Background: Assembly of the 26 S proteasome is a complex process involving formation of ring structures from homologous subunits.
Results: Mutations in the Walker A and Walker B motifs and C-terminal tails of Rpt subunits differentially affect cellular proteasome assembly.
Conclusion: The cellular assembly of the regulatory particle requires nucleotide binding and the C termini of Rpt subunits.
Significance: This study provides insight into the mechanism of the cellular proteasome assembly.
Keywords: ATPases, Chaperone Chaperonin, Proteasome, Protein Assembly, Protein Degradation, Rpt Subunits
Abstract
The 26 S proteasome is a large multi-subunit protein complex that degrades ubiquitinated proteins in eukaryotic cells. Proteasome assembly is a complex process that involves formation of six- and seven-membered ring structures from homologous subunits. Here we report that the assembly of hexameric Rpt ring of the 19 S regulatory particle (RP) requires nucleotide binding but not ATP hydrolysis. Disruption of nucleotide binding to an Rpt subunit by mutation in the Walker A motif inhibits the assembly of the Rpt ring without affecting heterodimer formation with its partner Rpt subunit. Coexpression of the base assembly chaperones S5b and PAAF1 with mutant Rpt1 and Rpt6, respectively, relieves assembly inhibition of mutant Rpts by facilitating their interaction with adjacent Rpt dimers. The mutation in the Walker B motif which impairs ATP hydrolysis does not affect Rpt ring formation. Incorporation of a Walker B mutant Rpt subunit abrogates the ATPase activity of the 19 S RP, suggesting that failure of the mutant Rpt to undergo the conformational transition from an ATP-bound to an ADP-bound state impairs conformational changes in the other five wild-type Rpts in the Rpt ring. In addition, we demonstrate that the C-terminal tails of Rpt subunits possessing core particle (CP)-binding affinities facilitate the cellular assembly of the 19 S RP, implying that the 20 S CP may function as a template for base assembly in human cells. Taken together, these results suggest that the ATP-bound conformational state of an Rpt subunit with the exposed C-terminal tail is competent for cellular proteasome assembly.
Introduction
The 26 S proteasome is a large proteolytic complex that functions in the ATP-dependent degradation of ubiquitinated proteins in eukaryotic cells (1). The 26 S proteasome consists of a 20 S core particle (CP)2 and a 19 S regulatory particle (RP, also known as PA700) (2). The 20 S CP has a barrel-shaped structure composed of two outer α rings and two inner β rings, where each ring contains seven homologous members (3). The central β rings possess proteolytically active sites, whereas the outer α rings provide attachment sites for the 19 S RP and control the access of substrates to the catalytic chamber by serving as a gated channel (4, 5). The 19 S RP can be further divided into the base and the lid subcomplexes. The base subcomplex contains three non-ATPase subunits and six homologous ATPase subunits, Rpt1-Rpt6, which assemble into a six-membered ring with the specific order of Rpt1-Rpt2-Rpt6-Rpt3-Rpt4-Rpt5 (6). The hexameric Rpt ring directly touches the heptameric α ring of the 20 S CP and opens the α ring channel. ATP binding to the Rpt subunits is required for the association of the RP to the CP and the opening of the gated channel (7, 8). ATP hydrolysis by the Rpt ring drives the unfolding of the substrate prior to its passage through the α ring channel. The lid subcomplex is located distally to the CP and contains nine non-ATPase subunits, where Rpn11 removes ubiquitin from the substrate for recycling of ubiquitin molecules (9, 10).
The assembly of the proteasome appears to involve chaperone-assisted formation of CP and ATPase ring structures as well as self-association between specific subunits. Thus, CP assembly is mediated by dedicated chaperones Pba1–4 and Ump1 in yeast and by PAC1–4 and UMP1/POMP in mammalian cells (11, 12). Recent studies have revealed that the assembly of the base subcomplex is supported by four specific chaperones, Hsm3, Rpn14, Nas6, and Nas2, in yeast, and by their respective orthologs, S5b, PAAF1, p28/gankyrin, and p27, in mammalian cells (13–20). The assembly of the base subcomplex starts with the formation of ATPase heterodimers, Rpt1-Rpt2, Rpt6-Rpt3, and Rpt4-Rpt5, each of which binds at least one specific chaperone. Structural analysis of the archaeal RP, the hexameric ATPase ring with identical subunits called proteasome-activating nucleotidase (PAN), has indicated that the archaeal RP has a trimer of dimers structure (21). PAN and Rpt subunits contain a coiled coil (CC) domain and an oligonucleotide/oligosaccharide-binding (OB) domain in the N terminus. Dimerization is promoted partly by a dimeric coiled coil formed by the CC domains of two subunits. PAN and three Rpt subunits, Rpt2, Rpt3, and Rpt5, contain a critical proline residue between CC and OB domains, which can assume a cis configuration enabling dimer formation with a partner subunit possessing a residue in a trans configuration at the corresponding site (21). The ATPase domain of proteasomal ATPases is composed of a nucleotide-binding subdomain followed by a smaller C-terminal helical subdomain called the C-domain, to which RP chaperones bind (13, 14, 22, 23). How each RP chaperone functions in the assembly of the RP ATPase ring is not fully understood.
C-terminal tails of Rpt subunits have been shown to play important roles in binding of 19 S RP to 20 S CP and gate opening (24–26). Rpt2, Rpt3, and Rpt5 contain hydrophobic-tyrosine-X (HbYX) motifs at their C-terminal ends (27). Recent studies have shown that the HbYX motif-containing C-terminal peptides of Rpt2 and Rpt5 bind to the 20 S proteasome and stimulate its activity (27, 28). Deletion of the HbYX motifs in Rpt2 and Rpt5 by carboxypeptidase A inhibited the association of the 19 S RP to the 20 S CP and caused the loss of proteasome activation (27, 28). In addition, the C-terminal HbYX motif of Rpt3 was shown to be required for the association of the 19 S RP with the 20 S CP (29). The C termini of Rpt1, Rpt4, and Rpt6 do not possess HbYX motifs and have no appreciable binding affinity to the 20 S CP. Although HbYX motifs have an important function in the stable association of the RP and CP, it is not clear whether they play any role in 19 S RP assembly in mammalian cells. In yeast, deletion of the last C-terminal residue from Rpt4 or Rpt6 resulted in severe defects in proteasome assembly (30). Recently, CP-assisted base assembly was proposed based on biochemical and genetic studies in yeast and mammalian cells (12, 30, 31, 33).
In this study, to better understand the proteasome assembly pathway, we investigated the effects of nucleotide binding and ATP hydrolysis of Rpt subunits on the cellular formation of the proteasome. We also evaluated the role of the C termini of Rpt subunits in the proteasome assembly process. We show that inhibition of nucleotide binding to Rpts resulted in impairment of ATPase ring assembly, which was rescued by the overexpression of base-specific chaperones in some cases. In addition, we demonstrate that the C-terminal tails of Rpt2, Rpt3, and Rpt5 are required for the efficient cellular assembly of the 19 S RP.
EXPERIMENTAL PROCEDURES
Plasmids
The cDNAs encoding human proteasome subunits were obtained from MRC geneservice (Cambridge, UK). To construct plasmids for the expression of epitope-tagged proteins, cDNAs were amplified by PCR with appropriate primers and ligated into pcDNA3.1 (Invitrogen, Carlsbad, CA) or pYR vectors (34).
Site-directed Mutagenesis
The lysine residues (K222 of Rpt1, K232 of Rpt2, K212 of Rpt3, K194 of Rpt4, K233 of Rpt5, and K196 of Rpt6) in the Walker A motif and the glutamic acid residues (E276 of Rpt1, E286 of Rpt2, E276 of Rpt3, E248 of Rpt4, E287 of Rpt5, and E250 of Rpt6) in the Walker B motif of Rpt subunits were mutagenized to serine and lysine, respectively. The mutations were introduced with the QuikChange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol using appropriate mutagenesis primers. All constructs were sequenced to ensure correctness.
