Background: Hydroperoxide detoxification in African trypanosomes relies on a cascade composed of trypanothione, trypanothione reductase, tryparedoxin, and tryparedoxin peroxidases.
Results: A library screening against the peroxidase system unraveled trypanocidal compounds that inactivate tryparedoxin in vitro and in the intact parasite.
Conclusion: Tryparedoxin is a druggable target.
Significance: Detection of novel target molecules is a crucial step to overcome the unsatisfactory chemotherapy of sleeping sickness.
Keywords: Drug Discovery, Enzyme Inhibitors, Parasite, Thiol, Thioredoxin, Trypanosoma brucei, Trypanothione, Tryparedoxin
Abstract
In African trypanosomes, the detoxification of broad spectrum hydroperoxides relies on a unique cascade composed of trypanothione (T(SH)2), trypanothione reductase, tryparedoxin (Tpx), and nonselenium glutathione peroxidase-type enzymes. All three proteins are essential for Trypanosoma brucei. Here, we subjected the complete system to a high throughput screening approach with nearly 80,000 chemicals. Twelve compounds inhibited the peroxidase system. All but one carried chloroalkyl substituents. The detailed kinetic analysis showed that two compounds weakly inhibited trypanothione reductase, but none of them specifically interacted with the peroxidase. They proved to be time-dependent inhibitors of Tpx-modifying Cys-40, the first cysteine of its active site WCPPC motif. Importantly, gel shift assays verified Tpx as a target in the intact parasites. T(SH)2, present in the in vitro assays and in the cells in high molar excess, did not interfere with Tpx inactivation. The compounds inhibited the proliferation of bloodstream T. brucei with EC50 values down to <1 μm and exerted up to 83-fold lower toxicity toward HeLa cells. Irreversible inhibitors are traditionally regarded as unfavorable. However, a large number of antimicrobials and anticancer therapeutics acts covalently with their target protein. The compounds identified here also interacted with recombinant human thioredoxin, a distant relative of Tpx. This finding might even be exploited for thioredoxin-based anticancer drug development approaches reported recently. The fact that the T(SH)2/Tpx couple occupies a central position within the trypanosomal thiol metabolism and delivers electrons also for the synthesis of DNA precursors renders the parasite-specific oxidoreductase an attractive drug target molecule.
Introduction
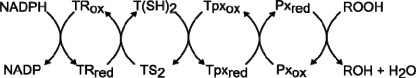
Trypanosomes and Leishmania are the causative agents of a variety of tropical diseases. Chemotherapy of diseases caused by these parasitic protozoa can hardly be rated satisfactory. The very few drugs available suffer from high toxicity, the need of hospitalization, and increasing resistance development. One approach toward the development of novel antimicrobial agents is the identification of pathways that do not occur or are substantially different in the mammalian host. In this context, the unique hydroperoxide metabolism of trypanosomatids is an attractive target. Trypanosoma brucei, the causative agent of African sleeping sickness, lacks catalase and classical glutathione peroxidases. Detoxification of hydroperoxides relies on 2-Cys-peroxiredoxins (Prxs)2 and nonselenium glutathione peroxidase-type (Px) enzymes, both of which act as tryparedoxin (Tpx) peroxidases (Scheme) (1, 2). The Prx-type enzymes exert a strong preference for hydrogen peroxide as substrate, whereas the Px-type peroxidases accept fatty acid-derived hydroperoxides and protect the parasite from membrane damage (3).
Trypanothione (T(SH)2, bis(glutathionyl)spermidine) is the main low molecular mass thiol of these parasites. The dithiol is kept reduced by NADPH and the flavoenzyme trypanothione reductase (TR), the system replacing the glutathione/glutathione reductase couple of the mammalian host (4). TR is the most thoroughly studied enzyme of the trypanothione metabolism, and a large number of inhibitors has been identified (5–9). Tpx is a distant member of the thioredoxin (Trx) protein family. The parasite-specific protein has a WCPPC active site motif instead of the canonical WCGPC sequence in Trxs, and with 16,000 Da, it is larger than human Trx (12,000 Da) (10, 11). Tpx is kept in the dithiol state by the spontaneous reaction with T(SH)2 (11, 12), whereas the host Trx is reduced by thioredoxin reductase, an enzyme also missing in trypanosomatid organisms. T. brucei Tpx plays a central role in most of the T(SH2)-dependent parasite pathways (1). Examples are the detoxification of hydroperoxides and, as shown recently, the reduction of protein-bound methionine sulfoxide residues (13). Most importantly, the T(SH)2/Tpx system delivers the reducing equivalents for the synthesis of DNA precursors catalyzed by ribonucleotide reductase and thus is involved in parasite replication (14).
Previous high throughput screening (HTS) approaches against the parasite trypanothione system mainly focused on the detection of TR inhibitors. Different chemotypes were identified that showed selectivity for TR over human glutathione reductase (15). Another approach revealed compounds with potent antiparasitic activity, but only moderate correlation with TR inhibition (6). All enzymes establishing the parasite peroxidase system, namely TR, Tpx, and both types of tryparedoxin peroxidases, have been shown to be essential for T. brucei (16–19) and thus fulfill a crucial prerequisite of a putative drug target molecule. Recently the effect of the antitumor quinol PMX 464 on the parasite peroxidase systems has been studied (20). In mammalian and yeast cells, the quinol inhibits thioredoxin (21–23). Toward T. brucei, PMX 464 showed cytocidal activity and caused depletion of cellular T(SH)2. In vitro, the compound interacted with T(SH)2 and both the Prx- and Px-type enzymes but neither with TR nor Tpx (20). Here, we present an HTS approach toward the peroxidase system of African trypanosomes followed by the in vitro and in vivo identification of the target protein. Primary goal was to identify putative lead compounds for a drug design directed against the Px-type enzyme. Interestingly, the analysis resulted in compounds that specifically inactivated Tpx. Importantly, Tpx could be demonstrated to be targeted in the intact parasite.
EXPERIMENTAL PROCEDURES
Materials
NADPH was purchased from AppliChem; t-bOOH, TCEP, Tween 20 and DMSO were from Sigma; and MAL-PEG (5272 Da) was from Iris Biotech. Recombinant tag-free T. brucei Px (24), T. brucei Prx (3), wild type T. brucei His6-Tpx, C40S-Tpx-His6, and C43S-Tpx-His6 (25), T. cruzi TR (26), T(SH)2, and trypanothione disulfide (27) were prepared as described. To obtain tag-free Tpx, the coding region was amplified by PCR from pQE-60-tpx (11), cloned into the pETtrx_1b vector (kindly provided by G. Stier, EMBL), and overexpressed in Escherichia coli. The protein was purified following the procedure for Px (24). A sample of human Trx was a kind gift of Dr. R. H. Schirmer (Biochemie-Zentrum der Universität Heidelberg). The compound library used is a collection of commercial chemicals, purchased from three vendors (AMRI, ENAMINE, and ChemBridge). The compounds were selected around carefully picked scaffolds that represent a subset of analogs. The library represents an optimal coverage of available chemical space around each scaffold, and the compounds fulfill the Lipinski criteria (28).
HTS Assay
The assays were performed in a total volume of 20 μl of 20 mm Na-HEPES, 150 mm NaCl, 1 mm EDTA, 0.05% Tween 20, pH 7.5 (screening buffer), in black 384-well plates with a round clear bottom (Corning Glass). The plates were prepared with 2 μl of 400 μm compounds (4% DMSO) as described by Sehr et al. (29). 10 μl of 600 μm t-bOOH in screening buffer was added. The reaction was started immediately by adding 8 μl of a premixture containing NADPH, TR, T(SH)2, Tpx, and Px, which resulted in a final concentration of 280 μm NADPH, 40 nm TR, 100 μm T(SH)2, 8 μm Tpx, 20 nm Px, 300 μm t-bOOH, and 0.4% DMSO. After 4 min of centrifugation at 1000 × g to remove air bubbles, the plates were transferred to an Envision multilabel plate reader (PerkinElmer Life Sciences), and NADPH consumption was recorded at 340 nm and 25 °C. The first data point was taken after 15 min. In total, nine reads (one data point every 19 min) were monitored. The absorption decrease between the second and seventh data point was used to calculate the peroxidase activity. Columns 1 and 2 of each plate contained 0.4% DMSO corresponding to full activity (0% inhibition). In columns 23 and 24, the reaction mixtures lacked Px and thus represented the spontaneous reaction of t-bOOH with T(SH)2 (100% inhibition). The data were evaluated using Activity Base (IDBS). The percentage of inhibition was calculated as 100 × (1 −((x − μ−)/(μ+ − μ−))), where x is the slope of the absorption decrease/time in the presence of inhibitor; μ− is the mean slope of the negative controls (0% inhibition), and μ+ is the mean slope of the positive control (100% inhibition). The Z′ factor as quality parameter of the assays was calculated from the controls in column 1, 2, 23, and 24 (30).
