Background: HLA-DQ2/8 heterozygous individuals have the highest risk for development of type 1 diabetes.
Results: The disease-associated HLA-DQ2/8 transdimer exhibits unique peptide binding features compared with other HLA-DQ2/8 dimers.
Conclusion: This newly identified binding motif predicts islet autoantigen-derived peptides as candidate T cell epitopes.
Significance: Predicting new HLA-DQ2/8 transdimer-specific candidate T cell epitopes sets the stage for testing candidate diabetogenic epitopes.
Keywords: Autoimmune Diseases, Diabetes, Major Histocompatibility Complex (MHC), Peptides, T Cell, HLA-DQ Transdimer, Binding Motif, Susceptibility, Type 1 Diabetes
Abstract
HLA-DQ2 and HLA-DQ8 are strongly predisposing haplotypes for type 1 diabetes (T1D). Yet HLA-DQ2/8 heterozygous individuals have a synergistically increased risk compared with HLA-DQ2 or HLA-DQ8 homozygote subjects that may result from the presence of a transdimer formed between the α-chain of HLA-DQ2 (DQA1*05:01) and the β-chain of HLA-DQ8 (DQB1*03:02). We generated cells exclusively expressing this transdimer (HLA-DQ8trans), characterized its peptide binding repertoire, and defined a unique transdimer-specific peptide binding motif that was found to be distinct from those of HLA-DQ2 and HLA-DQ8. This motif predicts an array of peptides of islet autoantigens as candidate T cell epitopes, many of which selectively bind to the HLA transdimer, whereas others bind to both HLA-DQ8 and transdimer with similar affinity. Our findings provide a molecular basis for the association between HLA-DQ transdimers and T1D and set the stage for rational testing of potential diabetogenic peptide epitopes.
Introduction
Type 1 diabetes (T1D)2 is a chronic autoimmune disease characterized by activation of CD4 and CD8 T cells and subsequent destruction of the insulin-secreting beta cells in the pancreas (1–3). The disease process involves autoreactive T cells against epitopes found within a diversity of islet auto-antigens, of which the best studied are insulin (4–7), islet tyrosine phosphatase insulinoma-associated 2 (IA-2) (8, 9), and glutamic acid decarboxylase (65-kDa isoform; GAD65) (10, 11).
In T1D the risk associated with the HLA-DQ2/8 heterozygous haplotype was found to be increased compared with homozygous HLA-DQ2 or HLA-DQ8 individuals, suggesting an epistatic or synergistic effect (12–15). Moreover, it has been shown that this high risk is associated with the presence of HLA-DQ transdimers formed by the α-chain of HLA-DQ2 (DQA1*05:01) and the β-chain of HLA-DQ8 (DQB1*03:02, termed HLA-DQ8trans hereafter) or by the α-chain of HLA-DQ8 (DQA1*03:01) and the β-chain of HLA-DQ2 (DQB1*02:01; termed HLA-DQ2trans hereafter). In particular the HLA-DQ8trans heterodimer confers the highest risk for development of T1D (13, 14, 16, 17). Consequently, this transdimer may present a unique repertoire of autoantigen-derived peptides triggering a pathogenic response that ultimately leads to beta cell destruction. However, the binding characteristics of the HLA-DQ transdimers are poorly understood.
In this study we have generated HLA-DQ2trans HLA-DQ8trans molecules and determined which peptides are naturally bound to these transdimers. Moreover, we designed transdimer-specific peptide binding assays and employed these to determine the requirements for peptide binding to transdimers in detail. The results demonstrate that both transdimers have vastly different peptide binding properties. Finally, we have used the novel binding motifs to identify potential beta cell autoantigen-derived epitopes specifically presented by HLA-DQ8trans.
EXPERIMENTAL PROCEDURES
Lentivirus Production and Transduction
A set of bicistronic lentiviral vectors containing the α-chain (DQA1*05:01 or DQA1*03:01) or the β-chain (DQB1*03:02, DQB1*02:01) coding sequences and, respectively, a GFP- or a puromycin-resistant gene were generated. Third generation lentiviruses were produced following standard procedures (18). Briefly, lentiviral vectors (pRRL-CMV-GOI-GFP/puro) were co-transfected with “helper” plasmids, encoding HIV-1 gag-pol (HIV-1 rev and HIV-1 VSV-G envelope) into 293T cells. Virus-containing supernatants were harvested after 48 and 72 h, passed through a 0.45-μm filter, and stored until use at −80 °C. Virus titers were estimated by ELISA for HIV p24 level (ZeptoMetrix Corp.). For cotransductions of HEK293 cells, virus-containing supernatants were added to fresh media supplemented with 8 μg/ml Polybrene (Sigma), and cells were incubated overnight. Transduced cells were cell-sorted for high DQ expression by FACS using GFP and an anti-DQ antibody (SPV-L3). By this, HEK293 cells expressed only one type of HLA-DQ molecule; DQA1*05:01/DQB1*02:01 (HLA-DQ2cis), DQA1*03:01/DQB1*03:02 (HLA-DQ8cis), DQA1*03:01/DQB1*02:01 (HLA-DQ2trans), or DQA1*05:01/DQB1*03:02 (HLA-DQ8trans).