Western Blot Analysis
Transfection was performed with Lipofectamine 2000 (Invitrogen). After 36 h, cells were lysed in 50 mm Tris-HCl (pH 7.4) buffer containing 150 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 0.2 mm PMSF, and 1.0% Nonidet P-40. Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes (Whatman, Kent, UK), and visualized by Western blotting with enhanced chemiluminescence reagents (Amersham Biosciences Pharmacia, Uppsala, Sweden). For Western blotting, we used antibodies against FLAG (Sigma), hemagglutinin (HA) (Roche, Basel, Switzerland), T7 (Novagen, Gibbstown, NJ), and proteasome subunits (Biomol, Plymouth Meeting, NJ).
Affinity Purification of the Proteasome from Cell Lines Expressing a FLAG-tagged Proteasome Subunit
Cells derived from a HeLa Tet-Off cell line (Clontech, Mountain View, CA) that stably expressed EBNA-1 were transfected with the episomal vector pYR for expression of FLAG-tagged proteasome subunits. These vectors contained the gene of interest under a tetracycline-regulated promoter, oriP for episome replication, and a selection marker for hygromycin B. The cells were selected and maintained in Dulbecco modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (GIBCO BRL, Carlsbad, CA), 100 μg/ml G418 (Sigma), 300 μg/ml hygromycin B (Clontech), 100 units/ml penicillin, 100 μg/ml streptomycin, 1 mm l-glutamine, and 2 μg/ml tetracycline (Sigma). Cells were grown without tetracycline for 3 days to induce the expression of the FLAG-tagged proteins. The cells were lysed in buffer A containing 20 mm Tris-HCl at pH 7.4, 150 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 0.2 mm PMSF, and 0.5% Nonidet P-40. The lysates were centrifuged at 20,000 × g for 15 min to remove cell debris. The supernatant was incubated with anti-FLAG M2 agarose (Sigma) overnight at 4 °C. After extensive washes with buffer A without Nonidet P-40, the proteins were eluted with 0.3 mg of FLAG peptide per ml in buffer A without Nonidet P-40.
Peptidase Activity Assay
All proteasome peptidase assays were performed in 200 μl reaction mixtures containing 50 mm Tris-HCl, pH 7.5, 40 mm KCl, 5 mm MgCl2, 0.5 mm ATP, 1 mm dithiothreitol (DTT), 100 μm fluorogenic substrates, and 1 μg of proteasome. Proteasome samples were assayed for chymotrypsin-like activity using the 7-amino-4-methylcoumarin (AMC)-labeled peptide Suc-LLVY-AMC as a fluorogenic substrate. Peptidase activity was measured at 37 °C for 30 min by continuously monitoring AMC production with a Gemini EM Microplate spectrofluorometer (Molecular Devices, Sunnyvale, CA) using 380 nm excitation and 460 nm emission filters and quantified with reference to an AMC calibration curve.
Native Polyacrylamide Gel Electrophoresis and Fluorescence Substrate Overlay Assay
Non-denaturing PAGE was conducted with 3.5% polyacrylamide gels as described (35). Proteasome samples were visualized with 50 μm Suc-LLVY-AMC in developing buffer containing 50 mm Tris-Cl (pH 7.4), 5 mm MgCl2, and 1 mm ATP for 20 min at 37 °C.
ATPase Assay
ATPase activity was assayed by a modified Malachite Green method (36). Proteasome samples were incubated in 80 μl of reaction buffer (20 mm HEPES, pH 7.2, 1 mm DTT, and 5 mm MgCl2) containing 200 μm ATP for 30 min at 37 °C. After incubation, the reactions were quenched with 20 μl of Malachite Green reagent (0.034% Malachite Green oxalate, 1.1% ammonium molybdate, 1 m HCl, and 0.04% Tween 20). The quenched reaction mixtures were incubated for 5 min at room temperature, and samples were measured at 655 nm with a Benchmark microplate reader (Bio-Rad).
In Vitro Ubiquitination and Degradation Assay
Auto-ubiquitination of the BIR domain-deleted C-terminal half of cIAP2 (cIAP2-C) was performed in a reaction buffer containing 25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm ATP, 10 mm MgCl2, 1 mm DTT, 1 μg of ubiquitin, 250 ng of Uba1, 500 ng of UbcH5a, and 100 ng of T7-cIAP2-C (37). After incubation at 25 °C for 1 h, the reaction was terminated by the addition of SDS sample buffer and analyzed by Western blotting analysis with anti-T7 antibody. The in vitro degradation assay was performed using polyubiquitinated cIAP2-C as a substrate. Polyubiquitinated cIAP2-C was incubated with the purified proteasome in a reaction buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm DTT, 2 mm ATP, and 5 mm MgCl2 at 37 °C. The reaction was terminated by adding SDS sample buffer and analyzed by Western blotting with anti-T7 antibody to assess the degree of degradation.
RESULTS
Rpt Ring Formation Requires Nucleotide Binding but Not ATP Hydrolysis
To examine the effects of nucleotide binding and ATP hydrolysis of Rpt subunits on the cellular formation of the proteasome, we have generated point mutants of Rpt subunits that are incapable of nucleotide binding or ATP hydrolysis. Because the invariable lysine residue of the Walker A motif and the conserved glutamate residue of the Walker B motif are crucial in nucleotide binding and ATP hydrolysis in other ATPases, respectively, we replaced the Walker A lysine and Walker B glutamate in Rpt subunits with serine and lysine by site-directed mutagenesis. We then tested whether the Walker A and B mutations have the expected effect on nucleotide binding and ATP hydrolysis by analyzing purified recombinant wild-type and mutant Rpt6 proteins. As shown in supplemental Fig. S1, wild-type and Walker B mutant Rpt6 were capable of binding radiolabeled ATP, whereas Walker A mutant Rpt6 was not. Recombinant wild-type Rpt6 as well as two mutant forms of Rpt6 did not display any ATPase activities (supplemental Fig. S1B), indicating that the isolated Rpt subunit by itself does not function as an ATPase. This finding is consistent with the recent study from DeMartino group showing that RP subcomplexes containing two different Rpt subunits does not show any ATPase activity although the reconstituted RP from the three subcomplexes possesses ATPase activity (38). Although we were not able to directly show the effect of Walker B mutation on ATPase activity of the isolated Rpt subunit, the data obtained with the RP containing a Walker B mutant Rpt strongly suggested that Walker B mutation renders the Rpt subunit nonfunctional in ATP hydrolysis as described below (Fig. 3).
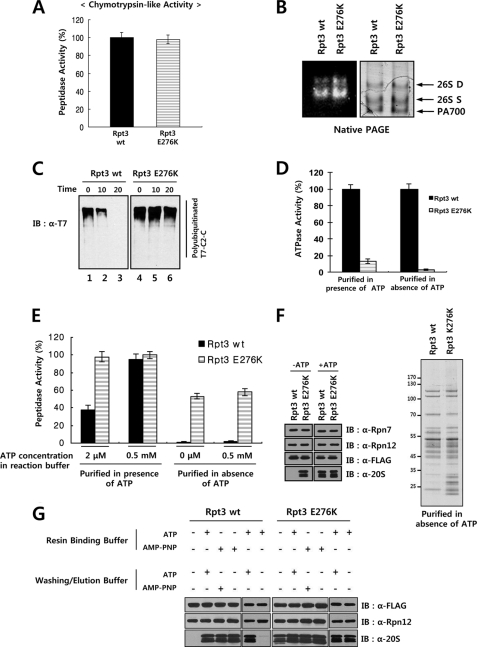
FIGURE 3.