IC50 Determinations
Compounds that in the HTS revealed >20% inhibition were re-ordered. The assays were conducted as described above using an 11-point titration from 200 μm to 200 nm. The final concentration of DMSO was 2%. The percentage of inhibition was plotted against the compound concentration, and IC50 values were calculated.
EC50 Determinations
Bloodstream T. brucei (strain 449) were grown as described (18). 10 mm stock solutions of the compounds were prediluted to 500 μm and then serially 1:5 (7 point titration) with DMSO. Aliquots of 10 μl were spotted on a 24-well plate (Greiner), and 990 μl of trypanosome culture (5 × 105 cells/well) was added. Cells cultured in the presence of 1 and 9% DMSO served as negative and positive control, respectively. After 24 h, living cells were counted using a hemocytometer. Cell density was plotted against the compound concentration, and EC50 values were calculated. For the determination of EC50 values after 72 h, the compound stock solutions were diluted with medium to 500 μm and then 1:1 with 5% DMSO in HMI-9 medium (10-point titration). 10-μl aliquots were spotted on a 96-well plate (PerkinElmer Life Sciences), and 90 μl of trypanosome suspension (250 cells) was added resulting in 2500 cells/ml and a final concentration of 0.5% DMSO. HeLa (Kyoto) cells (provided by Dr. R. Pepperkok, EMBL) were cultured as described by Erfle et al. (31). 1000 cells/well (90 μl) were seeded in a 96-well plate. The next day, 10 μl of compound solution (diluted in DMEM) was added to the adherent cells. After 72 h, proliferation of triplicate cultures (T. brucei and HeLa cells) was quantified using ATPLite one-step (PerkinElmer Life Sciences) following the protocol of the manufacturer. The luminescence of each sample, expressed as percentage of that of the control cultures, was plotted against the compound concentration, and EC50 values were calculated. Columns 1 and 12 of each plate with 0.5 and 9% DMSO served as negative and positive controls, respectively.
TR Inhibition
Inhibition of TR was measured in a 384-well plate. Plates were prepared with 2 μl of an 11-point titration of the compounds. 10 μl of a premixture of NADPH and TR in screening buffer was added, and the reaction was started by 8 μl of trypanothione disulfide in screening buffer. The final assay volume was 20 μl containing 280 μm NADPH, 0.3 nm TR, 150 μm trypanothione disulfide, and 2% DMSO. The Z′ factor was calculated from the controls in columns 1 (0% inhibition, with DMSO) and 24 (100% inhibition, without TR), respectively. The assay was conducted as described above for the IC50 determinations with nine data points (one read every 10 min).
Cuvette-based Peroxidase Assays
In a total volume of 200 μl, a premixture of NADPH, TR, T(SH)2, Tpx, and Px in screening buffer was mixed with the compounds, resulting in final concentrations of 140 μm NADPH, 220 nm TR, 100 μm T(SH)2, 10 μm Tpx, 90 nm Px, and 40, 20, or 10 μm inhibitor or 2% DMSO. The reaction was started either directly or after 40 min of preincubation by adding 100 μm H2O2, and the absorption decrease at 340 nm was followed at 25 °C. To evaluate if a compound specifically targeted the peroxidase, Px was replaced by 150 nm Prx. To analyze if Tpx was the target protein, premixtures containing all components except either Tpx or Px were incubated with the compounds. After 40 min, Tpx or Px was added immediately before starting the assay with H2O2.
ESI-MS Analysis of T. brucei Tpx and Human Trx
10 μm Tpx (His6-tagged or tag-free) was treated with 100 μm T(SH)2 for 10 min at 25 °C in screening buffer without Tween 20. Reduced human Trx was obtained by incubating 10 μm protein with 100 μm DTT. In a final volume of 100 μl, the reduced and oxidized protein species were incubated with 40 μm of compounds 1–6 for 40 min at 25 °C. The final concentration of DMSO was 2%. The samples were either directly subjected to ESI-MS or stored at −20 °C. The spectra presented in Fig. 3 and supplemental Fig. S1, a–h, were obtained from the Core Facility for Mass Spectrometry and Proteomics, Zentrum für Molekularbiologie der Universität Heidelberg, Heidelberg, Germany. The total protein masses were determined by ESI-MS on an API-QSTARTM Pulsar instrument (Applied Biosystems) with an HPLC (Agilent) on-line-coupled to the ESI-QTOF instrument. The reaction mixtures were diluted to 1 ml with 0.1% TCA, and 100 μl were loaded onto a 50 Poros R1 trapping column. After 1.5 min of washing with 0.1% TFA (0.4 ml/min), the protein was eluted into the electrospray ion source with 80% acetonitrile, 0.1% TFA at 10 μl/min as described by Rist et al. (32). The spectra depicted in supplemental Figs. S1, i–p, and S2 were recorded at the Proteomics Core Facility, EMBL Heidelberg. The protein samples were desalted using C18 ZipTips (Millipore). After wetting in 50% (v/v) acetonitrile in water, equilibrating in 0.1% TFA, binding, and washing (0.1% TFA), the samples were eluted in 10 μl of 50% acetonitrile containing 1% (v/v) formic acid. The purified protein samples were injected into a Q TOF II mass spectrometer (Micromass) using static nanospray borosilicate needles (PicoTip). Both instruments were calibrated with apomyoglobin (Sigma), and their mass accuracy was better than 100 ppm.
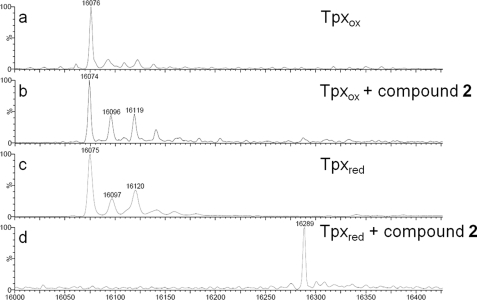
FIGURE 3.
Deconvoluted ESI-MS spectra of Tpx after reaction with compound 2. Tpx was incubated with compound 2 in the absence (Tpxox) or presence of T(SH)2 (Tpxred) and subjected to ESI-MS analysis. a, oxidized Tpx. The peak at 16,076 Da corresponds to the calculated mass (16,074.2 Da) of the full-length tag-free protein with an artificial N-terminal GAG tripeptide. b, oxidized Tpx incubated with compound 2. No mass shift was observed. c, reduced Tpx. d, reduced Tpx incubated with compound 2 showed a new peak at 16,289 Da. The mass increase by 214 Da corresponds to Tpx modified by compound 2 after elimination of HCl (expected mass difference 213 Da). The peaks at 16,096 (16,097) Da and 16,119 (16,120) Da with 23 and 45 Da higher masses probably represent mono- and di-sodium adducts of Tpx. The accuracy of the measurement was ± 2 Da.
Reaction of Tpx with MAL-PEG
In 40 μl of screening buffer without Tween 20, 125 μm wild type His6-Tpx, C40S-Tpx-His6, or C43S-Tpx-His6 was treated for 40 min at 25 °C with 500 μm of compounds 1–6 in the presence of 3.1 mm DTT. 10 μl of MAL-PEG (50 mm in screening buffer without Tween) and 50 μl of screening buffer were added. After 1 h at 37 °C, 25 μl of ice-cold TCA (100% (w/v) in 20 mm HCl) was added, and the mixture was kept for 10 min at 4 °C. After centrifugation, the pellet was washed three times with 100 μl of ice-cold acetone, and the denatured protein was resuspended in screening buffer. 20 μg of protein was subjected to SDS-PAGE on a 15% gel followed by Coomassie staining.
