Background: RAD51 and RecA homologs form a nucleoprotein filament (NPF) that includes an ATP cap, which sandwiches the adenosine nucleotide.
Results: The HsRAD51(D316K) ATP cap substitution dramatically enhances recombinase and NPF stability.
Conclusion: The HsRAD51(Asp-316) residue forms a salt bridge with ATP that allows more rapid protein turnover.
Significance: HsRAD51(D316K) provides a useful reagent for the study of recombinase function in physiological conditions.
Keywords: DNA Recombination, DNA Repair, Enzyme Turnover, Protein-DNA Interaction, Protein Turnover
Abstract
RAD51 mediates homologous recombination by forming an active DNA nucleoprotein filament (NPF). A conserved aspartate that forms a salt bridge with the ATP γ-phosphate is found at the nucleotide-binding interface between RAD51 subunits of the NPF known as the ATP cap. The salt bridge accounts for the nonphysiological cation(s) required to fully activate the RAD51 NPF. In contrast, RecA homologs and most RAD51 paralogs contain a conserved lysine at the analogous structural position. We demonstrate that substitution of human RAD51(Asp-316) with lysine (HsRAD51(D316K)) decreases NPF turnover and facilitates considerably improved recombinase functions. Structural analysis shows that archaebacterial Methanococcus voltae RadA(D302K) (MvRAD51(D302K)) and HsRAD51(D316K) form extended active NPFs without salt. These studies suggest that the HsRAD51(Asp-316) salt bridge may function as a conformational sensor that enhances turnover at the expense of recombinase activity.
Introduction
Failure to repair DNA double-stranded breaks may result in genomic instability and tumorigenesis (1–3). HR2 is an error-free pathway that utilizes a template strand to restore DSB ends (4). A key component in HR-mediated repair is RAD51 (5). Deletion of RAD51 in mice results in embryonic lethality, whereas RAD51 knockdown in chicken DT40 cell lines results in increased chromosomal instability (6, 7). RAD51 forms a presynaptic NPF that catalyzes homologous pairing and strand exchange. In addition, RAD51 associates with RAD52, RAD54, and BRCA2 during recombinational repair (8). Eukaryotic HR is further tuned by the presence of several RAD51 paralogs, where some have been shown to enhance human RAD51 (HsRAD51) functionality in vitro (9–12).
Despite functional conservation with the prototypical bacterial homolog RecA, RAD51 requires unusual salt conditions for an efficient strand exchange in vitro (13–15). For example, the ammonium (NH4+) cation appears to be most efficient at enhancing RAD51 recombinase activity, yet it is unlikely to occur at physiologically relevant conditions (13–15). Other cations such as potassium (K+) confer significantly reduced activity (14, 15). Cations that enhance recombinase activity appear to induce an NPF that mimics the active extended RecA NPF (15). Recombinase-enhancing cations may also promote preferential binding of single-stranded DNA (ssDNA) over double-stranded DNA (dsDNA), which influences RAD51 ATPase activity (14–16). Structural analysis of the Methanococcus voltae RAD51 (MvRAD51) has revealed cation-induced protein conformational rearrangements at the inter-subunit region that results in an active NPF (17, 18).
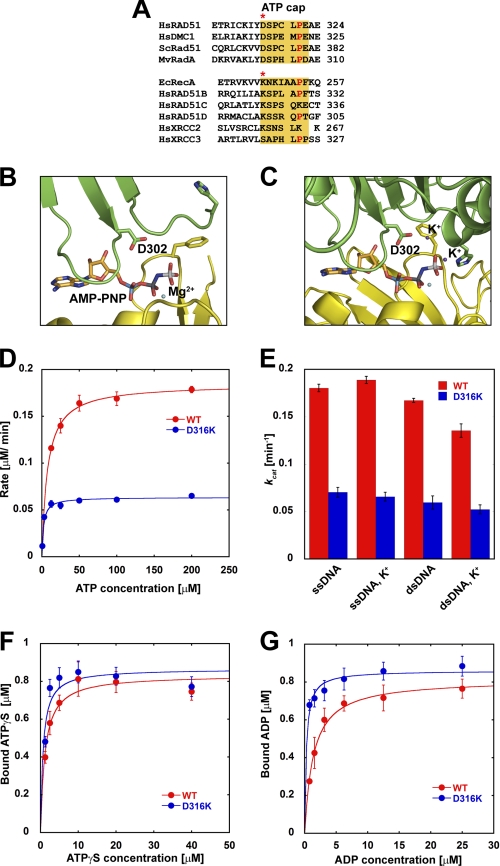
An evolutionarily intriguing feature of the RAD51/RecA NPF is the ATP cap located at the adenosine nucleotide-binding interface within the inter-subunit region (17, 18). A proline residue that is conserved in all RecA/RAD51 homologs appears to sandwich the adenine nucleotide at the ATP-binding interface (Fig. 1A). Five amino acids N-terminal to the conserved proline, the RAD51 homologs possess a conserved aspartate residue that forms a salt bridge with the γ-phosphate of ATP. In contrast, RecA homologs possess a conserved lysine at the analogous position that forms direct hydrogen bonds with the γ-phosphate (Fig. 1A, asterisk). Interestingly, four of the five HsRAD51 paralogs contain lysine at this corresponding position (Fig. 1A, asterisk).
FIGURE 1.
ATP binding and hydrolysis by HsRAD51 ATP cap substitution mutations. A, sequence alignment of the ATP cap region of Homo sapiens (Hs), Saccharomyces cerevisiae (Sc), M. voltae (Mv), and E. coli (Ec) recombinases. Sequence alignment of the ATP cap region of HsRAD51 paralogs is indicated below. The analogous position of HsRAD51(D302) is shown with an asterisk. B, ATP cap region of MvRadA (MvRAD51) structure in the absence of potassium cation (PDB code 1T4G). C, ATP cap region of MvRAD51 structure in the presence of potassium cation (PDB code 1XU4). Structural figures were generated using PyMOL. D, steady-state ATPase activity of HsRAD51 wild type or HsRAD51(D316K) with ssDNA in the presence of 150 mm KCl. E, ATP turnover values (kcat) with ssDNA and dsDNA in the presence and absence of KCl (K+). kcat values were calculated by Michaelis-Menten analysis. F, ATPγS binding by HsRAD51 wild type or HsRAD51(D316K) in the presence of ssDNA and 150 mm KCl. G, ADP binding by HsRAD51 wild type or HsRAD51(D316K) in the presence of ssDNA and 150 mm KCl. Error bars indicate standard deviation from at least three independent experiments.
Here, we examined the structure and function of the ATP cap lysine residue by constructing a substitution mutation where the conserved HsRAD51(Asp-316) was replaced by lysine (HsRAD51(D316K)). We found that the HsRAD51(D316K) substitution enabled the uncomplicated formation and maintenance of an active NPF. As a functional consequence, HsRAD51(D316K) displayed reduced ATP hydrolysis (ATPase), enhanced discrimination of ssDNA versus dsDNA, and significantly enhanced recombinase functions. Crystallographic and EM structural analysis indicate that MvRAD51(D302K) and HsRAD51(D316K) form a stable extended NPF in the absence of salt, which mimics salt-induced conformations of the wild type protein. Our results are consistent with the conclusion that the conserved aspartate in the ATP cap functions as a regulatory switch that enhances HsRAD51 NPF turnover and predict that analogous lysine-containing HsRAD51 paralogs may function to increase NPF stability. The enhanced stability and recombinase activity of hRAD51(D316K) in physiologically relevant conditions should provide a useful reagent for biochemical studies of HR.