Incorporation of a single Walker B mutant Rpt subunit abrogates the ATPase activity of 19 S RP. A, comparable chymotrysin-like activities of purified proteasome samples containing the wild-type or Walker B mutant Rpt3. One microgram each of the wild-type and Walker B mutant Rpt3 samples shown in Fig. 2C was assayed for chymotrypsin-like peptidase activity using the Suc-LLVY-AMC peptide as a fluorogenic substrate. B, native PAGE analysis and substrate overlay assay of proteasome samples containing wild-type or Walker B mutant Rpt3. Affinity-purified samples were subjected to native PAGE followed by Coomassie staining or by substrate overlay assay as described under “Experimental Procedures.” C, impaired degradation of a polyubiquitinated protein substrate by proteasome containing Walker B mutant Rpt3. Polyubiquitinated T7-cIAP2-C (C2-C) was prepared as described under “Experimental Procedures.” The polyubiquitinated substrate was incubated with indicated proteasome preparations and protein degradation was analyzed by Western blotting with anti-T7 antibody. D, impaired ATPase activity of proteasome containing Walker B mutant Rpt3. FLAG-Rpt3 samples purified in the presence or absence of ATP were assayed for ATPase activity as described under “Experimental Procedures.” E, chymotrysin-like activities of proteasome samples purified with or without ATP. Wild-type and Walker B mutant FLAG-Rpt3 samples purified in the presence or absence of ATP were assayed for chymotrypsin-like peptidase activity using Suc-LLVY-AMC as a fluorogenic substrate in buffers containing the indicated concentrations of ATP. Data are presented as the mean ± S.D. from three independent experiments. F, immunoblotting and SDS-PAGE analyses of proteasome samples purified with or without ATP. Wild-type and Walker B mutant FLAG-Rpt3 samples purified in the presence or absence of ATP were analyzed by immunoblotting with indicated antibodies. SDS-PAGE analysis of proteasome samples purified in the absence of ATP is shown in the right panel. G, increased stability of 26 S proteasome during purification by AMP-PNP. Wild-type and Walker B mutant FLAG-Rpt3 samples were purified using resin binding and washing/elution buffers with or without 2 mm ATP or AMP-PNP as indicated. Samples were analyzed by immunoblotting with indicated antibodies.
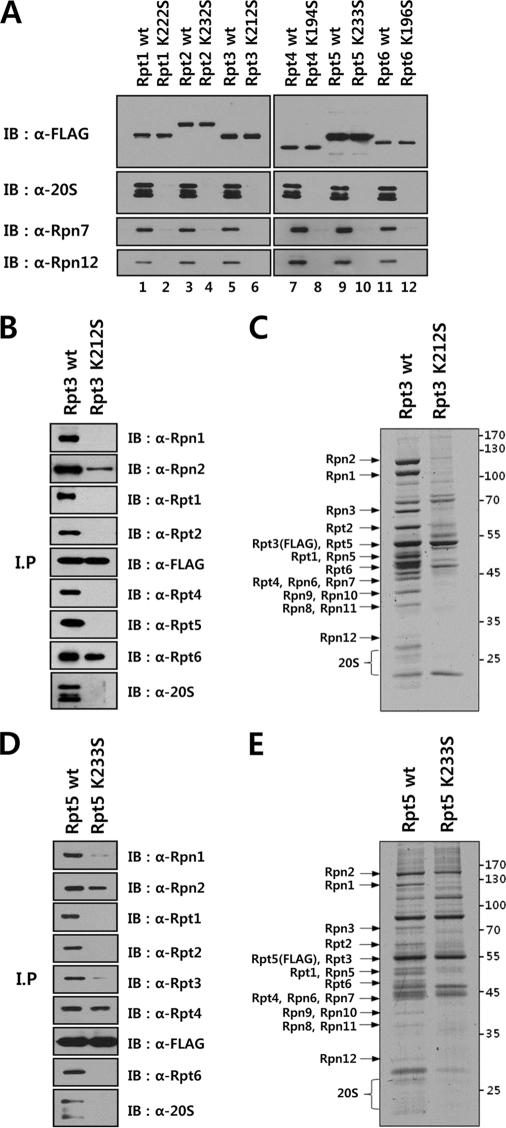
To investigate the incorporation of Rpt subunits into 26 S proteasome, FLAG-tagged versions of wild-type or Walker A mutant Rpt subunits were transiently expressed in HeLa cells and immunoprecipitated with anti-FLAG antibody. Western blotting analysis of immunoprecipitated samples revealed that the 20 S proteasome and lid subunits Rpn7 and Rpn12 were copurified with wild-type Rpt subunits but not with the Walker A mutant subunits (Fig. 1A), suggesting that the lysine to serine mutation in the Walker A motif impaired the incorporation of the mutant Rpts into the intact 26 S proteasome. To further study defects of Walker A mutant Rpts in the assembly of the proteasome, HeLa-derived cell lines were established which conditionally expressed FLAG-tagged wild-type or Walker A mutant Rpt subunits, and the proteasome was affinity purified from these cells. SDS-PAGE and immunoblotting analysis of purified FLAG-Rpt3 and FLAG-Rpt5 samples confirmed that the assembly of mutant Rpts into the proteasome was impaired (Fig. 1, B–E). Although mutant Rpt3 and Rpt5 were readily associated with their dimer partner subunits Rpt6 and Rpt4, respectively, they did not interact with other Rpt subunits as efficiently as wild-type ones, indicating that the hexameric Rpt ring was not formed properly. Taken together, these results suggest that nucleotide binding to Rpt subunits is a prerequisite for stable assembly of the proteasomal ATPase ring.
FIGURE 1.
Nucleotide binding to Rpt subunits is required for the assembly of proteasome. A, impaired incorporation of Walker A mutant Rpt subunits into proteasome. HeLa cells were transiently transfected with the expression constructs of FLAG-tagged wild-type or Walker A mutant Rpt subunits as indicated. Aliquots of FLAG-Rpt samples purified in the presence of 5 mm ATP were separated by SDS-PAGE and analyzed by immunoblotting with indicated antibodies. B–C, immunoblotting (B) and SDS-PAGE (C) analyses of affinity-purified FLAG-tagged wild-type or Walker A mutant Rpt3. HeLa cell lines that conditionally expressed FLAG-tagged wild-type or Walker A mutant Rpt3 were established, and the proteasome was affinity purified as described under “Experimental Procedures.” Ten microliters each of Rpt3 samples purified in the presence of 5 mm ATP were analyzed by immunoblotting with the indicated antibodies and by SDS-PAGE followed by Coomassie staining. D and E, immunoblotting (D) and SDS-PAGE (E) analyses of affinity-purified FLAG-tagged wild-type or Walker A mutant Rpt5.