Identification of Tpx as Intracellular Target
1 × 107 trypanosomes (1 × 106 cells/ml) were incubated with different concentrations of the compounds for 0 or 4 h at 37 °C, harvested, washed twice with 6 ml of PBS, resuspended in 15 μl of screening buffer in the presence or absence of 5 mm TCEP, and lysed by three cycles of freezing-thawing followed by 30 min of incubation at 37 °C. 5 μl of 50 mm MAL-PEG was added, and the reaction mixture was incubated for another 30 min. Lysates corresponding to 5 × 106 cells were subjected to SDS-PAGE as described above, followed by Western blot analysis with the polyclonal rabbit antiserum against T. brucei Tpx (1:2000) (19). Horseradish peroxidase-conjugated anti-rabbit IgGs served as secondary antibody (1:20000; Santa Cruz Biotechnology). Bands were visualized by chemiluminescence using the Super Signal west pico kit (Pierce).
RESULTS
Assay Principle, Miniaturization, and Optimization
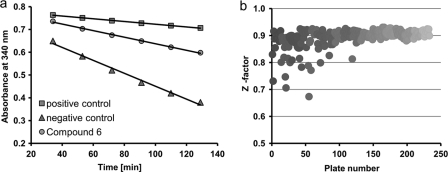
Tryparedoxin peroxidase activity is measured in a coupled assay following NADPH consumption (18, 33) (see Scheme 1). The cuvette-based assay was adapted for the HTS approach of 78,637 compounds. Although T. brucei Px has higher activity with hydrogen peroxide (3, 33), t-bOOH was used as substrate because of its much slower spontaneous reduction by T(SH)2 (34). Under the conditions chosen, the reaction rate was linear for at least 2 h and dependent on the Px activity. The slope of each graph was obtained by linear regression, and percentage of inhibition was calculated as outlined under “Experimental Procedures” and shown as an example for compound 6 (Fig. 1a). The fit of the curve to the data was calculated. For all compounds that were further characterized (see below), the R2 value was ≥0.92. With Z′-factors of ≥0.68, the assay performance and robustness proved to be very good (Fig. 1b). In comparison with a single end point determination, this kinetic data acquisition not only gives a more robust assay signal but also minimizes the probability of false-positive results, for instance in the case of compounds that absorb at 340 nm (35, 36).
SCHEME 1.
FIGURE 1.
Assay performance and Z′-factors of the coupled peroxidase assay. a, peroxidase activity was measured by following NADPH consumption at 340 nm with six reads every 19 min. A reaction mixture lacking Px represents 100% inhibition (positive control), whereas one containing Px but no inhibitor corresponds to 0% inhibition (negative control). The percentage of inhibition was calculated from the respective slopes as exemplified here for compound 6 (gray circles), which resulted in an inhibition of 65%. b, the full screening approach comprised 78,637 compounds distributed in 246 plates (384-well plates). The Z′-factor was calculated as described under “Experimental Procedures.”
HTS Results and Intracellular Activity
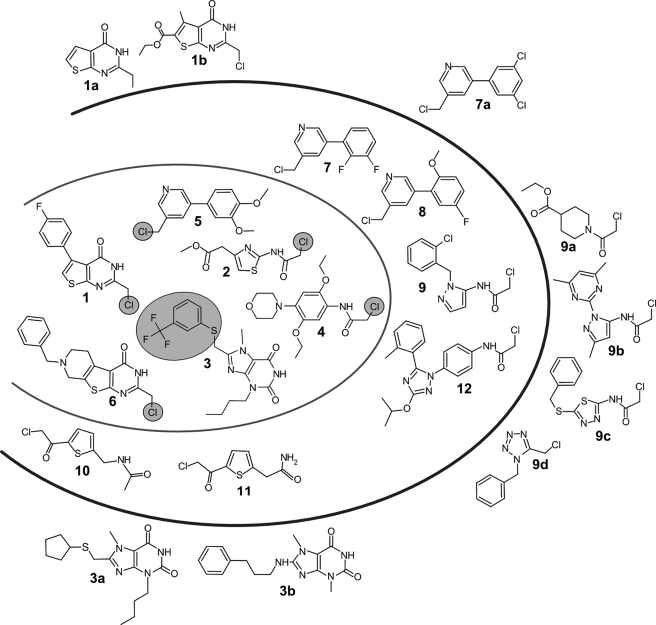
The HTS approach yielded 36 compounds that inhibited the peroxidase system by ≥20% at a concentration of 40 μm. This relatively low overall hit rate is comparable with HTSs reported for other parasite enzyme systems (6, 35). Subsequently, the library was virtually re-screened for derivatives with common core structures or similar side chains. The aim was to identify putative false-negatives and structure activity relationships. A total of 39 compounds (31 primary hits and 8 analogs) was reordered from the suppliers. Five chemicals were no longer available. For 25 of the 31 candidates, the inhibitory potency was confirmed by IC50 determinations yielding values between 3.5 and 66 μm. 12 Compounds with IC50 values ≤31 μm were further analyzed (Table 1). They grouped in chemicals with common core structure or with an identical reactive substituent (Fig. 2). None of the reordered analogs 1a, 1b, 3a, 3b, and 7a showed activity (supplemental Table S1). In the case of 1a, 3a, and 3b, this can be explained by the absence of the reactive leaving group. The analogs 9a–d revealed some inhibition but were less active when compared with the parent 9. The compounds 9 and 9a–c have in common the possession of a chloroacetamido substituent suggesting a similar mode of action. The core structure of 9d resembles those in 9 and 9b, but the compound carries a chloromethyl group as do compounds 1 and 5–8. In summary, except for compound 3, all compounds that significantly interfered with the peroxidase system possess a chloromethyl, chloroacetamido, or chloroacetyl group. The data also showed that, although necessary, the chloroalkyl substituents are not sufficient for the inhibitory potency indicating some specificity based on the core structures. The finding that none of the reordered analogs showed a pronounced effect confirmed the results of the HTS.
TABLE 1.
In vitro and cellular activity of compounds 1–12
|
In vitro inhibitiona |
EC50b |
Selectivity indexc | ||||
|---|---|---|---|---|---|---|
| Compound | HTS | IC50 |
T. brucei |
HeLa |
||
| 24 h | 72 h | 72 h | ||||
| % | μm | μm | μm | μm | ||
| 1 | 102 | 16.9 ± 2.6 | 0.7 ± 0.03 | 0.6 ± 0.1 | >50 | >83 |
| 2 | 103 | 4.8 ± 0.04 | 0.4 ± 0.1 | 0.8 ± 0.2 | 14.0 ± 4.5 | 18 |
| 3 | 100 | 13.7 ± 0.1 | 1.1 ± 0.04 | 4.2 ± 0.3 | >50 | >12 |
| 4 | 31 | 29.5 ± 0.2 | 3.1 ± 0.7 | 2.3 ± 0.1 | 22.1 ± 3.2 | 10 |
| 5 | 99 | 16.0 ± 0.1 | 1.3 ± 0.2 | 3.1 ± 0.5 | 28.4 ± 3.2 | 9 |
| 6 | 65 | 7.2 ± 0.2 | 0.4 ± 0.1 | 1.5 ± 0.9 | 10.0 ± 2.0 | 7 |
| 7 | 64 | 30 ± 0.3 | 0.2 ± 0.05 | 3.6 ± 0.6 | 16.3 ± 4.9 | 5 |
| 8 | 76 | 22 ± 0.1 | 0.5 ± 0.1 | 4.3 ± 1.1 | 12.2 ± 4.1 | 3 |
| 9 | 88 | 31 ± 0.8 | 0.5 ± 0.1 | 3.2 ± 0.1 | 8.0 ± 3.1 | 3 |
| 10 | 107 | 3.5 ± 0.02 | 3.1 ± 0.7 | 3.6 ± 0.6 | 8.0 ± 3.2 | 2 |
| 11 | 102 | 3.9 ± 0.06 | 5.1 ± 1.2 | 7.9 ± 1.7 | 12.3 ± 5.3 | 2 |
| 12 | 80 | 13.9 ± 2.6 | 2.2 ± 0.1 | ND | 8.3 ± 0.3 | ND |
a Percentage of inhibition of the peroxidase system in the initial HTS with 40 μm compound and IC50 value are shown, and both were measured in 384-well plate format.
b Bloodstream T. brucei and HeLa cells were cultured in the presence of different concentrations of the compounds. The parasites were counted (T. brucei, 24 h), and the ATP level of living T. brucei and HeLa cells was determined using ATPLite (72 h). The EC50 values are the inhibitor concentrations that resulted in 50% living cells compared with those grown in the presence of 0.5% DMSO.
c Ratio of the EC50 value for HeLa cells and T. brucei (72 h). The values are the means ± S.D. of three independent assays. ND means not determined, because of lack of material.