Peptide Elution and Isolation from Affinity-purified HLA-DQ Molecules
HLA-DQ2trans and HLA-DQ8trans molecules were isolated from the transduced HEK293 cells. Peptide elutions were performed as follows. Approximately 1010 cells were grown in Iscove's modified Dulbecco's medium supplemented with 10% FCS and l-glutamine. All purification steps were performed at 4 °C. The cells were harvested by centrifugation and washed with PBS, and the cell pellet was stored at −80 °C. Cells were lysed with lysis buffer (containing 50 mm Tris, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 10 mm iodoacetamide, and protease inhibitors (complete inhibitor mix, Roche Applied Science)) and subsequently centrifuged for 60 min at 10.000 × g to remove nuclei and insoluble material. Lysates were precleared with Sepharose beads and mixed with Sepharose beads coupled with a pan-DQ antibody (SPV-L3). After 60 min of incubation the beads were washed with 5 bed volumes lysis buffer subsequently followed by 4 bed volumes of low salt buffer (120 mm NaCl, 20 mm Tris-HCl, pH 8.0), 8 bed volumes of high salt buffer (1 m NaCl, 20 mm Tris-HCl, pH 8.0), 4 bed volumes of no salt buffer (20 mm Tris-HCl, pH 8.0), and 4 bed volumes of low Tris buffer (10 mm Tris-HCl, pH 8.0). The HLA-peptide complexes were eluted with 5 bed volumes of 10% acetic acid. To remove high molecular mass HLA molecules, a filtration step with a Centriprep filtration unit with a cutoff value of 10 kDa was used. The peptide fraction was freeze-dried and redissolved in 200 μl of 10% acetic acid. Fractionation was performed with an HPLC system (see below). The material was eluted using a gradient of 0–50% acetonitrile supplemented with 0.1%trifluoroacetic acid.
Peptide Identification by Mass Spectrometry (MS)
The complex MHC-peptide pool was prefractionated on a C18 reverse phase HPLC system (200 μm × 15 cm; Reprosil-C18-AQ 3 μm (Dr. Maisch GmbH, Ammerbuch, Germany). Fractions were reduced to near dryness, and 95/3/0.1 v/v/v water/acetonitrile/formic acid was added. Fractions were subsequently analyzed by tandem mass spectrometry. Peptides were characterized by nanoflow liquid chromatography using an Agilent 1100 HPLC system (Agilent Technologies) coupled on line to a 7-tesla LTQ-FT mass spectrometer (Thermo Electron, Bremen, Germany). The chromatographic system consisted of the following components. ReproSil-Pur C18-AQ3 μm was used as a resin for the analytical nano column, and AQUA-C18 5 μm was used as a resin for the trapping column. Columns were prepared in house. The end of the nanocolumn was drawn to a tip (ID ∼5 μm), from which the eluent was sprayed into the mass spectrometer. Peptides were trapped at 5 μl/min on a 1-cm column (100-μm internal diameter, packed in house) and eluted to a 15-cm column (50-μm internal diameter, packed in house) at 150 nl/min in a 60-min gradient from 0 to 50% acetonitrile in 0.1% formic acid. The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition. Full scan MS spectra were acquired in the FT-ICR-MS with a resolution of 25,000 at a target value of 5,000,000. The two most intense ions were then isolated for accurate mass measurements by a selected ion monitoring scan in FT-ICR with a resolution of 50,000 at a target accumulation value of 50,000. The selected ions were fragmented in the linear ion trap using collision-induced dissociation at a target value of 10,000. In a post analysis process, raw data were converted to peak lists using Bioworks Browser software, Version 3.1. For protein identification, MS/MS data were submitted to the human IPI data base using Mascot Version 2.4 (Matrix Science) with the following settings: 5 ppm and 0.8-Da deviation for precursor and fragment masses, respectively; no enzyme was specified. All reported hits were assessed manually, and peptides with MASCOT scores <45 were discarded.
HLA-DQ-Peptide Homology Modeling
Homology modeling of the complexes between the two HLA-DQtrans molecules and eluted antigenic peptides was performed as previously described (19). The crystal structures of HLA-DQ2cis (DQA1*05:01/DQB1*02:01) (20) and HLA-DQ8cis (DQA1*03:01/DQB1*03:02) (21) were used as base molecules, and the peptide coordinates in the latter were used for all peptides aligned. The HLA-DQ trans molecules were obtained by aligning the Cα of HLA-DQ2cis and HLA-DQ8cis in the β-pleated sheet regions of the α1β1 domains of these molecules and making the transdimers by selecting the α-chain and β-chain of the corresponding cis dimers. Energy minimization of each respective complex, unless otherwise noted, was carried out via the program Discover of Accelrys (San Diego, CA) on Silicon Graphics Fuel and Indigo instruments using 1000 cycles of the steepest gradient approach followed by 1000 cycles of the conjugate gradient approach. As outlined previously, homology modeling can be used to predict the physicochemical features of the pockets of the HLA-DQ2/8 trans molecules to a certain extent, and this information was used to identify the most likely nine-amino acid binding core in the eluted peptides. When alignment was equivocal, alternative alignments were tried and verified by energy minimization. All structures were examined for atomic clashes via the Discover program. Graphical representations were performed via the WebLabViewer (Version 3.5) and DSViewer Pro programs of Accelrys. The numbering of the residues in the amino acid sequences of the DQA and DQB molecules was done according to the scheme suggested by Fremont et al. (22) and extended by Bondinas et al. (23), as it ensures structural equivalence across MHC class II loci within the same and throughout different species. All modeled structures as Protein Data Bank coordinates are available in the supplemental material.
Peptide Synthesis
Peptides were synthesized according to standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry using a SyroII peptide synthesizer (MultiSynTech, Witten, Germany). The integrity of the peptides was checked using reverse phase HPLC and MS. Peptides to be tested as potential epitopes were synthesized as 13-mers containing two alanines both at the N and C termini.