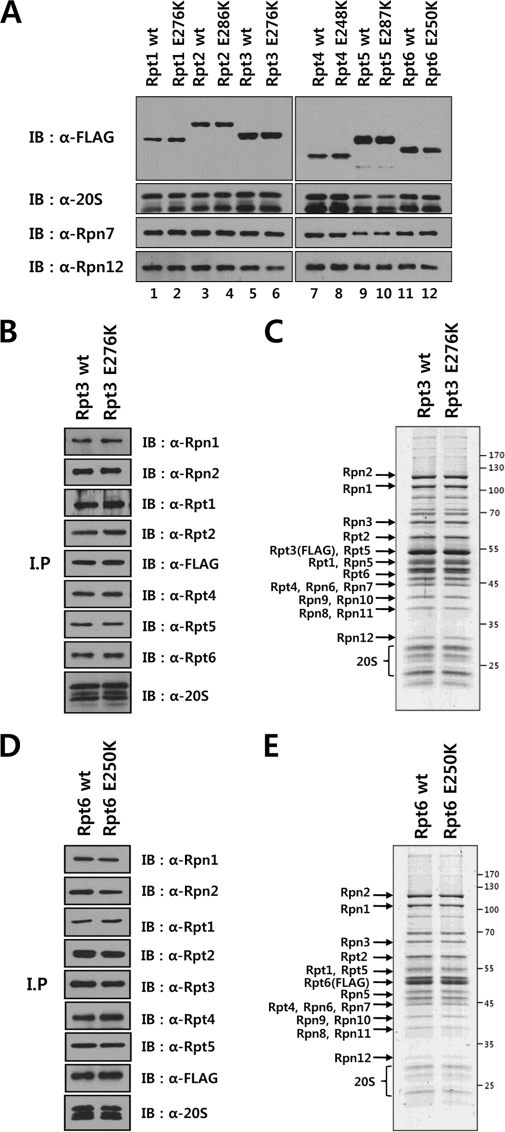
We next investigated the influence of ATP hydrolysis by Rpt subunits on the assembly of the proteasome. Walker B motif mutants of Rpt subunits were tested for their incorporation into the 26 S proteasome by transient transfection and immunoprecipitation of FLAG-tagged mutants. As shown in Fig. 2A, Western blotting analysis revealed that both the wild-type and the Walker B mutant Rpt subunits bound to the 20 S proteasome and lid subunits Rpn7 and Rpn12 to a similar extent, suggesting that Walker B mutations did not cause any negative effect on the assembly. To confirm the proper assembly of Walker B mutants into 26 S proteasome, we established tetracycline-inducible cell lines expressing FLAG-tagged Walker B mutant Rpts and analyzed purified FLAG-Rpt3 and FLAG-Rpt6 samples by SDS-PAGE and immunoblotting. The protein patterns of wild-type and Walker B mutant Rpt3 and Rpt6 were basically identical, showing that the mutant Rpts were readily incorporated into the intact 26 S proteasome (Fig. 2, B–E). These results indicated that ATP hydrolysis by Rpt subunits was not required for 26 S proteasome assembly.
FIGURE 2.
ATP hydrolysis by Rpt subunits is not essential for proteasome assembly. A, comparable incorporation of wild-type and Walker B mutant Rpt subunits into proteasome. HeLa cells were transiently transfected with the expression constructs of FLAG-tagged wild-type or Walker B mutant Rpt subunits as indicated. Aliquots of FLAG-Rpt samples purified in the presence of 5 mm ATP were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. B and C, immunoblotting (B) and SDS-PAGE (C) analyses of affinity-purified FLAG-tagged wild-type or Walker B mutant Rpt3. HeLa cell lines that conditionally expressed FLAG-tagged wild-type or Walker B mutant Rpt3 were established, and the proteasome was affinity purified as described under “Experimental Procedures.” Ten microliters each of Rpt3 samples purified in the presence of 5 mm ATP was analyzed by immunoblotting with indicated antibodies and by SDS-PAGE followed by Coomassie staining. D and E, immunoblotting (D) and SDS-PAGE (E) analyses of affinity-purified FLAG-tagged wild-type or Walker B mutant Rpt6.
Incorporation of a Single Walker B Mutant Rpt Subunit Completely Abrogates ATPase Activity of the 19 S RP
To study the effect of incorporation of a Walker B mutant Rpt subunit on proteasome function, we analyzed activities of purified proteasome samples containing wild-type or Walker B mutant FLAG-Rpt3 shown in Fig. 2C. Peptidase activities of proteasome samples were assessed by solution assays using Suc-Leu-Leu-Val-Tyr-AMC as a substrate. The magnitudes of peptidase activities of wild-type and mutant proteasomes were comparable to each other when measured in the presence of 0.5 mm ATP (Fig. 3A). Native gel electrophoresis of the purified proteasomes revealed that both wild-type and mutant proteasome preparations contained similar amounts of singly and doubly capped 26 S proteasome and 19 S RP (Fig. 3B, right panel). Substrate overlay assays confirmed that the peptidase activity of wild-type proteasome was similar to that of mutant proteasome (Fig. 3B, left panel), showing that the Walker B mutation in Rpt3 did not impair the gate opening and peptidase activities of the 26 S proteasome. Since the major function of the proteasome is the degradation of polyubiquitinated proteins, we prepared a polyubiquitinated form of cIAP2-C as a proteasome substrate and measured protein degradation activities of wild-type and mutant proteasomes. As shown in Fig. 3C, whereas the wild-type proteasome completely digested the polyubiquitinated protein within 20 min, the proteasome containing the Walker B mutant Rpt3 failed to efficiently digest the substrate. To examine whether the impaired degradation of the polyubiquitinated substrate by the mutant proteasome was caused by its decreased ATPase activity, we analyzed the ATPase activities of the wild-type and Walker B mutant proteasome. The ATPase activity of the proteasome containing the Walker B mutant Rpt3 was 13% of that of wild-type proteasome (Fig. 3D). Because the affinity-purified proteasome samples prepared in the presence of 2 mm ATP contained the doubly capped 26 S proteasome (Fig. 3B), it was likely that some of the purified proteasome complex possessed endogenous untagged Rpt3 in addition to the FLAG-tagged mutant Rpt3. To prevent the co-purification of 19 S RP containing endogenous Rpt3, we purified FLAG-Rpt3 and its associated proteins in the absence of ATP and determined the ATPase activity. Remarkably, while the 19 S RP preparation containing wild-type Rpt3 was active in ATPase assays, the sample containing the Walker B mutant FLAG-Rpt3 did not show any ATPase activity (Fig. 3D), indicating that a Walker B mutation in a single Rpt subunit resulted in a complete loss of ATPase activity of the hexameric Rpt ring.
Interestingly, when we measured the peptidase activities of the purified 26 S proteasome samples in reaction buffer containing 2 μm ATP, the peptidase activity of wild-type proteasome was reduced to ∼40% of the activity obtained in buffer containing 0.5 mm ATP, whereas the peptidase activity of proteasome containing Walker B mutant Rpt3 was reduced only slightly (Fig. 3E). Given that the activation of peptidase activity by ATP is closely related to association of the 19 S RP with the 20 S CP, the higher activity of the mutant proteasome might reflect a more stable association of the CP with the mutant RP than with wild-type RP under low-ATP conditions. To test this possibility, we purified the proteasome containing either wild-type or Walker B mutant FLAG-Rpt3 in the presence and absence of ATP, and examined the peptidase activity (Fig. 3E) and the presence of the 20 S CP by Western blot analysis (Fig. 3F). In the presence of 2 mm ATP, the 20 S CP as well as the 19 S RP was efficiently co-purified with wild-type and mutant FLAG-Rpt3. When purified in the absence of ATP, the wild-type FLAG-Rpt3 sample possessed practically no peptidase activity and very little 20 S CP. In contrast, the mutant FLAG-Rpt3 sample had ∼60% of peptidase activity even without ATP in the reaction buffer and more than 60% of the amount of 20 S CP obtained with 2 mm ATP in the purification buffer (Fig. 3, E and F). Similar results were obtained with the mutant FLAG-Rpt6 sample (data not shown). To test whether impairment of ATP hydrolysis by Walker B mutation is the main cause of enhanced stability of 26 S proteasome during purification, we investigated the effect of a non-hydrolyzable ATP analog, AMP-PNP, on stability of 26 S proteasome. As shown in Fig. 3G, addition of AMP-PNP to the cell lysate resulted in more stable 26 S proteasome which survived extensive washing and elution steps during purification with the washing/elution buffer lacking ATP or AMP-PMP. In contrast, when the lysate was supplemented with ATP instead of AMP-PNP, ATP-containing buffers were required in washing and elution steps to maintain the CP-RP interaction. These results suggested that when purified in the absence of ATP, the association between the RP and the CP is more stable in the mutant 26 S proteasome bearing the Walker B mutation than in the wild-type 26 S proteasome because of the inability of the mutant RP to hydrolyze ATP.