FIGURE 2.
Compounds active against the peroxidase cascade in the HTS and analogs identified by virtual screening. The 12 compounds within the outer oval revealed IC50-values of ≤31 μm. Compounds 1–6 (inner oval) displayed higher toxicity against T. brucei compared with HeLa cells (Table 1). The leaving group upon inactivation of Tpx is circled. The thienopyrimidine-4-ones 1, 1a, 1b, and 6, the purine 2,6-diones 3, 3a, and 3b, the chloromethyl-phenylpyridines 5, 7, 7a, and 8, as well as the chloroacetyl-thiophenes 10 and 11 each have a common core structure. Compounds 2, 4, 9, 9b, 9c, and 12 carry a 2-chloro-acetamido substituent. None of the analogues (marked by a–d) revealed pronounced inhibitory potency.
The 12 compounds were then studied for their activity toward bloodstream T. brucei. The parasites were treated with different inhibitor concentrations, and after 24 h living cells were counted. In a second approach, a luciferase-based viability assay was used. T. brucei and HeLa cells (31) were incubated with the compounds for 72 h, and the cellular ATP level was measured (37). The antitrypanosomal effect of the compounds became apparent after 24 h and remained practically constant for 72 h. The mammalian HeLa cells revealed generally higher EC50 values, which resulted in selectivity indices of up to >83 (Table 1). For most compounds, the EC50 values were an order of magnitude lower than the IC50 values obtained in the in vitro peroxidase assay. As outlined below, this does not contradict the peroxidase system to be targeted but is probably due to the fact that these compounds cause a time-dependent inactivation of their target protein.
Target Identification
To identify the distinct protein affected within the peroxidase cascade, the compounds were first subjected to a TR assay. Most of them yielded IC50 values of ≥200 μm (Table 2). Only the chloromethyl-phenylpyrimidines 5, 7, and 8 revealed IC50 values of 123, 29, and 2.3 μm, respectively. Subsequent cuvette-based assays and spectroscopic and mass analyses showed for 7 and 8, but not 5, a time-dependent inhibition of TR suggesting modification of the active site cysteine(s).3 This is not due to a generally higher reactivity of these derivatives because in the complete peroxidase system compounds 7 and 8 were less efficient than 5 (Table 1). Detailed analyses are required to reveal putative specific interactions with TR, especially because the halogen and methoxy substituents occupy different positions at the phenyl ring. Taken together, TR can be ruled out as target molecule of compound 5, whereas in the case of compounds 7 and 8, inhibition of the enzyme may contribute to the effect of the compounds toward the peroxidase cascade.
TABLE 2.
Effect of 1–12 toward TR
TR activity was measured in 384-well plates as described under “Experimental Procedures.” The IC50 values are the means ± S.D. from three experiments.
| Compound | IC50 |
|---|---|
| μm | |
| 1 | >200 |
| 2 | >200 |
| 3 | >200 |
| 4 | >200 |
| 5 | 123 ± 16 |
| 6 | >200 |
| 7 | 29 ± 4 |
| 8 | 2.3 ± 0.3 |
| 9 | >200 |
| 10 | 146 ± 22 |
| 11 | >200 |
| 12 | >200 |
The remaining 10 compounds were further characterized in cuvette-based peroxidase assays. In the first series, the inhibitor was added to a reaction mixture composed of NADPH, TR, Tpx, T(SH)2, and Px (complete), and the assay was immediately started with H2O2. The percentage of inhibition was calculated from the reactions without compound (0% inhibition) and without Px (100% inhibition). At 10 μm, compounds 6, 10, and 11 revealed ≥71% inhibition (Table 3). For all other compounds, inhibition was significantly weaker when compared with the HTS results (Table 1). In the case of compounds 1 and 2, this may partially be due to the lower inhibitor concentration used (10 μm instead of 40 μm in the HTS). However, the main difference between both systems was that in the cuvette-based assay, the activity was determined from the absorption decrease directly after adding the inhibitor, whereas in the HTS, the activity was calculated from the data recorded 34 min after starting the reaction. In the next series of kinetics, the reactants were thus preincubated for 40 min before the reaction was started by H2O2. As expected, this procedure resulted in significantly stronger inhibition. With compound 2, the time-dependent inactivation already became visible in assays that were started directly (see legend of Table 3). All assays in the plate format (HTS, IC50 determination, and TR inhibition) were conducted under identical conditions to allow comparison of the time-dependent reactions.
TABLE 3.
Identification of the target protein(s)
Peroxidase activity was measured as described under “Experimental Procedures.” The reaction was started either directly or after 40 min preincubation of the reaction mixture (complete, without either Px or Tpx) by adding the missing protein component immediately followed by H2O2. Values are the means ± S.D. of three assays. ND means not determined.
| Compound | Inhibition |
|||||
|---|---|---|---|---|---|---|
| No Preincubation, complete |
Preincubation, complete |
Preincubation-Px |
Preincubation-Tpx |
|||
| Px | Prx | Px | Prx | Px | Px | |
| % | ||||||
| 1 | 29 ± 4a | 40 ± 3b | 84 ± 1a | ND | 87 ± 1a | 13 ± 2a |
| 2 | 3 ± 1a,d | 9 ± 0.2c,d | 85 ± 4a | ND | 21 ± 4a | 3 ± 1a |
| 3 | 29 ± 1c | ND | 102 ± 2c | 102 ± 2c | 100 ± 1c | 21 ± 2c |
| 4 | 3 ± 1c | ND | 78 ± 3c | 75 ± 2c | 90 ± 1c | 11 ± 3c |
| 5 | 10 ± 2c | ND | 93 ± 1c | ND | 100 ± 1c | 13 ± 1c |
| 6 | 97 ± 1a | 88 ± 2a | 105 ± 0.3a | ND | 99 ± 2a | 70 ± 2a |
| 9 | 2 ± 0.4c | ND | 75 ± 1c | 85 ± 2c | 78 ± 1c | 10 ± 3c |
| 10 | 84 ± 3a | 74 ± 3a | 99 ± 2a | ND | 96 ± 1a | 6 ± 4a |
| 11 | 71 ± 1a | 64 ± 1a | 102 ± 1a | ND | 101 ± 2a | 12 ± 2a |
| 12 | 19 ± 3c | ND | 101 ± 2c | 102 ± 1c | 99 ± 1c | 17 ± 2c |
a The assay contained 10 μm of the respective inhibitor.
b The assay contained 20 μm of the respective inhibitor.
c The assay contained 40 μm of the respective inhibitor.
d The values refer to the activity calculated for the 1st min. In the presence of compound 2, NADPH oxidation declined rapidly during the course of the assay. This resulted after 3 min in 32% and 45% inactivation in the Px and Prx assay, respectively.