EXPERIMENTAL PROCEDURES
HsRAD51 Protein Expression and Purification
The HsRAD51(D316K) was constructed using PCR mutagenesis using primers 5′-ATC TGC AAA ATC TAC AAA TCT CCC TGT CT and its complement for mutagenesis (lysine encoding codon in boldface type), and for cloning into the pET24d expression vector (Novagen) primers 5′-TAT ACC ATG GCA ATG CAG ATG CAG CTT GAA and 5′-TTC GGA TCC TTA TCA GTC TTT GGC ATC TCC CA were used that contain NcoI and BamHI restriction sites, respectively. For untagged native protein expression, stop codons were introduced upstream of the BamHI restriction site. Mutation was confirmed by DNA sequencing. HsRAD51 wild type and HsRAD51(D316K) proteins were expressed and purified following previously published protocols (16, 19). Briefly, HsRAD51 was expressed in Escherichia coli BLR strain and precipitated using spermidine-HCl. Resuspended pellet was purified using Reactive-Blue-4-agarose (Sigma), heparin-Sepharose (GE Healthcare), hydroxyapatite (Bio-Rad), and Mono Q (GE Healthcare) column chromatography. Purity of the fractions was verified by SDS-PAGE analysis. HsRPA was expressed in BL21(AI) cells using pET11d-tRPA purified as described previously (20), and except for resuspension of cells, HI buffer containing 30 mm HEPES (pH 7.5), 1 mm DTT, 0.25 mm EDTA, 0.25% (w/v) inositol, and 0.01% (v/v), Nonidet P-40 was supplemented with 100 mm KCl.
DNA Substrates
φX174 single-stranded (ss) virion DNA, replicative form I (RFI), was purchased from New England Biolabs. φX174 RFIII was obtained by linearizing RFI with ApaLI restriction enzyme and gel-purifying with QIAquick gel extraction kit (Qiagen). For surface plasmon resonance (SPR) analysis, a 5′-biotinylated oligo(dT)50 was used as ssDNA, and for dsDNA, 5′-biotinylated 50-mer 5′-TCG AGA GGG TAA ACC ACA-ATT ATT GAT ATA AAA TAG TTT TGG GTA GGC GA was annealed with its complement and purified by HPLC on a Gen-Pak FAX column (Waters). For competition DNA-binding experiments, an oligo(dT)50 ssDNA and a 50-bp dsDNA were used, made by annealing the 50-mer 5′-AGA TCT ATA AAC GCA CCT TTG GAA GCT TGG AAG TGG GCC GAA TCT CCC CA with its complement followed by HPLC purification as above. D-loop assay substrates were prepared as described previously (21).
ATPase Assay
ATP hydrolysis was measured as described previously (16). Reactions (10 μl) were performed in Buffer A containing 20 mm HEPES (pH 7.5), 10% glycerol, 100 μg/ml BSA, 1 mm DTT, and 1 mm MgCl2 and, when indicated, 150 mm KCl. Each reaction mixture contained HsRAD51 (1 μm) and 6 μm (nt or bp) of φX174 ssDNA or dsDNA. Reactions were initiated by the addition of protein and were incubated at 37 °C for 1 h. The reaction was stopped by the addition of 400 μl of 10% activated charcoal supplemented with 10 mm EDTA and incubated on ice for 2 h. Following centrifuging for 10 min, 50-μl duplicate aliquots were taken for counting free [32P]phosphate by the Cerenkov method. Kinetic parameters were obtained by fitting data into the Michaelis-Menten equation using Kaleidagraph software (Synergy).
ATPγS/ADP Binding Assay
Experimental procedure was as described previously (16). Reactions were performed in Buffer A and 150 mm KCl, when indicated. HsRAD51 (1 μm) and φX174 ssDNA (6 μm nt) were used with the indicated amount of [γ-35S]ATPγS or [3H]ADP. Following a 30-min incubation at 37 °C, reactions were kept on ice until filtered. The reaction mix was added into 4 ml of ice-cold reaction buffer and filtered through HAWP nitrocellulose filters (Millipore) presoaked in the same buffer. Another 4 ml of buffer was used to wash the membrane, and the filter was dried for 2 h. Radioactivity was counted after an overnight incubation in liquid scintillation fluid.
ADP-ATP Exchange Assay
Reactions were performed in Buffer A with 150 mm KCl. HsRAD51 (1 μm) and φX174 ssDNA (6 μm nt) were incubated with 3 μm [3H]ADP in a 60-μl reaction volume at 37 °C for 10 min. ADP → ATP exchange was initiated by addition of cold ATP to a 5 mm final concentration. Aliquots (10 μl) were withdrawn at indicated time points, filtered, and analyzed as in ADP binding.
SPR Analysis
Biotinylated DNA was immobilized on a streptavidin-coated chip (GE Healthcare) for the analysis. Protein binding and dissociation were analyzed at 25 °C with a 5 μl/min flow rate on a Biacore 3000 (GE Healthcare). Reactions were performed in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, and 1 mm MgCl2 supplemented, 0.005% surfactant P-20 (GE Healthcare), 2.5 mm of the indicated nucleotide, and 150 mm KCl when indicated. For experiments with Ca2+, 1 mm CaCl2 was used instead of MgCl2. DNA binding was determined by injecting 100, 200, 400, and 800 nm and 1.6 μm of protein simultaneously into ssDNA and dsDNA containing flow channels to ensure saturated binding.
SPR data were performed at five different concentrations for each mutant under each set of conditions, and fit to a single exponential decay to obtain rate constants for dissociation (koff). The decay curve, foff(t), is given by Equation 1,
where A(C) is the amplitude, which depends on the concentration (C) of HsRAD51, and t is the time. In some cases, binding is insufficient and/or dissociation occurs very rapidly and cannot be accurately determined by SPR. For these cases we simply report not determinable for the value.
Gel Mobility Shift Assay
HsRAD51 wild type or HsRAD51(D316K), at indicated concentrations, was incubated with 32P-labeled oligo(dT)50 (81 nm nt) in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm ATP, and 150 mm KCl at 37 °C for 15 min. Samples were then kept on ice until resolved by 5% nondenaturing PAGE at 10 V/cm in 0.3× TBE buffer at 4 °C. Gels were dried and exposed to PhosphorImager (GE Healthcare) screens for imaging.
Competition DNA Binding Analysis
HsRAD51 wild type or HsRAD51(D316K) (730 nm) was incubated with 32P-labeled oligo(dT)50 (81 nm nt) and the indicated amounts (in bp) of 50-bp cold dsDNA in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm ATP and, when indicated, 150 mm KCl, at 37 °C for 15 min. Samples were kept on ice until resolved by 5% nondenaturing PAGE at 10 V/cm in TBE buffer at 4 °C. Dried gels were exposed to PhosphorImager (GE Healthcare) screens for quantification using ImageQuant software (GE Healthcare).
D-loop Assay
Labeled 90-mer (2.4 μm nt) was incubated with HsRAD51 (0.8 μm) in Buffer A with 1 mm ATP supplemented with the indicated amounts of MgCl2 or CaCl2 at 37 °C for 10 min. Reaction was initiated by adding supercoiled pBS-SK(−) DNA (35 μm bp) and incubating for an additional 15 min. Samples were deproteinized by addition of 1% SDS and 1 mg/ml proteinase K (final concentrations) and incubated for 15 min at 37 °C, mixed with 0.2 volume of gel loading dye and analyzed on 0.9% agarose gel in TAE buffer, run at 4 V/cm at 25 °C. D-loops were quantified using ImageQuant software (GE Healthcare) after drying and exposure to PhosphorImager screens.