Overexpression of S5b and PAAF1 Can Overcome the Assembly Defect of Walker A Mutant Rpt1 and Rpt6, Respectively, by Facilitating the Interaction between Rpt Dimers
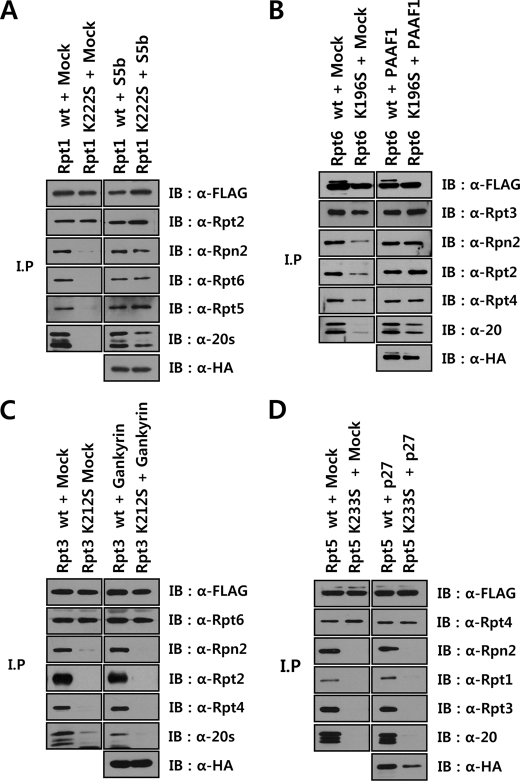
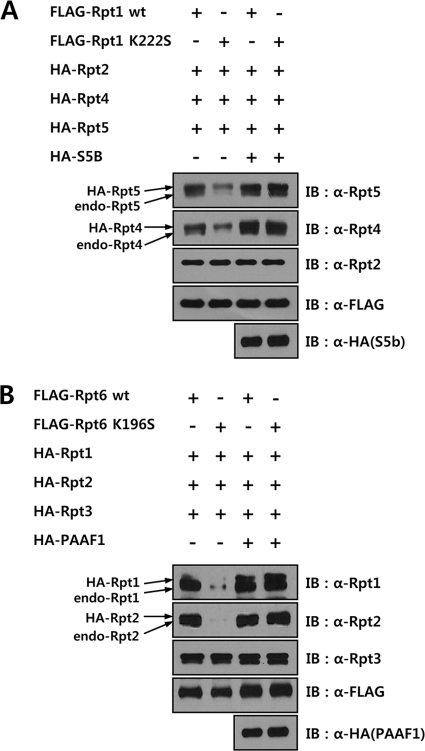
Because the defect in the proteasome assembly pathway due to Walker A mutations in Rpt subunits appeared to be at steps downstream of Rpt heterodimer formation, at which RP assembly chaperones may function, we investigated whether co-expression of RP assembly chaperones with Walker A mutant Rpts could overcome the assembly defect of mutant Rpts. Each of the RP assembly chaperones was co-expressed with the cognate wild-type or mutant Rpt subunit in HeLa cells and the immunopurified Rpt samples prepared in the presence of ATP were analyzed for proteasome assembly by Western blotting with antibodies against proteasome subunits. S5b, PAAF1, and gankyrin bound as efficiently to their cognate mutant Rpt subunits as to wild-type (Fig. 4, A–C), whereas p27 interacted less strongly with the mutant Rpt5 than with wild-type Rpt5 (Fig. 4D). Although a Walker A mutation in Rpts did not impair their association with their specific partner Rpt subunits, the interactions of the mutant Rpts with 20 S CP and 19 S RP subunits were greatly reduced. Remarkably, co-expression of S5b and PAAF1 with the mutant Rpt1 and Rpt6, respectively, enhanced the association of these Rpt mutants with the 20 S CP and 19 S RP subunits, almost to levels obtained with wild-type Rpt subunits (Fig. 4, A and B). In contrast, gankyrin and p27 did not increase the interaction of their cognate mutant Rpts with the proteasome subunits (Fig. 4, C and D). Rather, the binding of 20 S CP to 19 S RP containing the wild-type Rpt3 appeared to be reduced by gankyrin (Fig. 4C). These results suggested that the binding of S5b and PAAF1 to the Walker A mutant Rpt1 and Rpt6, respectively, led to efficient incorporation of the mutants into proteasome.
FIGURE 4.
Differential effects of base-specific chaperones on proteasome assembly with Walker A mutant Rpts. A–D, immunoblotting analysis of FLAG-Rpt samples following coexpression of base-specific chaperones. HeLa cells expressing wild-type or Walker A mutant FLAG-Rpt1 (A), Rpt6 (B), Rpt3 (C), and Rpt5 (D) were transfected with the expression constructs of HA-tagged S5b, PAAF1, Gankyrin, and p27, respectively. After affinity purification of FLAG-tagged proteins in the presence of 5 mm ATP, the samples were analyzed by immunoblotting with indicated antibodies.
To test whether suppression of the assembly defects of the Walker A mutants in Rpt1 and Rpt6 by cognate chaperones could be due to the increased stability of Rpts by the chaperones, we investigated the stability of the wild-type or mutant Rpt1 and Rpt6 with or without co-expression of S5b and PAAF1, respectively. As shown in supplemental Fig. S2, the mutation of the Walker A motif in Rpt1 or Rpt6 did not appreciably change the stability of the mutant proteins. Moreover, co-expression of S5b and PAAF1 did not noticeably affect the stability of the wild-type or mutant Rpt1 and Rpt6, respectively. These data suggested that suppression of the assembly defects of Walker A mutants by the cognate chaperones did not come from the increased stability of the Walker A mutants by the chaperones. We next examined whether PAAF1 and S5b enhanced the incorporation of the Walker A mutant Rpts into the proteasome by facilitating the interaction between the appropriate Rpt dimers. To test the effect of PAAF1 on mutant Rpt6, FLAG-tagged wild-type or mutant Rpt6, and HA-tagged Rpt1, Rpt2, and Rpt3 were transiently expressed in HeLa cells with or without HA-PAAF1. FLAG-Rpt6 immunoprecipitates were analyzed for the binding of the Rpt6-Rpt3 dimer to the Rpt1-Rpt2 dimer by Western blotting. As shown in Fig. 5A, the Walker A mutation in Rpt6 did not affect its interaction with Rpt3 but impaired its binding to Rpt1 and Rpt2. Expression of PAAF1 greatly enhanced the interaction of mutant Rpt6 with Rpt1 and Rpt2, while it had a minimal effect on wild-type Rpt6. Similarly, S5b caused augmentation in the interaction of mutant Rpt1 with Rpt4 and Rpt5 (Fig. 5B). A significant increase caused by S5b was also observed in the binding of wild-type Rpt1 to Rpt4 and Rpt5. Taken together, these results suggested that the binding of the RP specific chaperones PAAF1 and S5b to Rpt6 and Rpt1, respectively, facilitates proteasome assembly by enhancing the interaction between adjacent Rpt dimers.
FIGURE 5.