The time-dependent mode of inhibition indicated a covalent modification of the target protein. In the next series of kinetics, the Px was replaced by the Prx-type enzyme. The degree of inhibition was comparable with that in the corresponding Px assay suggesting that none of the peroxidases was the main target. Finally, the compounds were studied as inhibitors of Tpx. Reaction mixtures that lacked either Px (control) or Tpx were preincubated with the inhibitors for 40 min. Immediately before starting the reaction with H2O2, the missing protein was added. With the exception of compound 2 (see below), preincubation of the reaction mixtures lacking Px resulted in 78–100% inhibition as it was observed for the complete mixtures and strongly suggested Tpx as target protein. In contrast, the preincubation of reaction mixtures that lacked Tpx resulted in minimal inactivation. Only compound 6 caused 70% inhibition under these conditions. This can be explained by the high reactivity of the compound toward Tpx, which already resulted in 97% inactivation without any preincubation and does not reflect inhibition of Px. T(SH)2 is not an efficient direct reductant of the peroxidase (33). Thus, in the absence of Tpx, Px should be mainly in the inactive form, characterized by an intramolecular disulfide (38). In the case of compound 2, only 21% inhibition was observed when the preincubation mixture lacked Px in comparison with 85% for the complete system, which indicates that both Tpx and Px react with this compound.
Characterization of Tpx after Treatment with Compounds 1–6
10 μm recombinant Tpx was treated with 40 μm compound 2 for 40 min in the presence and absence of 100 μm T(SH)2 and subjected to ESI-MS analysis as described under “Experimental Procedures.” Oxidized Tpx, independent of the presence of the compound, as well as untreated reduced Tpx revealed the mass of the unmodified protein (Fig. 3). In contrast, when Tpx was incubated with compound 2 in the presence of T(SH)2, a new peak with a 214-Da higher mass appeared in accordance with covalent modification of Tpx under elimination of HCl. Table 4 and the supplemental Fig. S1 provide the respective analyses for compounds 1–6 using His-tagged Tpx. As shown by HPLC MS/MS, all compounds showed a purity of 99%, except for compound 6 with a content of 84% (supplemental Fig. S3). Treatment of reduced, but not oxidized, Tpx resulted in a mass increase that corresponded to the mass of the inhibitor minus HCl. An exception was compound 3. Here, the mass of the leaving group corresponded to the trifluoromethyl benzenethiol moiety of the compound (Fig. 2). Remarkably, under the conditions chosen, Tpx became completely modified by these compounds despite the presence of a 10-fold molar excess of T(SH)2. The highly reactive small dithiol did not interfere with the reaction.
TABLE 4.
ESI-MS data of His6-Tpx after reaction with compounds 1–6
| Compound | Molecular mass | Observed massesa | Mass increase | Mass of the leaving group |
|---|---|---|---|---|
| Da | Da | Da | Da | |
| 17921.5, 18100.2 | ||||
| 1 | 294.7 | 18181.4, 18359.4 | 259.9, 259.2 | 34.8, 35.5 |
| 2 | 248.7 | 18135.0, 18313.3 | 213.5, 213.1 | 35.2, 35.6 |
| 3 | 412.4 | 18158.2, 18336.5 | 236.7, 236.3 | 175.7, 176.1 |
| 4 | 342.0 | 18229.8, 18408.0 | 308.3, 307.8 | 33.7, 34.2 |
| 5 | 263.7 | 18149.7, 18328.0 | 228.2, 227.8 | 35.5, 35.9 |
| 6 | 345.8 | 18232.5, 18410.5 | 311.0, 310.3 | 34.8, 35.5 |
a The calculated mass of reduced His6-Tpx without and with the N-terminal Met is 17,923.2 and 18,054.3 Da, respectively. The analysis revealed two masses, one for the protein lacking the N-terminal Met and another one that corresponded to the full-length protein with an additional mass of 48 ± 1 Da. The latter mass did not appear when tag-free Tpx was used (see Fig. 3) and probably represents an oxidation product. Both peaks showed an identical mass increase when Tpx was reacted with the respective compound. The spectra are depicted in the supplemental Fig. S1.
In Vitro, Compounds 1–6 Inactivate also Human Thioredoxin
Tpx is a distant relative of thioredoxins. Both proteins have in common that under physiological conditions the first cysteine of their active site motif is a solvent-exposed reactive thiol(ate). As expected, reduced but not oxidized recombinant human Trx also became covalently modified by compounds 1–6 (supplemental Fig. S2 and Table S2). Interestingly, the reaction of compounds 5 and 6 with Trx did not result in the complete modification of the protein, as observed with Tpx, which may indicate some selectivity or higher reactivity of the parasite protein. This is supported by the finding that compounds 1–6 were more cytotoxic toward the parasites compared with mammalian cells.
Compounds 1–6 Modify Cys-40 of Tpx
During catalysis, Tpx cycles between the oxidized state with Cys-40 and Cys-43 forming an intramolecular disulfide and the reduced dithiol state (10, 11). A MAL-PEG-based gel shift assay was established that should result in a mass increase of 5 kDa per cysteine reacting. Although the apparent mass shifts observed on the SDS gel were much larger, mono- and bis-modified Tpx species were clearly distinguishable. In the absence of DTT, Tpx did not react with MAL-PEG, independent of a prior treatment with the compounds, and the unmodified protein was the most prominent band (Fig. 4, a and b). In the presence of the reducing agent but no inhibitor, both cysteines reacted with MAL-PEG, and the double-modified protein was formed. When reduced Tpx was reacted with compounds 1–6 followed by MAL-PEG, the mono-modified protein was observed. Treatment of 125 μm Tpx with 500 μm inhibitor in the presence of 3.1 mm DTT resulted in almost 100% modification of one cysteine residue. Thus, as described above for T(SH)2, the presence of excess DTT did not interfere with Tpx inactivation.
FIGURE 4.
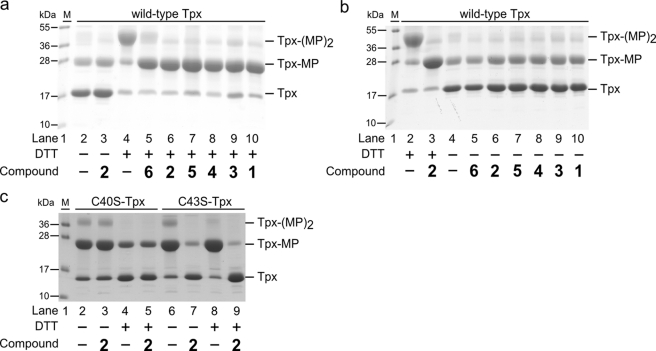
SDS-PAGE of Tpx inactivated by compounds 1–6. His-tagged Tpx was treated with the compounds in the presence or absence of DTT, and remaining free cysteines were modified by MAL-PEG (∼5 kDa) as described under “Experimental Procedures.” a, in the absence of DTT, Tpx did not react with MAL-PEG independent of prior incubation without (lane 2) or with compound 2 (lane 3). In the presence of DTT, but no inhibitor, both cysteines of Tpx reacted with MAL-PEG (Tpx-(MP)2; lane 4). In contrast, if reduced Tpx was incubated with the compounds prior to MAL-PEG treatment, the mono-modified protein was the main product (Tpx-MP; lanes 5–10). b, reduced Tpx, untreated (lane 2) or treated with 2 (lane 3) (corresponding to lanes 4 and 6 in a), served as controls. Oxidized Tpx either untreated (lane 4) or pretreated with compounds 1–6 (lanes 5–10) revealed the unmodified protein as main species. c, reaction of Tpx mutants with compound 2. In the absence of DTT, reaction of C40S-Tpx with MAL-PEG resulted in the mono-modified protein as main species independent of a prior treatment with compound 2 (lanes 2 and 3). In contrast, in the case of C43S-Tpx, treatment with compound 2 prevented the subsequent reaction with MAL-PEG (lanes 6 and 7). For further details see the text. In lane 1 the protein marker was loaded. Depicted are representative gels of at least three independent analyses.