DNA Strand Exchange Assay
HsRAD51 (5 μm) and φX174 circular ssDNA (30 μm nt) were preincubated in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm ATP at 37 °C for 5 min before addition of 150 mm of the indicated salt and linear φX174 dsDNA (15 μm bp). Following an additional 5-min incubation, HsRPA (2 μm) was added, and the incubation was continued. After 3 h, the samples were deproteinized by addition of 3 μl of stop buffer containing 2% SDS and 10 mg/ml proteinase K and analyzed on 0.9% agarose gels in TAE buffer. Electrophoresis was carried out at 4 V/cm at 25 °C with 0.1 mg/ml ethidium bromide. Gels were analyzed on a gel documentation station (Bio-Rad). For quantification of joint molecules ImageJ software (rsbweb.nih.gov) was utilized.
For strand exchange reaction similar to historical RecA conditions, φX174 circular ssDNA (5 μm nt) and linear φX174 dsDNA (5 μm bp) were preincubated with HsRAD51(5 μm) in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm ATP and 150 mm of the indicated salt (when added) at 37 °C for 5 min. After 5 min, HsRPA (2 μm) was added and analyzed as described above.
Electron Microscopy
The typical DNA binding reaction for electron microscopy was performed in 20 μl of strand exchange buffer containing either no salt, 100 mm ammonium sulfate, or 200 mm potassium chloride, and 5 ng/μl concentration of the substrate DNA (either ss or ds M13 DNA) and HsRad51 at three monomers per nucleotide or nucleotide base pair. The reactions were carried out at 37 °C for 15 min. For negative staining, an aliquot of the sample was diluted 1:4 in buffer, immediately adsorbed to glow-charged thin carbon foils for 3 min followed by staining with 2% unbuffered uranyl acetate for 5 min, and followed by air drying. For direct mounting, the remaining sample volume was passed over 2-ml columns of 6% agarose beads (ABT Inc., Burgos, Spain) equilibrated with TE buffer (10 mm Tris-HCl (pH 7.4), 0.1 mm EDTA/NaOH) to eliminate unwanted salts and unbound proteins, and fractions enriched for DNA-protein complexes were collected. Aliquots were mixed with a buffer containing spermidine and adsorbed onto copper grids coated with a thin carbon film glow-charged shortly before sample application. After adsorption of the samples for 2–3 min, the grids were dehydrated through a graded ethanol series and rotary shadowcast with tungsten at 10−7 torr. Samples were examined in an FEI T12 TEM and a Philips CM12 TEM equipped with Gatan 2kx2k SC200 CCD cameras at 40 kV (shadowcast samples) or at 80 kV (stained samples). Dimensions of particles in the images saved from the CCD cameras were analyzed using Digital Micrograph software (Gatan, Inc.). Adobe Photoshop software was used to arrange images into panels for publication.
MvRAD51 Protein Preparation and Crystallization
The recombinant protein was overexpressed in BL21 Rosetta2 (DE3) cells (Novagen). In addition to the D302K mutation, this recombinant protein also carried an N4G mutation and lacked the first three amino acid residues. The soluble protein was purified as reported for the wild type MvRAD51 protein (17, 22). In brief, the purification procedure involved steps of polymin P (Sigma) precipitation, high salt extraction, and three chromatography steps using heparin (Amersham Biosciences), hydroxyapatite (Bio-Rad), and Sephacryl S-300 gel filtration (GE Healthcare) columns. The purified MvRAD51(D302K) protein was concentrated to ∼30 mg/ml by ultrafiltration.
The hexagonal MvRAD51(D302K) crystals (P61 space group) were grown by the hanging drop vapor diffusion method. The maximum dimension was 0.2 × 0.2 × 0.3 mm. The sample contained ∼1 mm of concentrated protein and 2 mm AMP-PNP. The reservoir solutions contained 4–5% polyethylene glycol (PEG) 3,350, 50 mm MgCl2, 50 mm Tris-HCl (pH 7.9), 0.4 m NaCl, and 14% (w/v) sucrose. One crystal was transferred into a stabilizing solution composed of the reservoir solution supplemented with 28% (w/v) sucrose, looped out of the solution, and frozen in a nitrogen cryo-stream at 100 K. The diffraction dataset was acquired at the Canadian Light Source beamline 08ID-1 and was processed using the XDS program (23). The statistics of the diffraction data are listed in supplemental Table 1.
The previously solved wild type MvRAD51 model (PDB code 1T4G) was used as the starting model. After initial refinement, the electron density of the side chain of Lys-302 was clearly visible in the resulting 2Fo − Fc map. The model was then iteratively rebuilt using XtalView (24) and refined using CNS. The molecular figures were generated using PyMOL (25). The coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 3NTU).
RESULTS
HsRAD51(D316K) Substitution Affects ATPase but Not Adenosine Nucleotide Binding or Exchange
Comparison of RecA and RAD51 homologs identified a number of conserved residues (Fig. 1A). The HsRAD51(Asp-316) residue was examined to test the effect of lysine in the ATP cap that is conserved in RecA and most HsRAD51 paralogs (Fig. 1A, red asterisk). We purified the HsRAD51(D316K) protein (supplemental Fig. 1A) and examined the steady-state ATPase activity (Fig. 1D; Table 1). We found that the ATPase rate was ∼3-fold lower than the wild type protein. Our results are also consistent with previous studies that have suggested stimulatory cations may modestly enhance the discrimination between ssDNA- and dsDNA-stimulated ATPase (Fig. 1E, compare ssDNA with dsDNA ATPase with and without K+) (14, 15). However, we observe a reduced cation-induced difference in ssDNA- and dsDNA-stimulated ATPase with HsRAD51(D316K) compared with the wild type protein (Fig. 1E).
TABLE 1.
Summary of ATP hydrolysis and nucleotide binding data of HsRAD51 wild type and HsRAD51(D316K) mutant protein
| Kinetic parameter | WT | D316K |
|---|---|---|
| kcat (min−1) | ||
| ssDNA | 0.180 ± 0.004 | 0.070 ± 0.005 |
| ssDNA, KCI | 0.190 ± 0.004 | 0.063 ± 0.005 |
| dsDNA | 0.167 ± 0.002 | 0.060 ± 0.007 |
| dsDNA, KCI | 0.135 ± 0.007 | 0.052 ± 0.005 |
| Km (μm) | ||
| ssDNA | 17.48 ± 3.25 | 3.08 ± 0.10 |
| ssDNA, KCI | 9.34 ± 1.42 | 2.14 ± 0.002 |
| dsDNA | 6.54 ± 0.67 | 2.17 ± 0.15 |
| dsDNA, KCI | 5.52 ± 0.26 | 1.70 ± 0.003 |
| KD (μm) ATPγS | ||
| ssDNA | 2.49 ± 0.46 | 0.66 ± 0.21 |
| ssDNA, KCI | 1.18 ± 0.22 | 0.72 ± 0.21 |
| Bmax (μm) ATPγS | ||
| ssDNA | 1.02 ± 0.02 | 0.90 ± 0.03 |
| ssDNA, KCI | 0.83 ± 0.04 | 0.87 ± 0.05 |
| KD (μm) ADP | ||
| ssDNA, KCI | 1.24 ± 0.32 | 0.26 ± 0.06 |
| Bmax (μm) ADP | ||
| ssDNA, KCI | 0.82 ± 0.05 | 0.86 ± 0.02 |
The difference in HsRAD51(D316K) ATPase compared with the wild type HsRAD51 was not the result of altered ATP binding activity (Fig. 1F; Table 1), and there appeared to be no defect associated with ADP → ATP exchange, although kinetic differences at time points shorter than 30 s would not have been detected (supplemental Fig. 1B). We observed an ∼4-fold increase in KD values for ADP in the presence of ssDNA compared with the wild type HsRAD51 (Fig. 1G; Table 1). Importantly, the protein preparations appeared fully active because the ATP and ADP maximum binding (Bmax) as well as 1:1 stoichiometry appeared equivalent under a variety of conditions (Fig. 1, F and G; Table 1). These observations suggest that defects in adenosine nucleotide binding and exchange are unlikely to fully account for the reduced HsRAD51(D316K) ATPase activity.