Heat Map
A heat map attempts to visualize all the amino acid occurrences for all positions in one picture (24). The heat map represents a two-dimensional data matrix where every row is an amino acid and every column is a position and gives an overview of the preferred and non-preferred residues per position in the HLA-DQ binding core. One cell in the heat map matrix will be colored according to the calculated p value for that position and amino acid. The p value used for each analysis is 0.05. Only significantly up- and down-regulated residues according to the given p value 0.05 are colored in, respectively, a shade of green and red. The non-regulated elements are colored black.
HLA-Peptide Binding Assay
HLA-peptide binding assays were performed as described previously (25). In short, FluoroNunc 96-well plates were coated with the pan-DQ-antibody SPV-L3. Plates were washed, blocked with PBS, 0.5% BSA, and whole cell lysates (using Nonidet P-40 as detergent) of DQ-expressing HEK293 cells were incubated at 4 °C overnight. Titration ranges of the tested peptides (from 0 to 300 μm) were prepared with or without a fixed concentration (0.6 μm) of biotinylated indicator peptide depending on the type of assay (competition or direct). Subsequently, the peptide sample was applied to the wells containing binding buffer. After incubation, plates were washed, and europium-streptavidin in assay buffer was added to each well followed by incubation while shaking. After washing, wells were incubated with enhancement buffer. Plates were read using a time-resolved fluorometer (1234, Wallac). EC50 values were calculated based upon the observed binding of the tested peptides against the fixed concentration indicator peptide; the concentration of tested peptide required for half-maximal inhibition of binding of the reporter peptide indicate the EC50 value.
Construction of Data Base of Beta Cell-expressed Proteins
A tailored data base of all the known protein sequences of beta cell proteins was built by extraction of relevant entries from several databases at the European Bioinformatics Institute with the SRS system. These databases include: UniProtKB, UniProtKB/Swiss-Prot, UniProtKB/TrEMBL, UniRef100, UniRef90, UniRef50, uniParc, refSeq Proteome, IPI, Refseq Proteome (release), Refseq Proteome (updates), MHCBN, Swall(SPTR), PIR, RemTrEMBL. The following search terms (keywords) were included: organism: Homo sapiens AND beta cell, OR pancreatic islet, OR islet of Langerhans. This data base contained, but was not restricted to, all well known diabetogenic proteins, including islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP), glutamic acid decarboxylase, isoform 65 (GAD65), glutamic acid decarboxylase, isoform 67 (GAD67), IA-2, preproinsulin, phogrin, zinc transporter-8 (ZnT-8), and islet cell autoantigen 69-kDa (ICA69).
RESULTS
Generation of DQ2/8 Cis- and Transdimer-expressing Cells
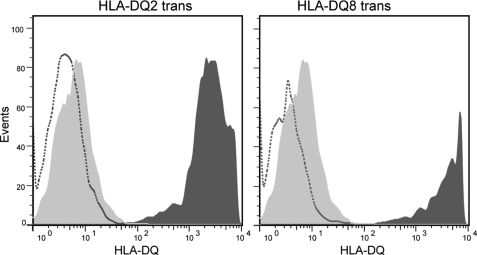
To generate cells that selectively express one kind of HLA-DQ molecule (HLA-DQ2cis, HLA-DQ8cis, HLA-DQ2trans, or HLA-DQ8trans), lentiviral constructs were generated encoding either the α- or β-chain from HLA-DQ2cis or HLA-DQ8cis. These constructs were used to infect HEK293 cells that do not naturally express HLA class II molecules. The resulting HEK293 cells stably expressed the desired HLA-DQ dimers. FACS sorting was used to isolate clones with high HLA-DQ expression. GFP expression correlated with expression of the HLA-DQA1 chains, and an antigen-processing cell (APC)-conjugated pan-HLA-DQ antibody (SPV-L3) was used to detect HLA-DQ expression (Fig. 1).
FIGURE 1.
Expression of HLA-DQ2/8 transdimers on viral-transduced HEK293 cells. Expression of the DQ transdimer molecules on HEK293 cells was examined by FACS analysis using a phosphatidylethanolamine-conjugated pan-HLA-DQ antibody (clone 1a3, Leinco Technologies). Shown is HLA-DQ expression on the surface of transduced cells (dark gray) compared with non-transduced HEK293 cells (dashed line), which do not naturally express MHC class II molecules. An isotype control antibody for SPV-L3 was included in the FACS experiments (light gray). Cells were single cell-sorted to obtain clones with high DQ expression. Clones obtained by this strategy displayed >95% DQ expression, which was stable in time (not shown).
Isolation and Sequencing of HLA-DQ Transdimer-bound Peptides
HLA-DQ2- and HLA-DQ8 transdimers were isolated from the appropriate HEK293 cells by affinity purification. Subsequently, the bound peptides were eluted with acid and separated from the high molecular weight material by filtration. The resulting peptide pools were fractionated by reversed phase HPLC, and the peptides present were analyzed by tandem mass spectrometry.
This resulted in the identification of 313 peptides for HLA-DQ2trans and 531 peptides for HLA-DQ8trans. Typically, and in agreement with previous studies, several length variants of most peptides were found. The 313 peptides eluted from HLA-DQ2trans represented 79 unique sequences, and the 531 HLA-DQ8trans peptides represented 180 unique sequences.