To elucidate which cysteine residue of the WCPPC active site motif was targeted, C40S-Tpx and C43S-Tpx mutants were studied (Fig. 4c). Incubation of C40S-Tpx with MAL-PEG resulted in the mono-modified protein independent of a pretreatment with compound 2. Thus, Cys-43 did not react with the compound but remained accessible to MAL-PEG modification. In both reactions, with or without inhibitor treatment, the presence of DTT resulted in a weaker modification of C40S-Tpx by MAL-PEG compared with the reaction in the absence of the reducing agent. This is probably due to the low reactivity of Cys-43 (12, 39), which allows DTT to compete for the reagent. In the absence of compound 2, C43S-Tpx reacted with MAL-PEG yielding the mono-modified protein as was the case with C40S-Tpx. However, pretreatment of C43S-Tpx with compound 2 prevented the subsequent modification by MAL-PEG. Here, the MAL-PEG reaction was not affected by DTT in accordance with the high reactivity of Cys-40. From the three-dimensional structure of C. fasciculata Tpx, it is known that Cys-40 is solvent-exposed, whereas Cys-43 is more buried (40, 41). Cys-40 is the catalytic thiolate that interacts with T(SH)2 as well as Prx or Px, its different substrates in the peroxidase cascade (see Scheme 1) (38–42). Taken together, compounds 1–6, disclosed by the HTS approach against the peroxidase cascade, target Tpx by covalently modifying Cys-40.
Modification of Tpx in the Intact Parasite
Bloodstream T. brucei were treated with 0.6 μm compound 1, 0.8 μm compound 2, 3.1 μm compound 5, or 3.6 μm compound 7, concentrations that corresponded to the respective EC50 value at 72 h (Table 1). After 4 h, the densities of the treated cultures were 80–100% of cells grown in the presence of DMSO, clearly proving viability of the parasites at this time point. Western blot analyses of total cell lysates from parasites that were treated with DMSO or with compounds 1, 2, 5, and 7, respectively, but immediately washed (0 h) followed by MAL-PEG treatment revealed the unmodified protein (Fig. 5a). Thus, in the cell lysates Tpx was present in oxidized form. When the parasites were cultured in the presence of the compounds for 4 h and then treated with MAL-PEG, mono-modified Tpx was the main species. Some remaining oxidized protein as seen here for compounds 5 and 7 was not observed in other series of experiments (see also Fig. 5b). In the case of compound 1, even without incubation (0 h), a small amount of Tpx appeared in mono-modified form probably because of the high reactivity or very rapid cellular uptake of the compound.
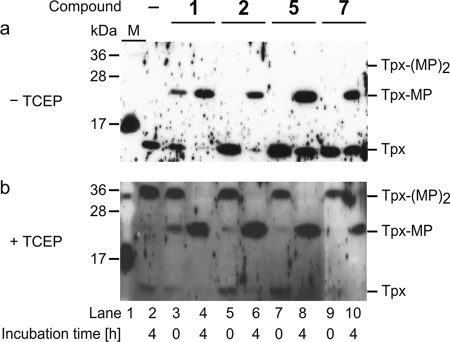
FIGURE 5.
Western blot analysis of bloodstream T. brucei cultured in the presence of compounds 1, 2, 5, or 7. a, cell lysates treated with MAL-PEG in the absence of reducing agent. In cells incubated with DMSO (4 h) or with compounds 2, 5, and 7 for 0 h, Tpx did not show any modification by MAL-PEG (Tpx). Treatment of the parasites with the compounds for 4 h, or in the case of compound 1 to a minor degree immediately, resulted in a mono-modification of Tpx by MAL-PEG (Tpx-MP). b, cell lysates treated with MAL-PEG in the presence of TCEP. In parasites cultured with DMSO for 4 h or the inhibitors for 0 h, MAL-PEG reacted with both cysteines of Tpx (Tpx-(MP)2). In contrast, if the parasites had been incubated with the compounds for 4 h, and the cell lysate was then reacted with MAL-PEG only Tpx-MP was obtained. Lane 1 of both gels shows the protein marker (M). Depicted are representative blots of at least three independent experiments.
Finally, the cells were incubated with the compounds as described above but reacted with MAL-PEG in the presence of TCEP. When the parasites were treated with either DMSO or with the compounds but then immediately washed, both cysteines of Tpx were susceptible to modification by MAL-PEG (Fig. 5b) confirming the successful removal of the inhibitors before cell lysis. In contrast, when the parasites were cultured for 4 h in the presence of compounds 1, 2, 5, or 7, the main product was again the mono-modified protein. These results clearly established that compounds 1, 2, 5, and 7 entered the cell and covalently modified Tpx. In addition, the finding that the protein reacted with the compounds in the intact parasites revealed that intracellular Tpx is present in the accessible reduced form. We cannot rule out that other proteins might be affected as well. The fact that the 4-h treatment resulted in 100% inactivation of Tpx but was not accompanied by a significant growth defect might indicate that the compounds have additional off-target effects. However, the phenotype observed can also be explained by Tpx being the main target. The delayed onset of growth impairment closely resembles that of parasites subjected to RNA interference against Tpx. Depletion of the mRNA in bloodstream T. brucei leads within 24 h to a down-regulation of the protein to concentrations of <1 μm, but a significant growth defect becomes obvious only after 48 h (19). The depletion of Tpx is accompanied by an up-regulation of the cellular low molecular mass thiols that transiently may compensate for the lack of the dithiol protein (19). Because Tpx delivers the reducing equivalents for the synthesis of DNA precursors by ribonucleotide reductase (14), the main consequence of Tpx inactivation is probably an impaired proliferation of the parasites.
DISCUSSION
Of the nearly 80,000 chemicals studied against the peroxidase system of African trypanosomes, 32 compounds displayed activity. All 12 compounds that were mechanistically further characterized proved to be time-dependent inhibitors. As shown for compounds 1, 2, and 4–6, the compounds reacted with Cys-40 of Tpx with the elimination of HCl. The finding that Tpx was not only targeted in vitro but also in the intact parasite is remarkable. It clearly shows that free T(SH)2 present in high molar excess does not interfere with the modification of the parasite oxidoreductase. The specificity for Tpx observed is further corroborated by the fact that all three proteins that form the peroxidase cascade, namely TR, Tpx, and Px, possess reactive cysteine residues.
The inhibitors identified can be classified according to their common core structures or reactive substituents. Compounds 10 and 11 are chloroacetyl-substituted thiophenes. Structurally related compounds have been described as irreversible inhibitors of human glycogen synthase kinase 3 (GSK3-β) (43). The cysteine residue that is putatively affected is conserved in the two GSK3 homologs of T. brucei (44). Both compounds 10 and 11 readily inhibited the peroxidase system in vitro but did not reveal any specificity for the parasite compared with HeLa cells in the cell-based assays. These derivatives are probably not specific toward the target protein nor the organisms and were not further analyzed. All compounds of the second group possess a chloromethyl substituent. Compounds 1, 2, and 6–8 reacted with Tpx, but only 7 and 8 displayed also activity toward TR. Thus, despite the common reactive substituent, the distinct core structures appear to play a critical role for target interaction. This is supported by the finding that 7a, in contrast to the related 7 and 8, did not reveal significant inhibition of the peroxidase system. The third group formed by compounds 2, 4, 9, and 12 had in common a chloroacetamido substituent. None of the compounds inhibited TR. Compound 2 may interact not only with Tpx but also with Px. Taken together, despite the presence of a chloroalkyl substituent, the compounds revealed distinct reactivities toward the three thiol redox proteins of the peroxidase system.
The parasite Tpx is a distant relative of the thioredoxin protein family (11, 19). Therefore, we studied the compounds also toward recombinant human Trx. The same type of inactivation was observed, namely modification of its redox-active cysteine residue. Nevertheless, some of the compounds reacted much slower with Trx compared with Tpx. Indeed, compounds that interact with Trx do not necessarily react with Tpx. The antitumor benzothiazole-substituted quinole PMX464 inhibits Trx (21–23). A recent study on the T. brucei peroxidase system revealed that the quinole reacts with T(SH)2 as well as Px and Prx but not with Tpx or TR (20). This supports the conclusion that a selective inhibition of these structurally related proteins should be possible.
The finding that the compounds described here inactivate human Trx in vitro and can affect the proliferation of HeLa cells may even be exploited for anticancer drug development approaches. Trx has been shown to be up-regulated in certain malignancies and is currently studied as a target for anticancer drugs (22). In the work presented here, we chose HeLa cells as controls because this cancer cell line expresses Trx at high levels (45), which may at least partially explain the rather low selectivity indices observed. Fibroblast MRC-5 cells that are often used as control cells in antiparasitic drug testing (see for instance Ref. 20) have been reported to have undetectable Trx1 protein levels (22) and might have resulted in higher selectivity indices.
