require a CBM to target the enzyme to its substrate. Indeed, xylobiose is small enough to enter the cell, presumably via an appropriate transport apparatus, and can be degraded internally into xylose units, so a CBM would be irrelevant for the enzyme. Consequently, as representative of the GH43 β-xylosidases, the two linked modules of T. fusca Xyl43A should thus be considered as an intact, synchronized functional unit, rather than a fortuitous pair of independent modules.
Acknowledgments
We are grateful for the critique and assistance of Drs. Ilit Noach and Bareket Dassa (Weizmann Institute of Sciences, Rehovot, Israel) throughout the stages of preparation of the manuscript.
This work was supported in part by grants from the United States-Israel Binational Science Foundation, Jerusalem, Israel, Israeli Centers of Research Excellence (I-CORE) Program (Center 152/11), Alternative Energy Research Initiative Bioenergy Consortium, Brazilian friends of the Weizmann Institute of Science Alternative Energy Research Initiative (research grants from Charles Rothschild, Mario Fleck, and Roberto and Renata Ruhman), Technion-Niedersachsen Research Cooperation Program, and Israel Science Foundation Grants 966/09, 159/07, and 24/11.
- pNPX
- p-nitrophenyl-β-d-xylopyranoside
- CBM
- carbohydrate-binding module.
REFERENCES
- 1. Kulkarni N., Shendye A., Rao M. (1999) Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23, 411–456 [DOI] [PubMed] [Google Scholar]
- 2. Beg Q. K., Kapoor M., Mahajan L., Hoondal G. S. (2001) Microbial xylanases and their industrial applications. A review. Appl. Microbiol. Biotechnol. 56, 326–338 [DOI] [PubMed] [Google Scholar]
- 3. Wilson D. B. (1992) Biochemistry and genetics of actinomycete cellulases. Crit. Rev. Biotechnol. 12, 45–63 [DOI] [PubMed] [Google Scholar]
- 4. Wilson D. B. (2004) Studies of Thermobifida fusca plant cell wall-degrading enzymes. Chem. Rec. 4, 72–82 [DOI] [PubMed] [Google Scholar]
- 5. Bachmann S. L., McCarthy A. J. (1991) Purification and cooperative activity of enzymes constituting the xylan-degrading system of Thermomonospora fusca. Appl. Environ. Microbiol. 57, 2121–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghangas G. S., Hu Y. J., Wilson D. B. (1989) Cloning of a Thermomonospora fusca xylanase gene and its expression in Escherichia coli and Streptomyces lividans. J. Bacteriol. 171, 2963–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Irwin D., Jung E. D., Wilson D. B. (1994) Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60, 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim J. H., Irwin D., Wilson D. B. (2004) Purification and characterization of Thermobifida fusca xylanase 10B. Can. J. Microbiol. 50, 835–843 [DOI] [PubMed] [Google Scholar]
- 9. Blanco J., Coque J. J., Velasco J., Martín J. F. (1997) Cloning, expression in Streptomyces lividans, and biochemical characterization of a thermostable endo-β-1,4-xylanase of Thermomonospora alba ULJB1 with cellulose-binding ability. Appl. Microbiol. Biotechnol. 48, 208–217 [DOI] [PubMed] [Google Scholar]
- 10. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moriyama H., Fukusaki E., Cabrera Crespo J., Shinmyo A., Okada H. (1987) Structure and expression of genes coding for xylan-degrading enzymes of Bacillus pumilus. Eur. J. Biochem. 166, 539–545 [DOI] [PubMed] [Google Scholar]
- 12. La Grange D. C., Pretorius I. S., van Zyl W. H. (1997) Cloning of the Bacillus pumilus β-xylosidase gene (xynB) and its expression in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 47, 262–266 [DOI] [PubMed] [Google Scholar]
- 13. Chun Y. C., Jung K. H., Lee J. C., Park S. H., Chung H. K., Yoon K. H. (1998) Molecular cloning and the nucleotide sequence of the Bacillus sp. KK1 beta-xylosidase gene. J. Mol. Microbiol. Biotechnol. 8, 28–33 [Google Scholar]
- 14. Whitehead T. R., Cotta M. A. (2001) Identification of a broad-specificity xylosidase/arabinosidase important for xylooligosaccharide fermentation by the ruminal anaerobe Selenomonas ruminantium GA192. Curr. Microbiol. 43, 293–298 [DOI] [PubMed] [Google Scholar]