HsRAD51(D316K) Exhibits RPA and Salt-independent Strand Exchange Activity
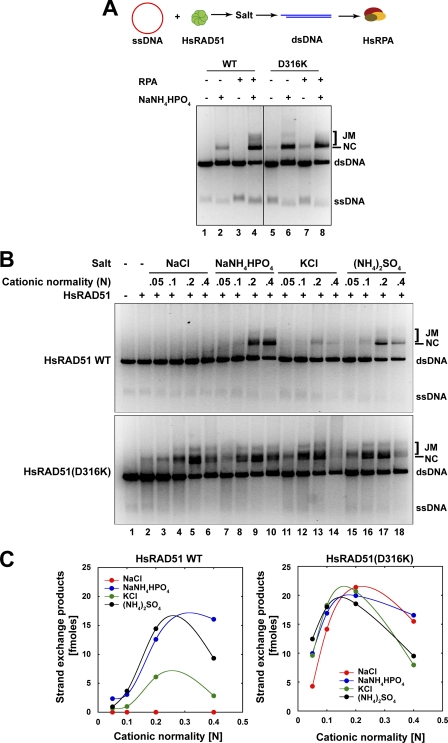
RPA facilitates RAD51-mediated strand exchange at both presynaptic and postsynaptic stages (21, 26, 27). Efficient strand exchange is facilitated by the NH4+ cation, which is a characteristic of RAD51 even though such conditions are unlikely to be physiologically relevant (13–15). We compared the strand exchange activity of wild type HsRAD51 and HsRAD51(D316K) in the presence of HsRPA and NH4+ cation (Fig. 2). Remarkably, HsRAD51(D316K) appeared significantly more active than wild type HsRAD51 and was capable of catalyzing efficient strand exchange in the absence of HsRPA (Fig. 2A). Moreover, we still observed strand exchange products in the absence of both HsRPA and NH4+ cation (Fig. 2A). These studies suggest that HsRAD51(D316K) may circumvent both the cation and ssDNA binding protein requirements of wild type HsRAD51.
FIGURE 2.
HsRAD51(D316K) exhibits salt and RPA-independent strand exchange activity. A, analysis of salt and RPA requirement for strand exchange. Reaction schematic shown at top, HsRAD51 wild type or HsRAD51(D316K) (5 μm) and φX174 circular ssDNA (30 μm nt) were preincubated in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm of ATP at 37 °C for 5 min prior to the addition of 150 mm NaNH4HPO4 (if indicated) and linear φX174 dsDNA (15 μm bp). After 5 min, HsRPA (2 μm) was added (if indicated), and the incubation was continued for 3 h. Samples were deproteinized and analyzed on 0.9% agarose gel with 0.1 μg/ml ethidium bromide. Single-stranded DNA substrate (ssDNA), double-stranded DNA substrate (dsDNA), nicked circular product (NC), and joint molecule product (JM) are labeled on the right. B, strand exchange activity of HsRAD51 and HsRAD51(D316K) as a function of cation normality. HsRAD51 (5 μm) and φX174 circular ssDNA (30 μm nt) were preincubated with 2.5 mm ATP and 1 mm MgCl2 at 37 °C for 5 min before addition of indicated amounts of salt and linear φX174 dsDNA (15 μm bp). After another 5 min of incubation HsRPA (2 μm) was added, and the incubation was continued. After 3 h, samples were deproteinized and analyzed on 0.9% agarose gel with 0.1 μg/ml ethidium bromide. ssDNA, dsDNA, nicked circular product (NC), and joint molecule product (JM) are labeled on the right. C, quantification of the nicked circular (NC) + joint molecules (JM) products in B.
We examined the cation requirements of HsRA51(D316K) and observed efficient strand exchange activity in the presence of sodium (Na+) and K+ cations (Fig. 2B). Moreover, peak strand exchange activity with HsRAD51(D316K) appears to occur with approximately half the NH4+ or K+ cation concentration compared with the wild type HsRAD51 (Fig. 2C).
The enhanced strand exchange activity and reduced ssDNA binding protein requirement of HsRAD51(D316K) appeared similar to bacterial RecA. The order-of-addition requirements for efficient RecA strand exchange are different from RAD51, the latter of which requires a series of steps that include the pre-formation of an NPF on the ssDNA substrate prior to the addition of salt and then the dsDNA donor. In contrast, all of the reaction components, including ssDNA and dsDNA substrates, may be included with RecA, and there is no unusual cation requirement. Remarkably, HsRAD51(D316K) appears to efficiently catalyze strand exchange under RecA order-of-addition in the presence of K+ and significantly less efficiently in the presence of NH4+ cation (Fig. 3A). These studies suggest that HsRAD51(D316K) may catalyze fundamental recombination reactions under more physiologically relevant conditions.
FIGURE 3.
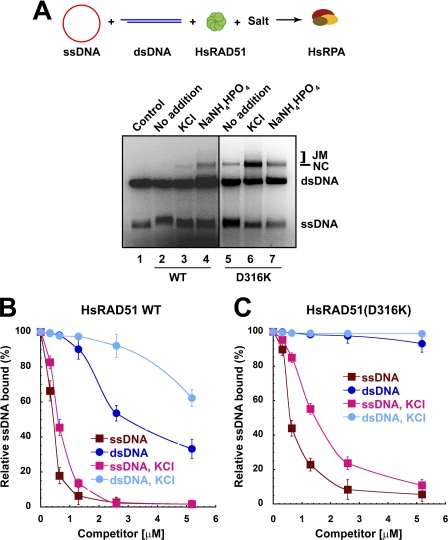
HsRAD51(D316K) preferentially binds to single-stranded DNA. A, strand exchange in RecA format. φX174 circular ssDNA (5 μm nt) and linear φX174 dsDNA (5 μm bp) were preincubated with HsRAD51 wild type or HsRAD51(D316K) (5 μm) in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm ATP and 150 mm KCl at 37 °C for 5 min. After 5 min, HsRPA (2 μm) was added, and the incubation was continued. After 3 h, samples were deproteinized and analyzed on 0.9% agarose gel with 0.1 μg/ml ethidium bromide. ssDNA, dsDNA, nicked circular product (NC), and joint molecule product (JM) are labeled on the right. B, quantification of competition DNA binding analysis (see supplemental Fig. 2) of HsRAD51 wild type and HsRAD51(D316K). HsRAD51 wild type or HsRAD51(D316K) (730 nm) was incubated with 32P-labeled oligo(dT)50 (81 nm nt) and the indicated amounts of 50-bp cold competitor dsDNA in buffer containing 20 mm HEPES (pH 7.5), 10% glycerol, 1 mm DTT, 1 mm MgCl2 supplemented with 2.5 mm of ATP and when indicated, 150 mm KCl, at 37 °C for 15 min. Samples were kept on ice until resolved by 5% nondenaturing PAGE at 10 V/cm in TBE buffer at 4 °C. Dried gels were exposed to PhosphoImager screens for quantification. C, competition DNA binding with oligo(dT)50 cold competitor DNA. Reaction conditions are similar to B.