Identification of Binding Cores of HLA-DQ Transdimers Deduced from Eluted Peptides
To identify the putative nine-amino acid binding cores in the eluted peptides, we made use of the previously reported peptide binding motifs of HLA-DQ2cis and HLA-DQ8cis (21, 26). HLA-DQ2cis prefers large hydrophobic and aromatic residues at positions p1/p9 and negatively charged residues at p4/p6/p7. HLA-DQ8cis has a preference for acidic residues at positions p1/p9 and secondary aliphatic/aromatic residues at p1 and mainly aliphatic residues at the central positions (p4/p6). The HLA-DQ transdimers are predicted to share some of the binding features of HLA-DQ2cis at the N terminus of the peptide and some of HLA-DQ8cis at the C terminus.
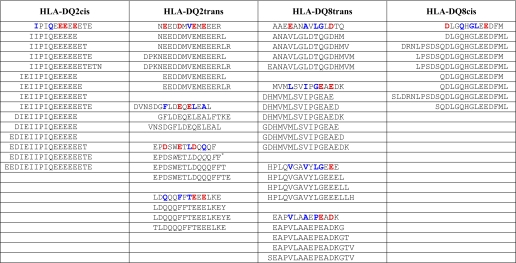
To confirm this notion, we used molecular simulations to verify that predictions made on the basis of the properties of HLA-DQ2cis and HLA-DQ8cis are likely to be valid for the HLA-DQ transdimers as well. Altogether, this yielded enough information on the preference of the putative binding pockets in the HLA-DQ transdimers to predict the 9-amino acid binding core for 132 HLA-DQ2trans-eluted peptides and 277 HLA-DQ8trans-eluted peptides. Table 1 shows examples of the alignment and core predictions of length variants of nucleobinding-2-eluted peptides from HLA-DQ2trans and thioredoxin eluted peptides from HLA-DQ8trans. For a number of eluted HLA-DQ2trans and HLA-DQ8trans peptides (<0.1% of the eluted unique peptides) core sequences could not be identified (data not shown). All unique eluted peptides and their predicted nine-amino acid core and anchor residues are shown in supplemental Table 1.
TABLE 1.
Alignment and core predictions of length variants of nucleobinding-2-eluted peptides from DQ2trans- and thioredoxin-eluted peptides from DQ8trans
Shown is an overview of alignments and binding core identifications of length variants eluted from DQ2trans (nucleobinding-2) and DQ8trans (thioredoxin). As a comparison, length variants eluted from HLA-DQ2cis (B-lymphocyte antigen CD20) and HLA-DQ8cis (secretory granule proteoglycan core protein) are also illustrated. Anchors are in bold. Acidic and non-acidic residues at the anchor positions are shown in red and blue, respectively.
* The residues in italics constitute an alternative binding register, although weaker because of p60.
Analysis of Binding of Selected Panel of HLA-DQ2trans- and HLA-DQ8trans-eluted Peptides
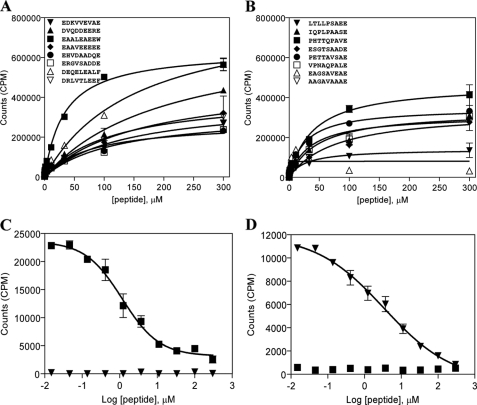
To verify the correctness of the predicted binding cores, we randomly selected a panel of eight HLA-DQ2trans- and eight HLA-DQ8trans-eluted peptides and tested these for binding to HLA-DQ2trans and HLA-DQ8trans in a cell-free binding assay. Peptides were synthesized as 13-mers containing the predicted 9-amino acid core flanked by two alanine residues at both the N and C termini of the peptide and a biotin label linked to the N terminus of the peptide. We observed that 15 of the 16 peptides tested bound to the respective HLA-DQ transdimer from which they had been isolated (Fig. 2, A and B). In contrast, only 1 of 8 HLA-DQ8trans peptides bound to HLA-DQ2trans, and 3 of 8 HLA-DQ2trans peptides bound to HLA-DQ8trans (data not shown). These data indicate that in the majority of the peptides the prediction of the binding core was successful. Moreover, the results suggest that HLA-DQ8trans, compared with HLA-DQ2trans, is less selective in binding peptides.
FIGURE 2.
Binding of a selected panel of HLA-DQ2trans and HLA-DQ8trans peptides representing the identified binding cores. Randomly selected panels of 8 DQ2trans- and 8 DQ8trans-eluted peptides containing the identified binding cores were synthesized as 13-mers containing two extra alanines, both N- and C-terminally necessary for proper DQ binding. Binding of a titration range (0–300 μm) of peptides was tested in a direct binding assay or in a competitive binding assay using a fixed concentration (0.6 μm) of biotinylated indicator peptide. A and B, shown is direct binding of a panel of 8 DQ2trans- and 8 DQ8trans- eluted peptides to DQ2trans (A) and DQ8trans (B). C and D, shown is competitive binding of the biotinylated DQ2trans indicator peptide AAEAALEAEEWAA (▼) and DQ8trans indicator peptide AAPHTTQPAVEAA (■) versus the same non-biotinylated peptide to DQ2trans (C) and DQ8trans (D). Data represent mean ± S.E. (n = 3).