A critical issue that could be raised is that all compounds identified here are covalent inhibitors of Tpx. Drugs that covalently attach to their target are traditionally considered as unfavorable. However, as outlined in an excellent recent review (46), covalent inhibitors have proved to be successful therapies for various indications and about one-third of all approved covalent drugs are anti-infectives. Current covalent drug discovery programs mainly try to target a noncatalytic nucleophile. However, this restriction may not be crucial when regarding antimicrobial agents. As the next step toward the development of Tpx inhibitors as putative novel antitrypanosomal drugs, work is in progress to test the most active compounds in the animal model.4 To our knowledge this is the first report on inhibitors of this parasite-specific dithiol/disulfide protein. Indeed, our data suggest that it should be possible to develop specific inhibitors of Tpx, a protein that occupies a central position in the trypanothione-based thiol redox metabolism of these parasites.
Supplementary Material
Acknowledgments
We are grateful to P. Sehr and V. Pande for technical support, analysis, and discussion of the data (Chemical Biology Core Facility, EMBL, Germany). We thank S. Leicht and J. Kirkpatrick (Proteomics Core Facility, EMBL), T. Ruppert (Core Facility for Mass Spectrometry and Proteomics, Zentrum für Molekularbiologie der Universität Heidelberg, Germany), and M. Muehlbaier (Elara Pharmaceuticals, Germany) for mass spectrometric analyses. We thank N. Dirdjaja and A. Dietl (Biochemie-Zentrum der Universität Heidelberg) for experimental help and R. H. Schirmer (Biochemie-Zentrum der Universität Heidelberg) for a sample of human thioredoxin. H. Budde and L. Flohé (Gesellschaft für Biotechnologische Forschung mbH, Braunschweig, Germany) are kindly acknowledged for providing us with Tpx expression plasmids.
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 544, Project B3 (to L. K.-S.).

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
A. Dietl and R. L. Krauth-Siegel, unpublished data.
M. Comini, D. Benitez, and G. Fernández (Pasteur Institut Montevideo), personal communication.
- Prx
- 2-Cys-peroxiredoxin
- HTS
- high throughput screening
- MAL-PEG
- α-methoxy-ω-ethylmaleimide poly(ethylene glycol)
- Px
- nonselenium glutathione peroxidase-type enzyme
- t-bOOH
- tertiary butyl hydroperoxide
- TCEP
- tris(2-carboxyethyl)phosphine
- Tpx
- tryparedoxin
- TR
- trypanothione reductase
- Trx
- thioredoxin
- T(SH)2
- trypanothione.
REFERENCES
- 1. Krauth-Siegel R. L., Comini M. A. (2008) Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim. Biophys. Acta 1780, 1236–1248 [DOI] [PubMed] [Google Scholar]
- 2. Castro H., Tomás A. M. (2008) Peroxidases of trypanosomatids. Antioxid. Redox. Signal. 10, 1593–1606 [DOI] [PubMed] [Google Scholar]
- 3. Diechtierow M., Krauth-Siegel R. L. (2011) A tryparedoxin-dependent peroxidase protects African trypanosomes from membrane damage. Free Radic. Biol. Med. 51, 856–868 [DOI] [PubMed] [Google Scholar]
- 4. Fairlamb A. H., Cerami A. (1992) Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol. 46, 695–729 [DOI] [PubMed] [Google Scholar]
- 5. Otero L., Vieites M., Boiani L., Denicola A., Rigol C., Opazo L., Olea-Azar C., Maya J. D., Morello A., Krauth-Siegel R. L., Piro O. E., Castellano E., González M., Gambino D., Cerecetto H. (2006) Novel antitrypanosomal agents based on palladium nitrofurylthiosemicarbazone complexes. DNA and redox metabolism as potential therapeutic targets. J. Med. Chem. 49, 3322–3331 [DOI] [PubMed] [Google Scholar]
- 6. Holloway G. A., Charman W. N., Fairlamb A. H., Brun R., Kaiser M., Kostewicz E., Novello P. M., Parisot J. P., Richardson J., Street I. P., Watson K. G., Baell J. B. (2009) Trypanothione reductase high throughput screening campaign identifies novel classes of inhibitors with antiparasitic activity. Antimicrob. Agents Chemother. 53, 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eberle C., Lauber B. S., Fankhauser D., Kaiser M., Brun R., Krauth-Siegel R. L., Diederich F. (2011) Improved inhibitors of trypanothione reductase by combination of motifs. Synthesis, inhibitory potency, binding mode, and antiprotozoal activities. Chem. Med. Chem. 6, 292–301 [DOI] [PubMed] [Google Scholar]
- 8. Krauth-Siegel R. L., Bauer H., Schirmer R. H. (2005) Dithiol proteins as guardians of the intracellular redox milieu in parasites. Old and new drug targets in trypanosomes and malaria-causing plasmodia. Angew. Chem. Int. Ed. Engl. 44, 690–715 [DOI] [PubMed] [Google Scholar]
- 9. Rivera G., Bocanegra-García V., Ordaz-Pichardo C., Nogueda-Torres B., Monge A. (2009) New therapeutic targets for drug design against Trypanosoma cruzi, advances and perspectives. Curr. Med. Chem. 16, 3286–3293 [DOI] [PubMed] [Google Scholar]
- 10. Nogoceke E., Gommel D. U., Kiess M., Kalisz H. M., Flohé L. (1997) A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol. Chem. 378, 827–836 [DOI] [PubMed] [Google Scholar]
- 11. Lüdemann H., Dormeyer M., Sticherling C., Stallmann D., Follmann H., Krauth-Siegel R. L. (1998) Trypanosoma brucei tryparedoxin, a thioredoxin-like protein in African trypanosomes. FEBS Lett. 431, 381–385 [DOI] [PubMed] [Google Scholar]
- 12. Gommel D. U., Nogoceke E., Morr M., Kiess M., Kalisz H. M., Flohé L. (1997) Catalytic characteristics of tryparedoxin. Eur. J. Biochem. 248, 913–918 [DOI] [PubMed] [Google Scholar]
- 13. Arias D. G., Cabeza M. S., Erben E. D., Carranza P. G., Lujan H. D., Téllez Iñón M. T., Iglesias A. A., Guerrero S. A. (2011) Functional characterization of methionine sulfoxide reductase A from Trypanosoma spp. Free Radic. Biol. Med. 50, 37–46 [DOI] [PubMed] [Google Scholar]
- 14. Dormeyer M., Reckenfelderbäumer N., Ludemann H., Krauth-Siegel R. L. (2001) Trypanothione-dependent synthesis of deoxyribonucleotides by Trypanosoma brucei ribonucleotide reductase. J. Biol. Chem. 276, 10602–10606 [DOI] [PubMed] [Google Scholar]
- 15. Martyn D. C., Jones D. C., Fairlamb A. H., Clardy J. (2007) High throughput screening affords novel and selective trypanothione reductase inhibitors with anti-trypanosomal activity. Bioorg. Med. Chem. Lett. 17, 1280–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krieger S., Schwarz W., Ariyanayagam M. R., Fairlamb A. H., Krauth-Siegel R. L., Clayton C. (2000) Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol. Microbiol. 35, 542–552 [DOI] [PubMed] [Google Scholar]
- 17. Wilkinson S. R., Horn D., Prathalingam S. R., Kelly J. M. (2003) RNA interference identifies two hydroperoxide-metabolizing enzymes that are essential to the bloodstream form of the African trypanosome. J. Biol. Chem. 278, 31640–31646 [DOI] [PubMed] [Google Scholar]
- 18. Schlecker T., Schmidt A., Dirdjaja N., Voncken F., Clayton C., Krauth-Siegel R. L. (2005) Substrate specificity, localization, and essential role of the glutathione peroxidase-type tryparedoxin peroxidases in Trypanosoma brucei. J. Biol. Chem. 280, 14385–14394 [DOI] [PubMed] [Google Scholar]