A gel-based competition analysis was used to examine the relative binding of HsRAD51 to ssDNA and dsDNA (Fig. 3, B and C). In this system HsRAD51 induces an ssDNA NPF gel shift that can be used in a competition analysis with cold ssDNA or dsDNA competitor (supplemental Fig. 2, A–C). As suggested previously for wild type HsRAD51, salt (KCl) dramatically decreases the ability of cold dsDNA to compete for labeled ssDNA binding while modestly affecting the ability of cold ssDNA to compete for labeled ssDNA binding (Fig. 3B, compare WT with WT-KCl) (14). In contrast, the HsRAD51(D316K) ssDNA NPF is largely stable to cold dsDNA competition regardless of salt (KCl), whereas the ssDNA binding appears further enhanced to cold ssDNA competition in the presence of salt (KCl). These results strongly suggest that the HsRAD51(D316K) naturally displays a substantial discrimination between ssDNA and dsDNA that the wild type HsRAD51 only displays in the presence of salt.
We also note that the HsRAD51(D316K) mutant protein is unable to catalyze strand exchange in the presence of ATPγS, yet it performs modest strand exchange in the presence of ADP (supplemental Fig. 3). Because ATPγS binding by wild type HsRAD51 and HsRAD51(D316K) appears identical, it is likely that this poorly hydrolysable ATP analog induces a conformation that is incompatible with strand exchange activity. Together, these results are consistent with the conclusion that the HsRAD51(D316K) substitution significantly alters the salt requirement for triggering an appropriate adenosine nucleotide-dependent conformational transition necessary for strand exchange (see Fig. 1C).
HsRAD51(D316K) Affects Protein Turnover
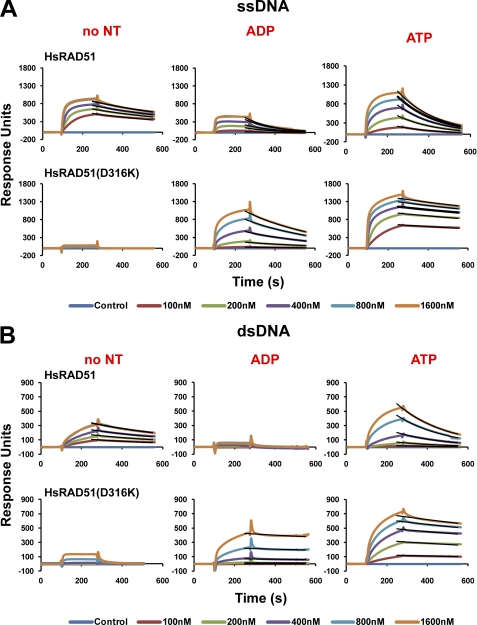
RAD51 DNA interaction(s) during pre- and postsynaptic stages is believed to play an important role during HR (28–30). We quantitatively examined the DNA binding and dissociation properties of the HsRAD51 substitution mutant proteins using SPR and biotin-streptavidin-immobilized dT50 ssDNA or 50-bp dsDNA. These studies were performed in physiologically relevant salt (150 mm KCl) with magnesium and ATP similar to strand exchange conditions (see Fig. 2). The protein was titrated between 100 nm and 1.6 μm to examine saturated binding on both ssDNA and dsDNA. After protein injection and NPF formation, the sample was analyzed for steady-state protein disassembly (Fig. 4).
FIGURE 4.
HsRAD51(D316K) displays a slow turnover from ssDNA and dsDNA. A, ssDNA binding analysis of wild type and HsRAD51(D316K) by SPR (Biacore) in the presence of the indicated nucleotide. Association and dissociation curves correspond to 100, 200, 400, and 800 nm and 1.6 μm of protein. Fitted cures are overlaid in black lines. B, dsDNA binding analysis of wild type and HsRAD51(D316K) in the presence of the indicated nucleotide. Protein concentrations are similar to A.
Previous studies have suggested a multimeric RAD51 binding that would significantly alter the simple concentration dependence of kon (31). A power dependence of concentration on kon would make any calculations of KD problematic. In contrast, the koff behaves as a simple single exponential decay and appears highly accurate for a variety of conditions.
The saturation and shape of the binding curves varied considerably for the wild type HsRAD51 as predicted by previous studies (14). Two general observations emerged. 1) HsRAD51 does not bind dsDNA in the presence of ADP; and 2) the HsRAD51 koff increases 2–4-fold in the presence of adenosine nucleotide with the koff ·ADP > koff·ATP (Fig. 4 and Table 2). These results are consistent with previous studies that suggested increased RecA and RAD51 turnover upon ATP hydrolysis (29, 32–35).
TABLE 2.
Dissociation rate constants for HsRAD51 WT and HsRAD51(D316K) from ssDNA and dsDNA
The statistical error of fitting was less that 0.00003 s−1 for each measure. However, binding drift suggested accuracy of ±10%.
| WT | D316K | |
|---|---|---|
| s−1 | s−1 | |
| ssDNA, no NTa | 0.0014 | NDb |
| ssDNA, ADP | 0.0070 | 0.0027 |
| ssDNA, ATP | 0.0053 | 0.0005 |
| dsDNA, no NT | 0.0017 | ND |
| dsDNA, ADP | ND | 0.0004 |
| dsDNA, ATP | 0.0041 | 0.0006 |
a NT means nucleotide.
b ND means not determinable as a result of low binding.
The HsRAD51(D316K) is completely deficient for both ssDNA and dsDNA binding in the absence of adenosine nucleotide (Fig. 4). Interestingly, RecA binds to ssDNA but does not bind to dsDNA in the absence of adenosine nucleotide, whereas yeast Rad51 binds to dsDNA but does not bind to ssDNA in the absence of adenosine nucleotide (36–39). In contrast, HsRAD51(D316K) binds strongly to ssDNA and dsDNA in the presence of both ADP or ATP (Fig. 4 and Table 2). Moreover, the stability of the NPF is increased 7–10-fold compared with the wild type HsRAD51 in the presence of ATP (koff·ATP·ssDNA = 0.0005 s−1; koff·ATP·dsDNA = 0.0006 s−1; see Table 2). Prolonged analysis confirmed the stability of the NPF (data not shown). The decreased koff as well as the ability to bind dsDNA in the presence of ADP suggests that the HsRAD51(D316K) mutant protein displays a significantly reduced adenosine nucleotide-induced DNA turnover compared with wild type HsRAD51.
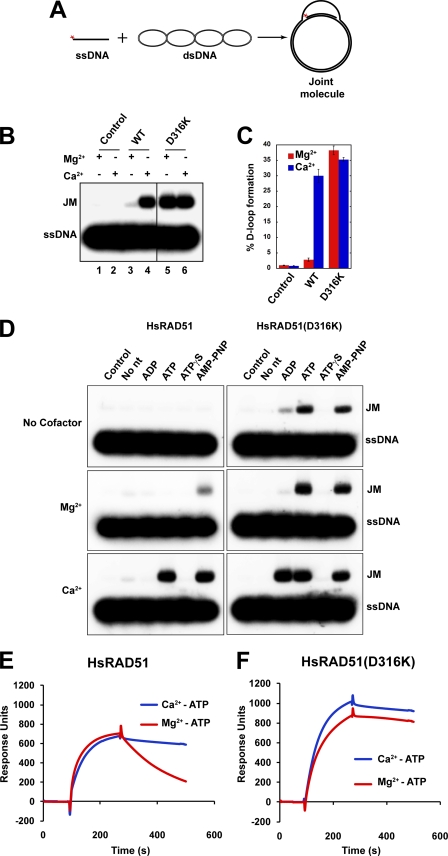
HsRAD51(D316K) Catalyzes Efficient D-loop Formation Independent of Divalent Cation
RAD51 catalyzes the formation of D-loop joint molecules in the presence of calcium (Ca2+) or nonhydrolyzable ATP analogs (29, 32). Moreover, the P-loop substitution mutation of HsRAD51(K133R) that binds ATP but renders the protein ATPase-deficient was found to catalyze efficient D-loop formation in the presence of magnesium (Mg2+) (29). Together, these results are consistent with the conclusion that the catalysis of D-loop joint molecules by RAD51 requires the formation and maintenance of an ATP-bound NPF, which can be enhanced by substitution of Ca2+ for Mg2+ or by using a hydrolysis-deficient HsRAD51 mutation. The intrinsic stability of the HsRA51(D316K) NPF suggested it might be more efficient at catalyzing D-loop formation.