The results of these direct peptide binding studies were used to select an indicator peptide for each HLA-DQ transdimer to establish a competitive peptide binding assay, which allows a more quantitative comparison of the affinity of HLA-peptide binding. As indicator peptides, we selected the peptides AAEAALEAEEWAA for HLA-DQ2trans and AAPHTTQPAVEAA for HLA-DQ8trans. Based on the results from the direct peptide binding assay, both are expected to be high affinity binding peptides (HLA-DQ2trans, 33.1 ± 3.5 μm; HLA-DQ8trans, 39.6 ± 4.8 μm), which minimizes the change of detecting false positive binders in a competitive peptide binding assay. Initially, these indicator peptides were examined for specific binding by testing them for self-competition in a competitive peptide binding assay with both HLA-DQ transdimers (Fig. 2, C and D). The results demonstrate that both indicator peptides bound to their respective HLA-DQ transdimer but not to the other transdimer, indicating that they are suitable for use in the competitive binding assay.
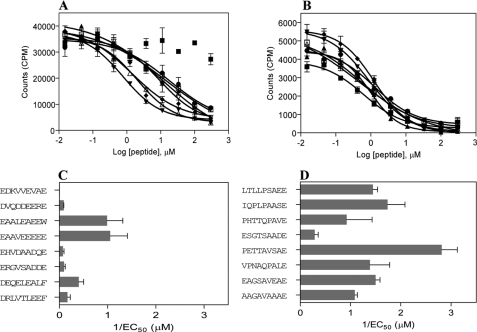
Next, we tested the two panels of selected peptides in the competitive binding assay to exclude potential false positive binders in the direct binding assay. The results show that only one HLA-DQ2trans peptide did not bind properly, as indicated by the lack of a correct S-shaped binding curve (EC50 > 100 μm) (Fig. 3). The calculated EC50 values obtained were in general lower for HLA-DQ2trans (Fig. 3C) than EC50 values for HLA-DQ8trans (Fig. 3D), but these values cannot be directly compared, precluding assigning differences in overall peptide binding affinity. In contrast to the direct binding assay, we observed proper binding to DQ8trans of the peptide harboring the core nonamer EAGSAVEAE (data not shown). This inconsistency might be caused by the presence of a biotin in the peptide used in the direct binding assay, whereas this is absent in the peptide used in the competitive assay.
FIGURE 3.
Binding of selected panels of HLA-DQ2trans and HLA-DQ8trans peptides in a competitive peptide binding assay. The two panels of 8 DQ2trans and DQ8trans peptides were tested in a titration range (0–300 μm) for binding in a competitive binding assay using a fixed concentration (0.6 μm) of the selected biotinylated indicator peptides AAEAALEAEEWAA (DQ2trans) and AAPHTTQPAVEAA (DQ8trans). A and B show competitive binding of the peptide panels to DQ2trans and DQ8trans, respectively. C and D show EC50 values were calculated based on the observed competition between the biotinylated indicator peptides and the test peptides for DQ2trans and DQ8trans, respectively. EC50 represents the concentration of the test peptide that is required for 50% inhibition of the binding of the indicator peptide. Data represent the mean ± S.E. (n = 3). Shown on the x axes is 1/EC50, thereby illustrating that large bars represent better binding.
Although the results support the notion that the nine-amino acid binding cores were generally correctly predicted, it is of note that peptide AAESGTSAADEAA displayed the lowest affinity for DQ8trans, whereas the residues at the anchor positions p1/p4/p6/p7/p9 fulfill the HLA-DQ8trans binding motif. Obviously, other factors also co-determine the binding of the peptide to the HLA-DQ transdimers.
Identification of Peptide Binding Motifs for HLA-DQ2trans and HLA-DQ8trans
Based on the identified binding cores of the eluted peptides, the frequency of the amino acids at the anchor and non-anchor positions was calculated for both transdimers (data not shown) and compared with the human proteome. Amino acids with a frequency 1.5 times higher than the same amino acid in the human proteome were considered as preferred residue. To visualize all the amino acid occurrences for all positions in one picture, heatmaps were generated (supplemental Fig. 1). For HLA-DQ2trans, the majority of amino acids at both anchor and non-anchor positions (p1–p9) showed similar frequencies compared with the human proteome. At positions p1, p4, p6, p7, and p9, the acidic amino acids Asp and Glu were dominantly present. In accordance with this strong preference, the basic amino acids Lys, Arg, and His were virtually absent at these positions (Table 2 and supplemental Fig. 1). Strikingly, acidic residues were also frequently found at the non-anchor positions in the peptides that were eluted from HLA-DQ2trans. The vast majority of identified peptides contained multiple acidic residues. One extreme example of this is the 15-mer peptide EDEEKDEEDEEDEDK (derived from calreticulin) harboring 13 acidic residues. Next to the strong preference for negative charges, minor preferred amino acids within the binding motif of HLA-DQ2trans are mostly (small) aliphatic residues like Gly, Ala, Val, and Leu. In contrast, HLA-DQ8trans showed a strong preference for acidic residues (Asp, Gly) at p9 only, whereas at the other anchor positions HLA-DQ8trans preferred aliphatic residues, like Gly, Ala, Val, and Leu. Similar to HLA-DQ2trans, HLA-DQ8trans did not accommodate basic residues, except at position p7.
TABLE 2.