- 19. Comini M. A., Krauth-Siegel R. L., Flohé L. (2007) Depletion of the thioredoxin homologue tryparedoxin impairs antioxidative defence in African trypanosomes. Biochem. J. 402, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. König J., Wyllie S., Wells G., Stevens M. F., Wyatt P. G., Fairlamb A. H. (2011) Antitumor quinol PMX464 is a cytocidal anti-trypanosomal inhibitor targeting trypanothione metabolism. J. Biol. Chem. 286, 8523–8533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bradshaw T. D., Matthews C. S., Cookson J., Chew E. H., Shah M., Bailey K., Monks A., Harris E., Westwell A. D., Wells G., Laughton C. A., Stevens M. F. (2005) Elucidation of thioredoxin as a molecular target for antitumor quinols. Cancer Res. 65, 3911–3919 [DOI] [PubMed] [Google Scholar]
- 22. Mukherjee A., Huber K., Evans H., Lakhani N., Martin S. (2007) A cellular and molecular investigation of the action of PMX464, a putative thioredoxin inhibitor, in normal and colorectal cancer cell lines. Br. J. Pharmacol. 151, 1167–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hall G., Bradshaw T. D., Laughton C. A., Stevens M. F., Emsley J. (2011) Structure of Mycobacterium tuberculosis thioredoxin in complex with quinol inhibitor PMX464. Protein Sci. 20, 210–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Melchers J., Krauth-Siegel L., Muhle-Goll C. (2008) 1H, 13C, and 15N assignment of the oxidized and reduced forms of T. brucei glutathione peroxidase-type tryparedoxin peroxidase. Biomol. NMR Assign. 2, 65–68 [DOI] [PubMed] [Google Scholar]
- 25. Budde H., Flohé L., Hecht H. J., Hofmann B., Stehr M., Wissing J., Lünsdorf H. (2003) Kinetics and redox-sensitive oligomerisation reveal negative subunit cooperativity in tryparedoxin peroxidase of Trypanosoma brucei brucei. Biol. Chem. 384, 619–633 [DOI] [PubMed] [Google Scholar]
- 26. Sullivan F. X., Walsh C. T. (1991) Cloning, sequencing, overproduction, and purification of trypanothione reductase from Trypanosoma cruzi. Mol. Biochem. Parasitol. 44, 145–147 [DOI] [PubMed] [Google Scholar]
- 27. Comini M. A., Dirdjaja N., Kaschel M., Krauth-Siegel R. L. (2009) Preparative enzymatic synthesis of trypanothione and trypanothione analogues. Int. J. Parasitol. 39, 1059–1062 [DOI] [PubMed] [Google Scholar]
- 28. Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 [DOI] [PubMed] [Google Scholar]
- 29. Sehr P., Pawlita M., Lewis J. (2007) Evaluation of different glutathione S-transferase-tagged protein captures for screening E6/E6AP interaction inhibitors using AlphaScreen. J. Biomol. Screen. 12, 560–567 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J. H., Chung T. D., Oldenburg K. R. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 [DOI] [PubMed] [Google Scholar]
- 31. Erfle H., Neumann B., Rogers P., Bulkescher J., Ellenberg J., Pepperkok R. (2008) Work flow for multiplexing siRNA assays by solid-phase reverse transfection in multiwell plates. J. Biomol. Screen. 13, 575–580 [DOI] [PubMed] [Google Scholar]
- 32. Rist W., Mayer M. P., Andersen J. S., Roepstorff P., Jørgensen T. J. (2005) Rapid desalting of protein samples for on-line microflow electrospray ionization mass spectrometry. Anal. Biochem. 342, 160–162 [DOI] [PubMed] [Google Scholar]
- 33. Hillebrand H., Schmidt A., Krauth-Siegel R. L. (2003) A second class of peroxidases linked to the trypanothione metabolism. J. Biol. Chem. 278, 6809–6815 [DOI] [PubMed] [Google Scholar]
- 34. Carnieri E. G., Moreno S. N., Docampo R. (1993) Trypanothione-dependent peroxide metabolism in Trypanosoma cruzi different stages. Mol. Biochem. Parasitol. 61, 79–86 [DOI] [PubMed] [Google Scholar]
- 35. Simeonov A., Jadhav A., Sayed A. A., Wang Y., Nelson M. E., Thomas C. J., Inglese J., Williams D. L., Austin C. P. (2008) Quantitative high throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade. PLoS Negl. Trop. Dis. 2, e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Järvinen P. P., Fallarero A., Gupta S., Mohan G. C., Hatakka A. I., Vuorela P. M. (2010) Miniaturization and validation of the Ellman's reaction-based acetylcholinesterase inhibitory assay into 384-well plate format and screening of a chemical library. Comb. Chem. High Throughput Screen. 13, 278–284 [DOI] [PubMed] [Google Scholar]
- 37. Mackey Z. B., Baca A. M., Mallari J. P., Apsel B., Shelat A., Hansell E. J., Chiang P. K., Wolff B., Guy K. R., Williams J., McKerrow J. H. (2006) Discovery of trypanocidal compounds by whole cell HTS of Trypanosoma brucei. Chem. Biol. Drug Des. 67, 355–363 [DOI] [PubMed] [Google Scholar]
- 38. Melchers J., Diechtierow M., Fehér K., Sinning I., Tews I., Krauth-Siegel R. L., Muhle-Goll C. (2008) Structural basis for a distinct catalytic mechanism in Trypanosoma brucei tryparedoxin peroxidase. J. Biol. Chem. 283, 30401–30411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schlecker T., Comini M. A., Melchers J., Ruppert T., Krauth-Siegel R. L. (2007) Catalytic mechanism of the glutathione peroxidase-type tryparedoxin peroxidase of Trypanosoma brucei. Biochem. J. 405, 445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alphey M. S., Leonard G. A., Gourley D. G., Tetaud E., Fairlamb A. H., Hunter W. N. (1999) The high resolution crystal structure of recombinant Crithidia fasciculata tryparedoxin-I. J. Biol. Chem. 274, 25613–25622 [DOI] [PubMed] [Google Scholar]
- 41. Hofmann B., Budde H., Bruns K., Guerrero S. A., Kalisz H. M., Menge U., Montemartini M., Nogoceke E., Steinert P., Wissing J. B., Flohé L., Hecht H. J. (2001) Structures of tryparedoxins revealing interaction with trypanothione. Biol. Chem. 382, 459–471 [DOI] [PubMed] [Google Scholar]
- 42. Budde H., Flohé L., Hofmann B., Nimtz M. (2003) Verification of the interaction of a tryparedoxin peroxidase with tryparedoxin by ESI-MS/MS. Biol. Chem. 384, 1305–1309 [DOI] [PubMed] [Google Scholar]
- 43. Perez D. I., Conde S., Pérez C., Gil C., Simon D., Wandosell F., Moreno F. J., Gelpí J. L., Luque F. J., Martínez A. (2009) Thienylhalomethylketones. Irreversible glycogen synthase kinase 3 inhibitors as useful pharmacological tools. Bioorg. Med. Chem. 17, 6914–6925 [DOI] [PubMed] [Google Scholar]
- 44. Ojo K. K., Gillespie J. R., Riechers A. J., Napuli A. J., Verlinde C. L., Buckner F. S., Gelb M. H., Domostoj M. M., Wells S. J., Scheer A., Wells T. N., Van Voorhis W. C. (2008) Glycogen synthase kinase 3 is a potential drug target for African trypanosomiasis therapy. Antimicrob. Agents Chemother. 52, 3710–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jeong W., Yoon H. W., Lee S. R., Rhee S. G. (2004) Identification and characterization of TRP14, a thioredoxin-related protein of 14 kDa. New insights into the specificity of thioredoxin function. J. Biol. Chem. 279, 3142–3150 [DOI] [PubMed] [Google Scholar]
- 46. Singh J., Petter R. C., Baillie T. A., Whitty A. (2011) The resurgence of covalent drugs. Nat. Rev. Drug Discov. 10, 307–317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.