As expected, we found that HsRAD51 catalyzes the formation of D-loops in the absence of salt but requires Ca2+ to induce an active ATP-bound NPF (33). The yield of D-loops by wild type HsRAD51 in the presence of Mg2+ divalent cation was 8.1 fmol (3%) and 81 fmol (30%) in the presence of Ca2+ (Fig. 5, B, lanes 3 and 4, and C). Importantly, HsRAD51(D316K) catalyzed the formation of D-loops in the absence of any divalent cation or in the presence of either Mg2+ or Ca2+ (107 fmol (38%) and 94 fmol (35%), respectively; Fig. 5, B, lanes 5 and 6, and C and D). Kinetic analysis revealed an increase in the initial rate of D-loop formation by HsRAD51(D316K) (data not shown). D-loop catalysis by HsRAD51(D316K) in the presence of the nonhydrolyzable nucleotide analog AMP-PNP appeared similar to the ATP-driven catalysis regardless of divalent cation cofactor (Fig. 5D). Moreover, in the presence of Ca2+ the HsRAD51(D316K) mutant protein appeared capable of efficiently catalyzing D-loop formation with ADP (Fig. 5D).
FIGURE 5.
HsRAD51(D316K) catalyzes D-loop formation in the presence of ATP and magnesium. A, reaction schematic. B, HsRAD51(0.8 μm) wild type or HsRAD51(D316K) and 32P-labeled ssDNA (90-mer; 2.4 μm nt) were preincubated for 10 min at 37 °C in the reaction buffer containing 1 mm ATP and 1 mm MgCl2 or CaCl2. Reactions were initiated by the addition of supercoiled pBS SK(−) plasmid DNA (35 μm bp). After 15 min, reactions were terminated by the addition of deproteinization solution and processes similar to Fig. 2. Joint molecules (JMs) were analyzed on a 0.9% agarose gel. C, quantification of joint molecules in B. Error bars indicate standard deviation from at least three independent experiments. D, D-loop formation analysis with different nucleotides. HsRAD51 (0.8 μm) wild type or HsRAD51(D316K) and 32P-labeled ssDNA (90-mer; 2.4 μm nt) were preincubated for 10 min at 37 °C in the reaction buffer containing 1 mm of the indicated nucleotide and 1 mm MgCl2, CaCl2, or without a cofactor. Reactions were initiated and analyzed as described in B. E, ssDNA binding analysis of wild type HsRAD51 using SPR in the presence of Mg2+-ATP and Ca2+-ATP. F, ssDNA binding analysis of HsRAD51(D316K) in the presence of Mg2+-ATP and Ca2+-ATP.
We examined the stability of the wild type HsRAD51 and HsRAD51(D316K) presynaptic NPF on ssDNA in the presence of ATP and either Mg2+ or Ca2+ (Fig. 5, E and F). A significant decrease in koff was observed for wild type HsRAD51 in the presence of Ca2+ as well as for HsRAD51(D316K) in the presence of both Ca2+ and Mg2+ (Fig. 5, E and F). These results strongly suggest that HsRAD51(D316K) inherently forms and maintains an active ATP-bound NPF regardless of associated divalent cations. Collectively, these results underline previous conclusions that the ability to form and maintain an active adenosine nucleotide-bound NPF conformation drives D-loop formation (33, 37, 40).
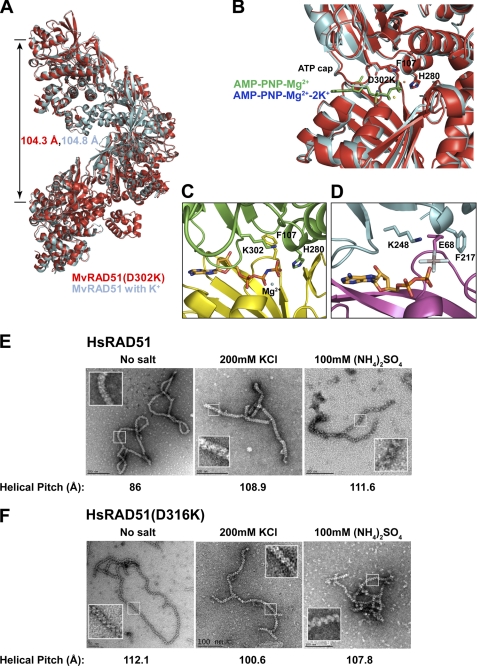
MvRAD51(D302K) Induces Active Nucleoprotein Filament
We determined the structure of the MvRAD51(D302K) at 1.9-Å resolution (supplemental Table 1). The MvRAD51(D302K) protein forms a filament with a helical pitch of 104.3 Å, similar to the helical pitch of active wild type MvRadA (104.8 Å; Fig. 6A). The L2 single strand binding region (Asp-256 to Arg-285) of MvRAD51(D302K) is largely ordered. The nonhydrolysable ATP analog, AMP-PNP, is buried between RadA monomers as expected (Fig. 6B). The adenosine nucleotide moiety is sandwiched between the ATP cap (residues Lys-302 to Asp-308) and the side chain of Arg-158. The amino group of the Lys-302 side chain forms a total of four hydrogen bonds as follows: one with the terminal phosphate of the ATP analog and three with the carbonyls of residues Gly-279, His-280, and Ala-282. These four groups stabilized a Ca2+ cation in a previously reported calcium-containing active form of MvRAD51 (41). The Lys-302 appears to provide further stabilization of the γ-phosphate in addition to the well known P-loop residues and Mg2+ cation (17). Importantly, the Lys-302 side chain replaces both K+ cations, including one that normally bridges the wild type Asp-302 with the γ-phosphate of ATP and additionally coordinates an ordered helix (residues Gly-275 to Ala-282) analogous to helix G of EcRecA (Fig. 6C) (42). The His-280 side chain makes a direct hydrogen bond with the γ-phosphate of the ATP analog, and Phe-107 is flipped away from the nucleotide binding pocket as is required to form an active MvRAD51 NPF. Interestingly, the Lys-248 of the E. coli RecA-active nucleoprotein filament is at the analogous position of Lys-302 (Fig. 6D) (43). The structure of MvRAD51(D302K) closely resembles the recurrent active form of M. voltae and Methanococcus maripaludis RAD51·AMP-PNP complexes (18, 44, 45) but differs from the recurrent inactive form of MvRAD51 (17), which has a largely disordered L2 region (compare Fig. 6C with Fig. 1, B and C).
FIGURE 6.