Amino acid preferences at anchor positions within peptide binding motifs of the DQ2/8 cis- and transdimers
Homology modeling and inspection of the five binding pockets of the DQ2/8 cis and trans molecules revealed their binding preferences (with main preferences in boldface).
| Molecule/pocket | p1 | p4 | p6 | p7 | p9 |
|---|---|---|---|---|---|
| DQA1*05:01/DQB1*02:01a | Aliphatic, Phe, Tyr | Acidic | Small aliphatic | Acidic | Trp, Phe, Tyr |
| (DQ2cis) | Gln, Asn, Glu, Pro | Aliphatic, Phe, Tyr | Asp, Glu, Pro | No Arg, Lys, His | Val, Leu, Ile, Met |
| DQA1*03:01/DQB1*02:01 | Acidic | Acidic | Acidic | Acidic | Acidic |
| (DQ2trans) | Aliphatic (Val, Leu) | Aliphatic (Gly, Val, Ile), Trp | Aliphatic (Val, Ala), Met, Pro | Aliphatic No Arg, Lys, His | Aliphatic (Leu), Phe |
| DQA1*05:01/DQB1*03:02 | Aliphatic, Pro | Aliphatic, Ser, Thr | Aliphatic, Pro | Aliphatic | Acidic |
| (DQ8trans) | Glu, Met | No Gln, Trp | Glu Including Arg, Lys, His | ||
| DQA1*03:01/DQB1*03:02b | Acidic | Aliphatic | Small aliphatic | Wide spectrum Including Arg, Lys | Acidic |
| (DQ8cis) | Aliphatic, Phe, Tyr | Phe, Tyr No Gln, Asn | Asp | ||
| All four DQ molecules | No Arg, Lys, His, Cys | No Arg, Lys, His, Cys | No Arg, Lys, His, Trp, Cys | No Arg, Lys, His, Cys |
In conclusion, HLA-DQ2trans and HLA-DQ8trans display largely distinct peptide binding requirements. Moreover, both binding motifs are distinct from those previously reported for HLA-DQ2cis and HLA-DQ8cis (Table 2 and supplemental Fig. 1).
Amino Acid Substitutions at Anchor and Non-anchor Positions
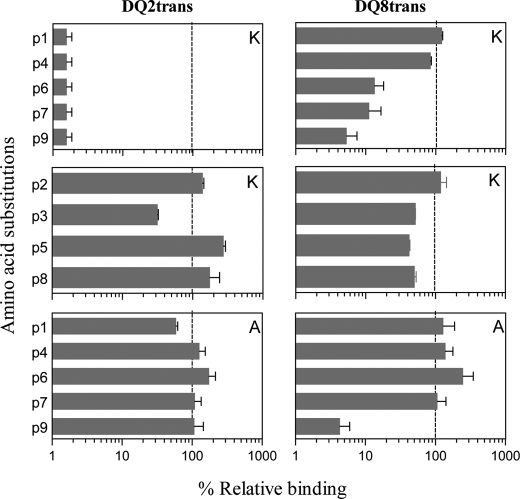
To verify the anchor positions p1, p4, p6, p7, and p9, we tested variant peptides in which the amino acids were systematically substituted for Lys (Fig. 4). Single Lys substitutions at the anchor positions in the HLA-DQ2trans indicator peptide EAALEAEEW abrogated binding to HLA-DQ2trans, whereas substitutions at the non-anchor positions had no such effect. In contrast, only single Lys substitutions at the p9 anchor position in the HLA-DQ8trans indicator peptide PHTTQPAVE almost completely abrogated binding to HLA-DQ8trans. Substitutions at p1 and p4 had no effect, whereas substitutions at p6 and p7 reduced binding. Single Lys substitutions at a non-anchor position had no or little effect. In addition, we tested variant peptides in which two anchor residues were substituted with Lys and observed that this abrogated binding in all cases regardless of the positions substituted (data not shown). Double Lys substitutions at non-anchor positions resulted in severe obstruction of peptide binding to HLA-DQ2trans in 5/6 cases and in 3/6 cases for HLA-DQ8trans. This is probably due to the many positive charges on the surface of the HLA class II allele, similar to HLA-DQ2cis (25). Together, these results indicate that the anchor positions were correctly identified and highlight the importance of anchor position p9 for HLA-DQ8trans binding peptides.
FIGURE 4.
Amino acid substitutions at anchor and non-anchor positions within the HLA-DQ2trans and HLA-DQ8trans indicator peptides. Binding of p1/p4/p6/p7/p9 (anchor) Lys-substituted DQ2trans indicator peptide AAEAALEAEEWAA and DQ8trans indicator peptide AAPHTTQPAVEAA to their respective transdimer is shown. Also, binding of p2/p3/p5/p8 Lys-substituted DQ2trans indicator peptide and DQ8trans indicator peptide was tested. Binding of A-substituted indicator peptides to DQ2trans and DQ8trans is shown. EC50 values were calculated on the basis of competition between the biotinylated indicator peptide and the non-biotinylated Lys-substituted test peptides. EC50 represents the concentration of the test peptide that is required for 50% inhibition of the binding of the indicator peptide. Data represent the mean ± S.E. (n = 3). Shown on the x axes is % relative binding of peptide without any substitutions (dashed line).