- 15. Shallom D., Leon M., Bravman T., Ben-David A., Zaide G., Belakhov V., Shoham G., Schomburg D., Baasov T., Shoham Y. (2005) Biochemical characterization and identification of the catalytic residues of a family 43 β-d-xylosidase from Geobacillus stearothermophilus T-6. Biochemistry 44, 387–397 [DOI] [PubMed] [Google Scholar]
- 16. Smaali I., Rémond C., O'Donohue M. J. (2006) Expression in Escherichia coli and characterization of β-xylosidases GH39 and GH-43 from Bacillus halodurans C-125. Appl. Microbiol. Biotechnol. 73, 582–590 [DOI] [PubMed] [Google Scholar]
- 17. Gosalbes M. J., Pérez-González J. A., González R., Navarro A. (1991) Two β-glycanase genes are clustered in Bacillus polymyxa. Molecular cloning, expression, and sequence analysis of genes encoding a xylanase and an endo-β-(1,3)-(1,4)-glucanase. J. Bacteriol. 173, 7705–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bourgois T. M., Van Craeyveld V., Van Campenhout S., Courtin C. M., Delcour J. A., Robben J., Volckaert G. (2007) Recombinant expression and characterization of XynD from Bacillus subtilis subsp. subtilis ATCC 6051. A GH 43 arabinoxylan arabinofuranohydrolase. Appl. Microbiol. Biotechnol. 75, 1309–1317 [DOI] [PubMed] [Google Scholar]
- 19. McKie V. A., Black G. W., Millward-Sadler S. J., Hazlewood G. P., Laurie J. I., Gilbert H. J. (1997) Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo-mode of action. Biochem. J. 323, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nurizzo D., Turkenburg J. P., Charnock S. J., Roberts S. M., Dodson E. J., McKie V. A., Taylor E. J., Gilbert H. J., Davies G. J. (2002) Cellvibrio japonicus α-l-arabinanase 43A has a novel five-blade β-propeller fold. Nat. Struct. Biol. 9, 665–668 [DOI] [PubMed] [Google Scholar]
- 21. Leal T. F., de Sá-Nogueira I. (2004) Purification, characterization, and functional analysis of an endo-arabinanase (AbnA) from Bacillus subtilis. FEMS Microbiol. Lett. 241, 41–48 [DOI] [PubMed] [Google Scholar]
- 22. Alhassid A., Ben-David A., Tabachnikov O., Libster D., Naveh E., Zolotnitsky G., Shoham Y., Shoham G. (2009) Crystal structure of an inverting GH 43 1,5-α-l-arabinanase from Geobacillus stearothermophilus complexed with its substrate. Biochem. J. 422, 73–82 [DOI] [PubMed] [Google Scholar]
- 23. Matsuo N., Kaneko S., Kuno A., Kobayashi H., Kusakabe I. (2000) Purification, characterization, and gene cloning of two α-l-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem. J. 346, 9–15 [PMC free article] [PubMed] [Google Scholar]
- 24. van den Broek L. A., Lloyd R. M., Beldman G., Verdoes J. C., McCleary B. V., Voragen A. G. (2005) Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 67, 641–647 [DOI] [PubMed] [Google Scholar]
- 25. Vandermarliere E., Bourgois T. M., Van Campenhout S., Strelkov S. V., Volckaert G., Delcour J. A., Courtin C. M., Rabijns A. (2007) Crystallization and preliminary x-ray analysis of an arabinoxylan arabinofuranohydrolase from Bacillus subtilis. Acta Crystallogr Sect. F Struct. Biol. Cryst. Commun. 63, 692–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ichinose H., Yoshida M., Kotake T., Kuno A., Igarashi K., Tsumuraya Y., Samejima M., Hirabayashi J., Kobayashi H., Kaneko S. (2005) An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280, 25820–25829 [DOI] [PubMed] [Google Scholar]
- 27. Caspi J., Irwin D., Lamed R., Shoham Y., Fierobe H. P., Wilson D. B., Bayer E. A. (2006) Thermobifida fusca family-6 cellulases as potential designer cellulosomes components. Biocat. Biotransform. 24, 3–12 [Google Scholar]
- 28. Kluepfel D., Vats-Mehta S., Aumont F., Shareck F., Morosoli R. (1990) Purification and characterization of a new xylanase (xylanase B) produced by Streptomyces lividans 66. Biochem. J. 267, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fierobe H. P., Mingardon F., Mechaly A., Bélaïch A., Rincon M. T., Pagès S., Lamed R., Tardif C., Bélaïch J. P., Bayer E. A. (2005) Action of designer cellulosomes on homogeneous versus complex substrates. Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280, 16325–16334 [DOI] [PubMed] [Google Scholar]