Structural comparison of MvRAD51 and RAD51 ATP cap Asp → Lys substitution mutation. A, superimposed filament assembly of active wild type MvRAD51 structure in the presence of K+ cations (cyan) (PDB code 1XU4) and MvRAD51(D302K) (red) (PDB code 3NTU). Helical pitch is indicated with the respective color. B, enlarged view of the intersubunit ATP binding region of active wild type MvRAD51 and MvRAD51(D302K). Relative location of AMP-PNP-Mg2+-2K+ of the wild type MvRAD51 (blue) and the AMP-PNP-Mg2+ of the MvRAD51(D302K) (green) are shown. C, ATP cap region of MvRadA(D302K). Phe-129 and His-280 are in active form conformation. γ-phosphate group of ATP analog forms a direct interaction with ϵ-amino group of Lys-302. D, active filament form of E. coli RecA. Lys-248 of the ATP cap forms a direct contact with the ALF4− group of the ATP analog (PDB code 3CMU). Electron microscopy analysis of nucleoproteins filaments of wild type HsRAD51 (E) and HsRAD51(D316K) (F) with ssDNA. Reaction conditions and product visualization are described under “Experimental Procedures.” Reaction conditions are indicated above each EM image, and the corresponding helical pitch is indicated below. Insets indicate enlarged images of the helical configuration.
HsRAD51(D316K) Maintains an Active Nucleoprotein Filament in the Absence of Salt
To confirm the structural analysis of MvRAD51(D302K) we examined the wild type HsRAD51 and HsRAD51(D316K) NPFs by electron microscopy (EM) (Fig. 6, E and F). These studies were performed in strand exchange conditions with ATP in the absence of salt or in the presence of equi-normal K+ or NH4+ cations. The wild type HsRAD51 protein exhibited a 30% increase in the helical pitch with visible striation of the filament in the presence of K+ or NH4+ (Fig. 6E). In contrast, HsRAD51(D316K) maintains an inherently extended conformation (helical pitch of 100–112Å) similar to the MvRadA(D302K) filament and the salt-induced wild type HsRAD51 filament under all conditions (Fig. 6F). These studies are consistent with the biochemical and structural analyses that suggest HsRAD51(D316K) forms and maintains an active adenosine nucleotide bound NPF under a wide range of conditions.
DISCUSSION
The ATP cap of RecA/RAD51 homologs is a structurally conserved region that overlays the ATP-binding site of an adjacent protomer within an NPF. It contains a highly conserved proline residue that functions to sandwich the adenine moiety (Fig. 1A). RAD51 homologs possess a conserved aspartate residue adjacent to the conserved proline in the ATP cap, whereas RecA homologs and a large majority of RAD51 paralogs have a lysine at the analogous position (Fig. 1A). Both the aspartate and lysine residues ultimately contact the γ-phosphate of ATP (18, 43). The lysine residue does this directly, whereas the cation of the aspartate salt bridge mediates the γ-phosphate contact. Conformational transition(s) of the MvRAD51 Phe-107 and His-280 residues have been shown to be associated with the development of the aspartate salt bridge, which together appear necessary to form an active NPF (18). The available data suggest that the cation that occupies this salt bridge must have a sufficient ionic radius to provoke the formation and maintenance of an active ATP-bound NPF (14). In its absence or under conditions in which ATP hydrolysis may readily occur, the RAD51 NPF appears largely inactive. Taken together, these results suggest that the aspartate salt bridge plays a significant role in the management and regulation of RAD51 recombinase functions.
In contrast, RecA homologs and RAD51 containing a lysine residue in the analogous position appear to more easily form and maintain an active NPF that translates to enhanced recombinase function(s) (43). The majority of RAD51 paralogs contain a lysine residue in this analogous position (Fig. 1A). The human RAD51 paralogs have been purified in a variety of heteromeric forms (9, 10, 12, 46). However, the quantities of these heteromeric forms and their tendency to aggregate have thus far inhibited a complete examination of their biophysical and structural role in HR. The HsRAD51(D316K) substitution mutation has allowed us to examine the effect of a RAD51 protein containing an ATP cap lysine on recombinase functions.
One possible prediction of this analysis is that RAD51 paralogs containing an ATP cap lysine may function to stabilize the RAD51 filament. Such a Rad51 NPF stabilization has recently been demonstrated for the yeast paralogs Rad55 to Rad57 (47). It is possible that near stoichiometric quantities of these RAD51 paralogs might be required to induce such stability because they would have to occupy a large fraction of the NPF. Of the human RAD51 paralogs, only HsRAD51B and HsRAD51C appear to be expressed in quantities equivalent or in excess of HsRAD51 (48). Interestingly, the HsRAD51B-HsRAD51C heterodimer appears prevalent in human cells, and both contain an ATP cap lysine (48). Moreover, both HsRAD51B and HsRAD51C display modest binding to ssDNA, and HsRAD51B contains the conserved proline in the ATP cap region (49).
Mechanism of ATP Cap Regulation
The mechanism associated with ATP cap regulation of RAD51 might be inferred from the MvRAD51 structure (18). In the MvRAD51 K+ cation-induced active NPF structure, the His-280 residue formed a hydrogen bond with the γ-phosphate of the ATP analog, whereas the Phe-107 residue was moved out of the ATP binding interface to accommodate two K+ cations (one forming the Asp-302 salt bridge). A similarly active MvRAD51 contains an offset calcium that occupies the space of both K+ cations but still forms a salt bridge with the γ-phosphate (41). Together, these observations suggest that activation of MvRAD51 requires salt occupation of the ATP cap and that cations, which disrupt the conformational changes associated with the ATP hydrolytic cycle, further enhance activity. The MvRAD51(Lys-302) and HsRAD51(Lys-316) residues appears to perform these functions naturally but without the ability to perform post-synaptic turnover.
RPA Independence
It has been suggested that RPA enhances strand exchange activity by disrupting ssDNA secondary structure formation and by stabilizing the newly formed joint molecule by binding to the newly released ssDNA (27, 50). Because HsRAD51(D316K) demonstrates RPA independence in catalyzing strand exchange, it is possible that this protein possesses an enhanced ability to disrupt secondary structures on ssDNA and/or it binds the ssDNA product more efficiently from the triplex joint molecule. Indeed, HsRAD51(D316K) binds ssDNA ∼15-fold better that the wild type HsRAD51 in the presence of ATP and ∼3-fold better than wild type HsRAD51 in the presence of ADP.
Evolution of Aspartate in ATP Cap
An intriguing evolutionary question is why RAD51 recombinases have selected an aspartate residue in the ATP cap region instead of a lysine found in bacterial RecA? One possibility is that the preservation of the aspartate residue and associated salt bridge fundamentally provides the turnover required during HR. Is the ability to efficiently recycle HsRAD51 from an expanding presynaptic filament during the construction of an active NPF or the release of HsRAD51 from a completed recombination product more important in eukaryotic genomes? In this scenario, the aspartate residue and associated salt bridge would function as a conformational sensor in delineating active and inactive protomers in the NPF before and/or after HR. Finally, the development of the HsRAD51(D316K) should significantly enhance biochemical studies of HR when physiologically relevant conditions are required.
Supplementary Material
Acknowledgments
We thank Michael Poirier and Charles Bell for helpful discussions. Crystallographic work was performed at the Canadian Light Source, which is supported by National Sciences and Engineering Research Counsel, National Research Counsel, Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University or Saskatchewan.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31819 and ES13773 (to J. G.) and GM080176 (to R. F.). This work was also supported by Canadian Institutes of Health Research Grant 63860 (to Y. L.) and National Science Foundation Grant DMR-0706002 (to R. B.).

This article contains supplemental Table 1 and Figs. 1–3.
The atomic coordinates and structure factors (code 3NTU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HR
- homologous recombination
- NPF
- nucleoprotein filament
- ssDNA
- single-stranded DNA
- PDB
- Protein Data Bank
- nt
- nucleotide
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- SPR
- surface plasmon resonance
- RPA
- replication protein A.