Increased interaction of an Rpt dimer containing a Walker A mutant with the adjacent dimer by S5b and PAAF1. A, immunoblotting analysis of wild-type and Walker A mutant FLAG-Rpt1 samples following expression of S5. HeLa cells were transfected with the expression constructs of wild-type or Walker A mutant FLAG-Rpt1 along with HA-tagged Rpt2, Rpt4, Rpt5, and S5B. Immunopurified FLAG-Rpt1 samples were analyzed by immunoblotting with indicated antibodies. B, immunoblotting analysis of wild-type and Walker A mutant FLAG-Rpt6 samples following the expression of PAAF1. Hela cells were transfected with the expression constructs of wild-type or Walker A mutant FLAG-Rpt6 along with HA-tagged Rpt1, Rpt2, Rpt3, and PAAF1. Immunopurified FLAG-Rpt6 samples were analyzed by immunoblotting with the indicated antibodies.
Deletion of HbYX Motifs Negatively Affects 19 S RP Assembly
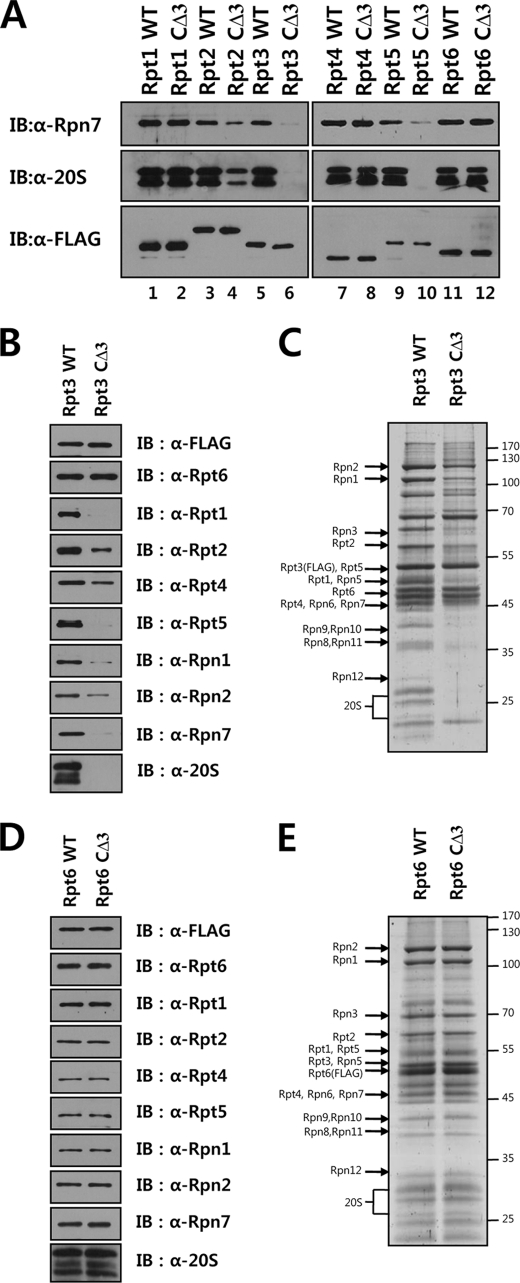
To investigate whether the C-terminal tails of Rpt subunits have any role in 19 S RP assembly in mammalian cells, we generated deletion mutants of Rpt subunits with three C-terminal residues truncated. Tetracycline-inducible cell lines were established expressing FLAG-tagged wild-type and mutant Rpts, from which FLAG-tagged proteins were affinity purified. The copurification of the 20 S CP and the lid subunit Rpn7 with FLAG-tagged proteins were examined by immunoblotting analysis with appropriate antibodies. As shown in Fig. 6A, the 20 S CP was not detected in Rpt3 or Rpt5 samples with the C-terminal deletion and was decreased in the mutant Rpt2 sample compared with wild-type, whereas similar levels of 20 S CP were purified with wild-type and mutant forms of Rpt1, Rpt4, and Rpt6. Remarkably, compared with their wild-type counterparts, mutant Rpt2, Rpt3, and Rpt5 samples contained reduced levels of Rpn7, suggesting that the C-terminal truncation of Rpt2, Rpt3, or Rpt5 inhibited the assembly of not only the 26 S proteasome but also the 19 S RP. To further examine defects in RP assembly caused by C-terminal deletions in Rpts, purified Rpt3 and Rpt6 samples were subjected to SDS-PAGE followed by Coomassie staining or Western blotting with antibodies against various proteasome subunits. The SDS-PAGE protein patterns and the intensity of bands on Western blots of wild-type and mutant Rpt6 preparations were basically identical (Fig. 6, D and E). In contrast, SDS-PAGE analysis of Rpt3 samples revealed that intensities of many of the RP subunits as well as CP subunits were decreased or absent in the mutant sample (Fig. 6B). Western blot analysis confirmed that all base subunits tested were reduced in intensity in the mutant Rpt3 sample except FLAG-Rpt3 and its partner subunit Rpt6, which were present at comparable levels in wild-type and mutant Rpt3 samples (Fig. 6C). These results indicated that although the C-terminal deletion in Rpt6 did not affect proteasome assembly, the deletion mutant Rpt3 failed to efficiently form the intact 19 S RP.
FIGURE 6.
Deletion of the HbYX motif negatively affects proteasome assembly. A, immunoblotting analysis of affinity purified proteasome samples containing wild-type or mutant Rpts with the deletion of the HbYX motif. Cell lines that conditionally expressed FLAG-tagged wild-type or C-terminal three-residue deletion mutant Rpt subunits were established. Aliquots of FLAG-Rpt samples purified in the presence of 5 mm ATP were separated by SDS-PAGE and analyzed by immunoblotting with the indicated antibodies. B and C, immunoblotting (B) and SDS-PAGE (C) analyses of affinity-purified FLAG-tagged wild-type or C-terminal deletion mutant Rpt3. The proteasome was affinity purified as described under “Experimental Procedures.” Ten microliters each of Rpt3 samples purified in the presence of 5 mm ATP was analyzed by immunoblotting with the indicated antibodies and by SDS-PAGE followed by Coomassie staining. D and E, immunoblotting (D) and SDS-PAGE (E) analyses of affinity-purified FLAG-tagged wild-type or C-terminal deletion mutant Rpt6.
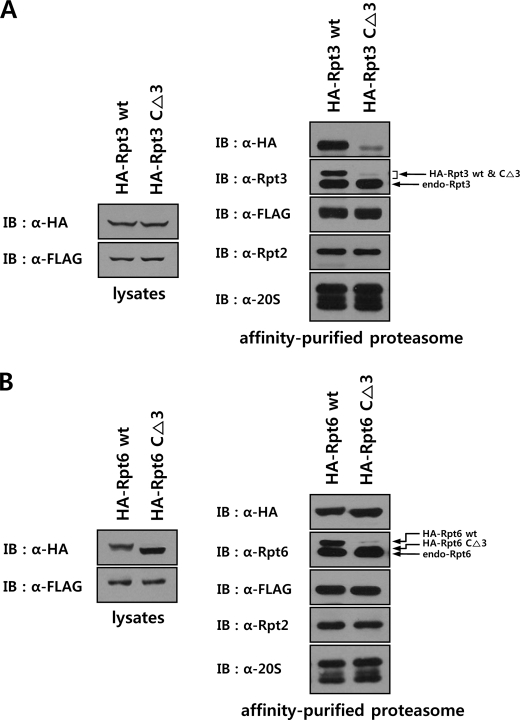
We next evaluated wild-type and mutant Rpt3 and Rpt6 for their relative efficiencies of incorporation into the proteasome, by purifying the proteasome after transient expression of Rpt subunits. HeLa cells expressing a FLAG-tagged version of the lid subunit Rpn12 were transfected with HA-tagged wild-type and mutant Rpt3 or Rpt6. Following affinity purification of the proteasome using anti-FLAG antibody resin, incorporation of Rpt subunits were analyzed by Western blotting. As shown in Fig. 7A, although comparable levels of wild-type and mutant Rpt3 were expressed, much less mutant Rpt3 was associated with FLAG-Rpn12 than wild-type Rpt3. In contrast, mutant Rpt6 was incorporated into the proteasome as efficiently as wild-type (Fig. 7B). Taken together, these results demonstrate that deletion of the HbYX motif impaired the efficient assembly of the ATPase ring in the 19 S RP, whereas C-terminal deletions in Rpts not possessing the HbYX motif did not have any deleterious effect on proteasome assembly.