- 30. Tabka M. G., Herpoël-Gimbert I., Monod F., Asther M., Sigoillot J. C. (2006) Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzyme Microb. Technol. 39, 897–902 [Google Scholar]
- 31. Kukolya J., Nagy I., Láday M., Tóth E., Oravecz O., Márialigeti K., Hornok L. (2002) Thermobifida cellulolytica sp. nov., a novel lignocellulose-decomposing actinomycete. Int. J. Syst. Evol. Microbiol. 52, 1193–1199 [DOI] [PubMed] [Google Scholar]
- 32. Brüx C., Ben-David A., Shallom-Shezifi D., Leon M., Niefind K., Shoham G., Shoham Y., Schomburg D. (2006) The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J. Mol. Biol. 359, 97–109 [DOI] [PubMed] [Google Scholar]
- 33. Price N. C., Stevens L. (2001) Fundamentals of Enzymology: The Cell and Molecular Biology of Catalytic Proteins, 3rd Ed., pp. 133–135, Oxford University Press, New York [Google Scholar]
- 34. Tina K., Bhadra R., Srinivasan N. (2007) PIC: Protein interactions calculator. Nucleic Acids Res. 35, 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakka K., Yoshikawa K., Kojima Y., Karita S., Ohmiya K., Shimada K. (1993) Nucleotide sequence of the Clostridium stercorarium xylA gene encoding a bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities, and properties of the translated product. Biosci. Biotechnol. Biochem. 57, 268–272 [DOI] [PubMed] [Google Scholar]
- 36. Utt E. A., Eddy C. K., Keshav K. F., Ingram L. O. (1991) Sequencing and expression of the Butyrivibrio fibrisolvens xylB gene encoding a novel bifunctional protein with β-d-xylosidase and α-l-arabinofuranosidase activities. Appl. Environ. Microbiol. 57, 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pons T., Naumoff D. G., Martínez-Fleites C., Hernández L. (2004) Three acidic residues are at the active site of a β-propeller architecture in glycoside hydrolase families 32, 43, 62, and 68. Proteins 54, 424–432 [DOI] [PubMed] [Google Scholar]
- 38. Davies G., Henrissat B. (1995) Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 39. Jordan D. B., Li X. L. (2007) Variation in relative substrate specificity of bifunctional β-d-xylosidase/α-l-arabinofuranosidase by single-site mutations. Roles of substrate distortion and recognition. Biochim. Biophys. Acta 1774, 1192–1198 [DOI] [PubMed] [Google Scholar]
- 40. Yoshida S., Hespen C. W., Beverly R. L., Mackie R. I., Cann I. K. (2010) Domain analysis of a modular α-l-arabinofuranosidase with a unique carbohydrate binding strategy from the fiber-degrading bacterium Fibrobacter succinogenes S85. J. Bacteriol. 192, 5424–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamaguchi A., Tada T., Wada K., Nakaniwa T., Kitatani T., Sogabe Y., Takao M., Sakai T., Nishimura K. (2005) Structural basis for thermostability of endo-1,5-α-l-arabinanase from Bacillus thermodenitrificans TS-3. J. Biochem. 137, 587–592 [DOI] [PubMed] [Google Scholar]
- 42. Brunzelle J. S., Jordan D. B., McCaslin D. R., Olczak A., Wawrzak Z. (2008) Structure of the two-subsite β-d-xylosidase from Selenomonas ruminantium in complex with 1,3-bis[tris(hydroxymethyl)methylamino]propane. Arch. Biochem. Biophys. 474, 157–166 [DOI] [PubMed] [Google Scholar]
- 43. Barker I. J., Petersen L., Reilly P. J. (2010) Mechanism of xylobiose hydrolysis by GH43 β-xylosidase. J. Phys. Chem. B 114, 15389–15393 [DOI] [PubMed] [Google Scholar]
- 44. Burstein T., Shulman M., Jindou S., Petkun S., Frolow F., Shoham Y., Bayer E. A., Lamed R. (2009) Physical association of the catalytic and helper modules of a family-9 glycoside hydrolase is essential for activity. FEBS Lett. 583, 879–884 [DOI] [PubMed] [Google Scholar]
- 45. Richards F. M. (1958) On the enzymic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 44, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]