REFERENCES
- 1. Bernstein K. A., Rothstein R. (2009) At loose ends. Resecting a double strand break. Cell 137, 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ciccia A., Elledge S. J. (2010) The DNA damage response. Making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jackson S. P., Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krogh B. O., Symington L. S. (2004) Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271 [DOI] [PubMed] [Google Scholar]
- 5. West S. C. (2003) Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4, 435–445 [DOI] [PubMed] [Google Scholar]
- 6. Sonoda E., Sasaki M. S., Buerstedde J. M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsuzuki T., Fujii Y., Sakumi K., Tominaga Y., Nakao K., Sekiguchi M., Matsushiro A., Yoshimura Y., Morita T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 6236–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 9. Masson J. Y., Stasiak A. Z., Stasiak A., Benson F. E., West S. C. (2001) Complex formation by the human RAD51C and XRCC3 recombination repair proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 8440–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masson J. Y., Tarsounas M. C., Stasiak A. Z., Stasiak A., Shah R., McIlwraith M. J., Benson F. E., West S. C. (2001) Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 15, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schild D., Lio Y. C., Collins D. W., Tsomondo T., Chen D. J. (2000) Evidence for simultaneous protein interactions between human Rad51 paralogs. J. Biol. Chem. 275, 16443–16449 [DOI] [PubMed] [Google Scholar]
- 12. Shim K. S., Schmutte C., Tombline G., Heinen C. D., Fishel R. (2004) hXRCC2 enhances ADP/ATP processing and strand exchange by hRAD51. J. Biol. Chem. 279, 30385–30394 [DOI] [PubMed] [Google Scholar]
- 13. Sigurdsson S., Trujillo K., Song B., Stratton S., Sung P. (2001) Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem. 276, 8798–8806 [DOI] [PubMed] [Google Scholar]
- 14. Shim K. S., Schmutte C., Yoder K., Fishel R. (2006) Defining the salt effect on human RAD51 activities. DNA Repair 5, 718–730 [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Stasiak A. Z., Masson J. Y., McIlwraith M. J., Stasiak A., West S. C. (2004) Conformational changes modulate the activity of human RAD51 protein. J. Mol. Biol. 337, 817–827 [DOI] [PubMed] [Google Scholar]
- 16. Tombline G., Fishel R. (2002) Biochemical characterization of the human RAD51 protein. I. ATP hydrolysis. J. Biol. Chem. 277, 14417–14425 [DOI] [PubMed] [Google Scholar]
- 17. Wu Y., He Y., Moya I. A., Qian X., Luo Y. (2004) Crystal structure of archaeal recombinase RADA. A snapshot of its extended conformation. Mol. Cell 15, 423–435 [DOI] [PubMed] [Google Scholar]
- 18. Wu Y., Qian X., He Y., Moya I. A., Luo Y. (2005) Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence. J. Biol. Chem. 280, 722–728 [DOI] [PubMed] [Google Scholar]
- 19. Baumann P., Benson F. E., Hajibagheri N., West S. C. (1997) Purification of human Rad51 protein by selective spermidine precipitation. Mutat. Res. 384, 65–72 [DOI] [PubMed] [Google Scholar]
- 20. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) Recombinant replication protein A. Expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 21. Van Komen S., Petukhova G., Sigurdsson S., Sung P. (2002) Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J. Biol. Chem. 277, 43578–43587 [DOI] [PubMed] [Google Scholar]
- 22. Qian X., He Y., Wu Y., Luo Y. (2006) Asp-302 determines potassium dependence of a RadA recombinase from Methanococcus voltae. J. Mol. Biol. 360, 537–547 [DOI] [PubMed] [Google Scholar]
- 23. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 24. McRee D. E. (1999) XtalView/Xfit. A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 [DOI] [PubMed] [Google Scholar]
- 25. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 26. Eggler A. L., Inman R. B., Cox M. M. (2002) The Rad51-dependent pairing of long DNA substrates is stabilized by replication protein A. J. Biol. Chem. 277, 39280–39288 [DOI] [PubMed] [Google Scholar]
- 27. Sugiyama T., Zaitseva E. M., Kowalczykowski S. C. (1997) A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 272, 7940–7945 [DOI] [PubMed] [Google Scholar]
- 28. Symington L. S., Heyer W. D. (2006) Some disassembly required. Role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 20, 2479–2486 [DOI] [PubMed] [Google Scholar]
- 29. Chi P., Van Komen S., Sehorn M. G., Sigurdsson S., Sung P. (2006) Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair 5, 381–391 [DOI] [PubMed] [Google Scholar]
- 30. Solinger J. A., Kiianitsa K., Heyer W. D. (2002) Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10, 1175–1188 [DOI] [PubMed] [Google Scholar]
- 31. Hilario J., Amitani I., Baskin R. J., Kowalczykowski S. C. (2009) Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bugreev D. V., Golub E. I., Stasiak A. Z., Stasiak A., Mazin A. V. (2005) Activation of human meiosis-specific recombinase Dmc1 by Ca2+. J. Biol. Chem. 280, 26886–26895 [DOI] [PubMed] [Google Scholar]
- 33. Bugreev D. V., Mazin A. V. (2004) Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bianco P. R., Tracy R. B., Kowalczykowski S. C. (1998) DNA strand exchange proteins. A biochemical and physical comparison. Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 35. Kowalczykowski S. C. (1991) Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 20, 539–575 [DOI] [PubMed] [Google Scholar]
- 36. Zaitseva E. M., Zaitsev E. N., Kowalczykowski S. C. (1999) The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 274, 2907–2915 [DOI] [PubMed] [Google Scholar]
- 37. Shibata T., Cunningham R. P., DasGupta C., Radding C. M. (1979) Homologous pairing in genetic recombination. Complexes of recA protein and DNA. Proc. Natl. Acad. Sci. U.S.A. 76, 5100–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pugh B. F., Cox M. M. (1987) Stable binding of recA protein to duplex DNA. Unraveling a paradox. J. Biol. Chem. 262, 1326–1336 [PubMed] [Google Scholar]
- 39. De Zutter J. K., Knight K. L. (1999) The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol. 293, 769–780 [DOI] [PubMed] [Google Scholar]
- 40. Cox M. M., Lehman I. R. (1981) RecA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc. Natl. Acad. Sci. U.S.A. 78, 3433–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qian X., He Y., Ma X., Fodje M. N., Grochulski P., Luo Y. (2006) Calcium stiffens archaeal Rad51 recombinase from Methanococcus voltae for homologous recombination. J. Biol. Chem. 281, 39380–39387 [DOI] [PubMed] [Google Scholar]
- 42. Story R. M., Steitz T. A. (1992) Structure of the recA protein-ADP complex. Nature 355, 374–376 [DOI] [PubMed] [Google Scholar]
- 43. Chen Z., Yang H., Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 44. Li Y., He Y., Luo Y. (2009) Conservation of a conformational switch in RadA recombinase from Methanococcus maripaludis. Acta Crystallogr. D Biol. Crystallogr. 65, 602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qian X., Wu Y., He Y., Luo Y. (2005) Crystal structure of Methanococcus voltae RadA in complex with ADP. Hydrolysis-induced conformational change. Biochemistry 44, 13753–13761 [DOI] [PubMed] [Google Scholar]
- 46. Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J. S., Sung P. (2001) Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 15, 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., Heyer W. D. (2011) Rad51 paralogs Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miller K. A., Yoshikawa D. M., McConnell I. R., Clark R., Schild D., Albala J. S. (2002) RAD51C interacts with RAD51B and is central to a larger protein complex in vivo exclusive of RAD51. J. Biol. Chem. 277, 8406–8411 [DOI] [PubMed] [Google Scholar]
- 49. Lio Y. C., Mazin A. V., Kowalczykowski S. C., Chen D. J. (2003) Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J. Biol. Chem. 278, 2469–2478 [DOI] [PubMed] [Google Scholar]
- 50. Mazin A. V., Kowalczykowski S. C. (1998) The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 17, 1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.