FIGURE 7.
Impaired incorporation of mutant Rpt3 with a deletion in the HbYX motif into the proteasome. A and B, immunoblotting analysis of the proteasome following transfection of wild-type or C-terminal three-residue deletion mutant Rpt3 (A) or Rpt6 (B). HeLa cells expressing FLAG-Rpn12 were transfected with the expression constructs of HA-tagged wild-type or C-terminal deletion mutant Rpt3 or Rpt6. After affinity purification of the proteasome in the presence of 5 mm ATP, the samples were analyzed by immunoblotting with the indicated antibodies.
DISCUSSION
Cellular assembly of the RP is a seemingly formidable task partly due to the formation of the Rpt ring structure of a defined sequence from six homologous ATPases. Knowledge of the functional motifs in Rpts and the roles of Rpt-interacting proteins involved in RP assembly will help understand the RP assembly mechanism. In this study we generated various Rpt mutants and analyzed their stable incorporation into proteasome using stable and transient transfection methods and IP techniques. We found that when compared with the wild-type Rpts, mutant proteins differentially affected proteasome formation in the in vitro analysis. The in vivo proteasome assembly of Rpt mutants in higher eukaryotes is beyond the scope of this report. However, because mutations at identical Rpt motifs in yeast were shown to display severe phenotypes related to proteasome assembly and function (30, 39), the results of in vitro analysis of Rpt mutants in human cell lines could be applied to the biology of the intact organism.
In this study, we demonstrate that the cellular assembly of the RP requires nucleotide binding but not ATP hydrolysis by Rpt subunits. Hexameric AAA ATPases such as the Rpt ring of RP use chemical energy from ATP hydrolysis to generate mechanical force through coordinated conformational changes of ATPase monomers. Recent studies on differential susceptibility to protease digestion and nucleotide binding properties of PAN indicated that the PAN monomer adopts three different conformations depending on nucleotide binding site occupancy and the bound nucleotide (40, 41). The ATP binding site in PAN is strategically positioned so that ATP binding and subsequent hydrolysis can induce conformational changes, especially affecting interactions between the ATPase core and the C-domain of the PAN monomer and monomer-monomer interfaces. The mutation in the Walker A motif of an Rpt subunit renders it unable to bind a nucleotide and likely keeps its conformation in the nucleotide-free state which appears incompatible with Rpt ring assembly. ATP binding to an Rpt subunit is thought to influence the relative position of the ATPase core and C-domain to induce the transition to a conformation suitable for hexameric ring formation. Since base-specific chaperones bind to the C-domains of Rpts, their binding may affect the equilibrium between different conformational states of Rpts. Indeed, our data show that overexpression of S5b and PAAF1 facilitated the assembly into the RP of Walker A mutant Rpt1 and Rpt6, respectively, by increasing the interaction of the mutants with adjacent Rpt dimers. We do not exclude the possibility that chaperone binding enhanced the interaction of the mutants with other RP subunits as well. Gankyrin and p27 did not appreciably affect the extent of incorporation of their cognate Rpt mutants into the proteasome, which might imply that they have a different mechanism of action from S5b and PAAF1.
Rpt subunits with mutations in the Walker B motif are incorporated into the proteasome as efficiently as wild-type subunits, allowing the purification of the mutant proteasome and subsequent measurement of its activity. ATPase activity of the RP containing mutant Rpt3 or Rpt6 was undetectable. The complete inhibition of ATPase activity of RP composed of five wild-type and one mutant Rpt subunits was unexpected, since the hexameric ATPase ClpX having one active and five inactive subunits still contained substantial ATPase activity (42). Recently, the Goldberg group proposed an elegant model for ATP hydrolysis by the proteasomal ATPases (41). In the model, the hexameric ring contains two ATP-bound, two ADP-bound, and two nucleotide-free subunits, with each pair of an identical conformational state present in para positions. In the ATP hydrolysis cycle, the conformational change of a subunit caused by ATP binding, ATP hydrolysis, or ADP release induces the conformational transition of adjacent subunits in concert. Our data imply that the six subunits of the Rpt ring are tightly coupled so that all Rpt subunits simultaneously change their conformations in the course of ATP hydrolysis. Failure of the conformational transition from the ATP-bound to ADP-bound state in the mutant subunit likely impairs conformational changes in the other five wild-type subunits, leading to the loss of ATPase activity of the Rpt ring. When purified in the absence of ATP, the wild-type proteasome easily dissociates into the RP and CP, whereas the RP containing one Walker B mutant Rpt subunit associates with CP quite stably. The RP-CP association depends on ATP binding to Rpt subunits, which induces exposure of the C-terminal tails for docking into CP pockets. The proposed model for ATP hydrolysis predicts that when there is no incoming ATP during proteasome purification, ATPase-active RP hydrolyzes its bound ATP thereby leaving no Rpts bound to ATP, which in turn leads to RP-CP dissociation. In contrast, an ATPase-dead RP cannot hydrolyze bound ATP and will stay associated with the CP until ATP dissociates from the Rpt subunits. Our data with Walker B mutants are consistent with this scenario and imply that dissociation of ATP from ATPase-dead RP is a slow process.
The C-terminal tails of Rpt2, Rpt3, and Rpt5 containing the HbYX motifs bind to the 20 S CP with varying affinities (28, 29). There appears a correlation between the CP binding affinity of the C-terminal tails and the deleterious effect of their deletion on RP assembly. Thus, the deletion of the Rpt3 or Rpt5 C terminus with a higher CP binding affinity had a greater effect on RP assembly than that of the Rpt2 tail, whereas the C-terminal truncation of Rpts not possessing the HbYX motif did not affect RP assembly. Data from the differential incorporation of wild-type and mutant Rpts into the proteasome shown here by both stable and transient transfections is consistent with the idea that CP may function as a template for base assembly. We note that although deletion of the HbYX motif inhibits RP assembly, it does not completely impair the incorporation of mutant Rpts into the proteasome. Therefore, CP-independent RP assembly must be in operation as well. Kim et al. (32) very recently reported that the HbYX motifs of Rpt3 and Rpt5 are required for 26 S proteasome assembly, which is in agreement with our data. However, they failed to detect the effect of the HbYX motif deletion on RP assembly. The basis of the discrepancy is not clear but may come from differences in experimental design; it could be due to the fact that conditional rather than constitutive expression of Rpt mutants was employed in our experiments which enabled expression of the mutants only for a short period to minimize potential cellular adaptation to the expression of mutant proteins. Unlike their human counterparts, the C termini of yeast Rpt4 and Rpt6 appear to play important roles in 26 S proteasome assembly, since deletion of the last residue from each Rpt impaired efficient formation of 26 S proteasome (30). The proteasome assembly pathway may have diverged between yeast and human. The data presented here suggest that RP assembly in human cells can be driven normally by two different pathways: the CP-assisted pathway requiring the HbYX motifs and the CP-independent pathway.
Supplementary Material
This work was supported in part by a mid-career Researcher Program through an NRF Grant funded by the MEST (No. 2009-0081317), by grants from the Korean Ministry of Science and Technology through the Studies on Ubiquitome Functions Project, and from the Translational Research Center for Protein Function Control, NSF.