Predictive Power of HLA-DQ8trans Binding Motif
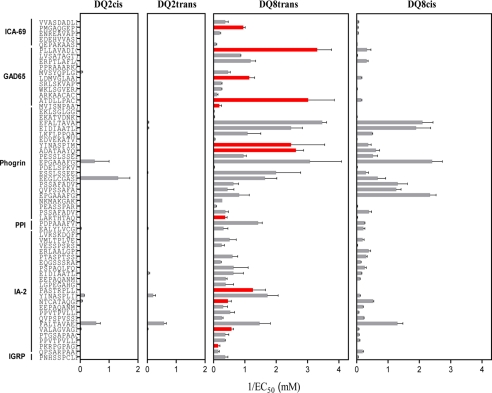
The genetic predisposition for development of T1D is significantly higher in individuals heterozygous for HLA-DQ2/8, and the HLA-DQ8 transdimer is suggested to be largely responsible for the increased risk in heterozygous individuals (13). Therefore, we tested if the HLA-DQ8trans binding motif could be used to identify peptides from islet autoantigens that would exclusively or preferentially bind to the HLA-DQ8 transdimer. For this purpose, a data base containing islet-expressed proteins (including the well known diabetogenic proteins GAD65, IA-2 and preproinsulin) was generated. We scanned this data base with our established HLA-DQ8trans binding motif, allowing for single (Table 3) or double mismatches (not shown) at anchor positions that yielded 192 hits. A number of predicted candidate epitopes were known T cell epitopes in the context of HLA-DQ2cis, HLA-DQ8cis, or its mouse equivalent, I-Ag7 (Table 3, sequences are in italics). From the 192 unique predicted candidate epitopes, we randomly selected 61 (with and without mismatches) and determined their binding to HLA-DQ8trans and/or HLA-DQ8cis (Fig. 5). The InsB13–21 epitope, which we published recently as an epitope presented by both HLA-DQ cis- and transdimers (19), was also a hit and tested for binding. With one exception, all predicted peptides bound to HLA-DQ8trans and the majority with EC50 values below 10 μm, indicative for high affinity binding. Several peptides were found to bind 100–1000 times more strongly to HLA-DQ8trans than to HLA-DQ8cis (examples PKRPGPAGE (IA-2131–139) and MVISNPAAT (GAD65568–576), whereas other peptides bound strongly to both HLA-DQ8trans and HLA-DQ8cis (for example, PSSAFADVE (phogrin540–548) and QPSARPAAE (IA-2461–469)). In contrast, only a few bound to HLA-DQ2cis or HLA-DQ2trans. These data support the accuracy of the HLA-DQ8trans binding motif for prediction of candidate epitopes in islet autoantigens that either selectively bind HLA-DQ8trans or both HLA-DQ8trans and HLA-DQ8cis.
TABLE 3.
DQ8trans predicted candidate epitopes within known diabetogenic proteins
Using the HLA-DQ8trans peptide binding motif with single mismatches at the anchor positions, candidate epitopes were predicted for HLA-DQ8trans using the algorithm program MOTIFS. Predictions were made in the known diabetogenic proteins. Sequences in italics are known human or mouse diabetogenic epitopes. The dashes indicate no hits. IGRP, islet-specific glucose 6 phosphatase catalytic subunit-related protein.
| Protein | Full motif | Mismatch p1 | Mismatch p4 | Mismatch p6 | Mismatch p7 | Mismatch p9 |
|---|---|---|---|---|---|---|
| IGRP | — | — | — | — | PnhSsPclE | — |
| GAD65 | — | — | — | — | — | MviSnPAat |
| GAD67 | — | — | — | — | — | MviSnPAat |
| IA-2 | — | qpsArPAaE | PkrpgPAgE | — | PpvTpVllE | PtgSaPAaq |
| falTaVAeEa | PsarpAAeE | EepAqAnmDa | EkpAsPAvq | |||
| PasTrPllD | VllTlVAla | |||||
| ValAgVAgl | ||||||
| PPI | — | — | — | — | — | — |
| Phogrin | — | falTaVAeE | — | LarThtAqD | EpgAaAfgE | PeaSsPArp |
| PssAfAdvE | PllApAApq | |||||
| EetAgVEnv | ||||||
| EeqSlPAga | ||||||
| PnsSfVAqr | ||||||
| PlpAtVAdf | ||||||
| ZnT-8 | — | — | — | — | — | — |
| ICA69 | — | qepAkAAsD | EdehvVAsD | PmgAqgEpD | VvaSdAdlD | — |
| PmkklVEkE | VgkTdkEhE | |||||
| EnreaVApE | ||||||
| PtagtPEpE |
a From Ref. 36.
FIGURE 5.
Binding of predicted HLA-DQ8trans candidate epitopes with double mismatches to all four HLA-DQ2/8 molecules. Prediction of potential DQ8trans epitopes within the top known diabetogenic proteins was performed with double mismatches at the anchor positions of the DQ8trans binding motif using the algorithm program MOTIFS. A selection of 46 potential DQ8trans epitopes was tested for binding all four DQ2/8 cis and trans molecules. EC50 values were calculated on the basis of competition between the biotinylated indicator peptide and the test peptides. Data represent the mean ± S.E. (n = 3). Shown on the x axes is 1/EC50, thereby illustrating that large bars represent better binding. Red bars represent HLA-DQ8trans-specific peptides. PPI, preproinsulin; IGRP, glucose-6-phosphatase catalytic subunit-related protein.
DISCUSSION
It is widely accepted that T1D is an autoimmune disease where auto-reactive T cells are the mediators of islet damage. Individuals carrying HLA-DQ2 (DQA1*05:01/DQB1*02:01) and/or HLA-DQ8 (DQA1*03:01/DQB1*03:02) have an increased risk for the development of celiac disease (CD) and T1D. The role of these HLA-DQ molecules with regard to binding and presentation of beta cell autoantigen-derived peptides in T1D is still unclear. It is well established that individuals heterozygous for HLA-DQ2 and HLA-DQ8 have by far the highest risk for development of T1D, and this has been linked to the formation of transdimers between the HLA-DQ2 α-chain and the HLA-DQ8 β-chain (HLA-DQ8trans) in particular (13, 14, 17, 27). This indicates that such HLA-DQ transdimers can bind and present a unique (set of) auto-antigen-derived peptide(s) that lead to beta cell destruction in the pancreas and the development of T1D. It is also conceivable that T cell responses to an exogenous antigen (e.g. viruses) are restricted by the transdimer leading to induction or aggravation of T1D through molecular mimicry. However, in this study we focus on identification of beta cell autoantigen-derived peptides. For the identification of such autoantigen derived peptides, it would thus be useful to know the peptide binding properties of such transdimers.