This article contains supplemental Figs. S1 and S2.
- CP
- core particle
- RP
- regulatory particle
- AMC
- 7-amino-4-methylcoumarin
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- PAN
- proteasome-activating nucleotidase
- CC
- coiled coil
- OB
- oligonucleotide/oligosaccharide-binding.
REFERENCES
- 1. Schwartz A. L., Ciechanover A. (2009) Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 49, 73–96 [DOI] [PubMed] [Google Scholar]
- 2. Finley D. (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Groll M., Ditzel L., Löwe J., Stock D., Bochtler M., Bartunik H. D., Huber R. (1997) Structure of 20 S proteasome from yeast at 2.4 A resolution. Nature. 386, 463–471 [DOI] [PubMed] [Google Scholar]
- 4. Whitby F. G., Masters E. I., Kramer L., Knowlton J. R., Yao Y., Wang C. C., Hill C. P. (2000) Structural basis for the activation of 20 S proteasomes by 11S regulators. Nature. 408, 115–120 [DOI] [PubMed] [Google Scholar]
- 5. Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 [DOI] [PubMed] [Google Scholar]
- 6. Tomko R. J., Jr., Funakoshi M., Schneider K., Wang J., Hochstrasser M. (2010) Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol. Cell 38, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu C. W., Li X., Thompson D., Wooding K., Chang T. L., Tang Z., Yu H., Thomas P. J., DeMartino G. N. (2006) ATP binding and ATP hydrolysis play distinct roles in the function of 26 S proteasome. Mol. Cell 24, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stadtmueller B. M., Hill C. P. (2011) Proteasome activators. Mol. Cell 41, 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., 3rd, Koonin E. V., Deshaies R. J. (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26 S proteasome. Science 298, 611–615 [DOI] [PubMed] [Google Scholar]
- 10. Yao T., Cohen R. E. (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 [DOI] [PubMed] [Google Scholar]
- 11. Murata S., Yashiroda H., Tanaka K. (2009) Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 10, 104–115 [DOI] [PubMed] [Google Scholar]
- 12. Kusmierczyk A. R., Hochstrasser M. (2008) Some assembly required: dedicated chaperones in eukaryotic proteasome biogenesis. Biol. Chem. 389, 1143–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roelofs J., Park S., Haas W., Tian G., McAllister F. E., Huo Y., Lee B. H., Zhang F., Shi Y., Gygi S. P., Finley D. (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459, 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saeki Y., Toh-E A., Kudo T., Kawamura H., Tanaka K. (2009) Multiple proteasome-interacting proteins assist the assembly of the yeast 19 S regulatory particle. Cell 137, 900–913 [DOI] [PubMed] [Google Scholar]
- 15. Funakoshi M., Tomko R. J., Jr., Kobayashi H., Hochstrasser M. (2009) Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137, 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaneko T., Hamazaki J., Iemura S., Sasaki K., Furuyama K., Natsume T., Tanaka K., Murata S. (2009) Assembly pathway of the mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell 137, 914–925 [DOI] [PubMed] [Google Scholar]
- 17. Le Tallec B., Barrault M. B., Guérois R., Carré T., Peyroche A. (2009) Hsm3/S5b participates in the assembly pathway of the 19 S regulatory particle of the proteasome. Mol. Cell 33, 389–399 [DOI] [PubMed] [Google Scholar]
- 18. Bedford L., Paine S., Sheppard P. W., Mayer R. J., Roelofs J. (2010) Assembly, structure, and function of the 26 S proteasome. Trends Cell Biol. 20, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park S., Tian G., Roelofs J., Finley D. (2010) Assembly manual for the proteasome regulatory particle: the first draft. Biochem. Soc. Trans. 38, 6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tomko R. J., Jr., Hochstrasser M. (2011) Order of the proteasomal ATPases and eukaryotic proteasome assembly. Cell Biochem. Biophys. 60, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang F., Hu M., Tian G., Zhang P., Finley D., Jeffrey P. D., Shi Y. (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell 14, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakamura Y., Umehara T., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26 S proteasome. Biochem. Biophys. Res. Commun. 359, 503–509 [DOI] [PubMed] [Google Scholar]
- 23. Nakamura Y., Nakano K., Umehara T., Kimura M., Hayashizaki Y., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26 S proteasome. Structure 15, 179–189 [DOI] [PubMed] [Google Scholar]
- 24. Rabl J., Smith D. M., Yu Y., Chang S. C., Goldberg A. L., Cheng Y. (2008) Mechanism of gate opening in the 20 S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu Y., Smith D. M., Kim H. M., Rodriguez V., Goldberg A. L., Cheng Y. (2010) Interactions of PAN's C termini with archaeal 20 S proteasome and implications for the eukaryotic proteasome-ATPase interactions. EMBO J. 29, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stadtmueller B. M., Ferrell K., Whitby F. G., Heroux A., Robinson H., Myszka D. G., Hill C. P. (2010) Structural models for interactions between the 20 S proteasome and its PAN/19 S activators. J. Biol. Chem. 285, 13–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith D. M., Chang S. C., Park S., Finley D., Cheng Y., Goldberg A. L. (2007) Docking of the proteasomal ATPases' carboxyl termini in the 20 S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell 27, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar B., Kim Y. C., DeMartino G. N. (2010) The C terminus of Rpt3, an ATPase subunit of PA700 (19 S) regulatory complex, is essential for 26 S proteasome assembly but not for activation. J. Biol. Chem. 285, 39523–39535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park S., Roelofs J., Kim W., Robert J., Schmidt M., Gygi S. P., Finley D. (2009) Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kusmierczyk A. R., Kunjappu M. J., Funakoshi M., Hochstrasser M. (2008) A multimeric assembly factor controls the formation of alternative 20 S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 [DOI] [PubMed] [Google Scholar]
- 32. Kim Y. C., DeMartino G. N. (2011) C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J. Biol. Chem. 286, 26652–26666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hendil K. B., Kriegenburg F., Tanaka K., Murata S., Lauridsen A. M., Johnsen A. H., Hartmann-Petersen R. (2009) The 20 S proteasome as an assembly platform for the 19 S regulatory complex. J. Mol. Biol. 394, 320–328 [DOI] [PubMed] [Google Scholar]
- 34. Min K. W., Hwang J. W., Lee J. S., Park Y., Tamura T. A., Yoon J. B. (2003) TIP120A associates with cullins and modulates ubiquitin ligase activity. J. Biol. Chem. 278, 15905–15910 [DOI] [PubMed] [Google Scholar]
- 35. Elsasser S., Schmidt M., Finley D. (2005) Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 398, 353–363 [DOI] [PubMed] [Google Scholar]
- 36. Hoffman L., Rechsteiner M. (1996) Nucleotidase activities of the 26 S proteasome and its regulatory complex. J. Biol. Chem. 271, 32538–32545 [DOI] [PubMed] [Google Scholar]
- 37. Park S. M., Yoon J. B., Lee T. H. (2004) Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. 566, 151–156 [DOI] [PubMed] [Google Scholar]
- 38. Thompson D., Hakala K., DeMartino G. N. (2009) Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Horwitz A. A., Navon A., Groll M., Smith D. M., Reis C., Goldberg A. L. (2007) ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J. Biol. Chem. 282, 22921–22929 [DOI] [PubMed] [Google Scholar]
- 41. Smith D. M., Fraga H., Reis C., Kafri G., Goldberg A. L. (2011) ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin A., Baker T. A., Sauer R. T. (2005) Rebuilt AAA + motors reveal operating principles for ATP-fueled machines. Nature 437, 1115–1120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.