In this study we took an approach previously applied to HLA-DQ2cis and HLA-DQ8cis (25, 28) to define the peptide binding properties of the HLA-DQ8- and HLA-DQ2 transdimers. We observed that the peptide binding properties of the two transdimers are distinct from each other and from the previously defined binding motifs for HLA-DQ2cis and HLA-DQ8cis (Table 2 and supplemental Fig. 1). HLA-DQ8trans disfavored basic amino acids and displayed a strong dependence for acidic amino acids at anchor position p9 and mainly aliphatic residues at the other anchor positions, clearly distinguishing it from HLA-DQ8cis, HLA-DQ2trans, and HLA-DQ2cis. Whereas it is known that HLA-DQ2cis shares a preference for acidic amino acids, particularly at positions p4, p6, and p7 (25, 29), our present results indicate that HLA-DQ2trans has an even stronger preference for acidic amino acids along the entire binding core. This may be attributed to the nature of the binding pockets in HLA-DQ2trans as well as the predicted presence of several basic amino acids surrounding the peptide binding groove in the HLA-DQ2transdimer.
Because genetic epidemiology defined HLA-DQ8trans as most strongly associated with the risk of development of T1D, we next investigated whether the HLA-DQ8trans peptide binding motif could be used to predict autoantigen-derived peptides that would (preferentially) bind to HLA-DQ8trans. Using an algorithm search engine, the beta cell data base was searched for peptides that (partially) fulfilled HLA-DQ8trans binding motif. Binding studies revealed that ∼20% of the predicted peptides preferentially bound HLA-DQ8trans, whereas most of the other peptides also bound to HLA-DQ8cis but rarely to HLA-DQ2cis and HLA-DQ2trans. This results from the fact that both DQ8cis and DQ8trans share the HLA-DQ8 beta chain (DQB1*03:02), which is predicted to encompass positions p5 to p9 in the transdimer and, thus, form the p9 pocket, which has a strong preference for acidic residues in both HLA-DQ8cis and HLA-DQ8trans. Nevertheless, the observation that 20% of the predicted candidate epitopes specifically bind HLA-DQ8trans implicates that this HLA-DQ transdimer has unique binding characteristics.
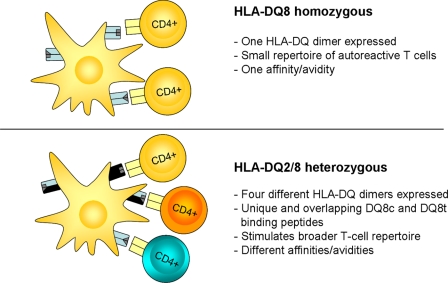
In a recent report we demonstrated that the HLA-DQ8transdimer is functional, as it could present gluten-derived peptides to CD4 T cell clones isolated from a small intestinal biopsy of a CD patient that expressed the HLA-DQ8transdimer but neither HLA-DQ2cis nor HLA-DQ8cis (30). In the same study we observed that several of such CD4 T cell clones specifically responded to stimulation with gliadin-derived peptides in either HLA-DQ8cis or HLA-DQ8trans, whereas a glutenin-specific CD4 T cell clone preferentially responded to stimulation with its cognate peptide in HLA-DQ8trans. These results underscore our present observations that a large percentage of the predicted epitopes bind to both HLA-DQ8cis and HLA-DQ8trans, whereas only a minority bind preferentially to HLA-DQ8trans. Together this provides a possible explanation for the observed high risk associated with the HLA-DQ2/8 heterozygous phenotype as compared with the lower risk associated with the HLA-DQ8cis-positive phenotype (Fig. 6). Our current and previous results demonstrate that HLA-DQ8cis-positive individuals potentially present a repertoire of autoantigen-derived peptides that could become the target of autoreactive T cells. HLA-DQ2/8 heterozygous individuals, on the other hand, can form four distinct HLA-DQ heterodimers, each with their own specific peptide binding properties (Table 2 and supplemental Fig. 1). We have strong evidence that HLA-DQ2/8 heterozygous APCs express both cis- and transdimers on their surface; in earlier studies we isolated gluten-specific HLA-DQ2cis- and HLA-DQ8cis-restricted CD4 T cell clones responding to their cognate gluten peptide in the presence of HLA-DQ2/8 heterozygous APCs (31, 32). Thus, next to the transdimer(s) (30), the cis dimers are also expressed on HLA-DQ2/8 heterozygous APCs. Such individuals can thus present a much more diverse repertoire of autoantigen-derived peptides that could trigger T cell responses. Although some of those T cells may be specific for autoantigen-derived peptides specifically binding to HLA-DQ8trans, other T cells may cross-react with other peptides bound to either HLA-DQ8cis or HLA-DQ8trans. Thus, there are multiple mechanisms that can act simultaneously.
FIGURE 6.
Proposed concept for the high risk association of HLA-DQ2/8 heterozygosity with development of T1D. HLA-DQ8 homozygous individuals only express a single HLA-DQ dimer on the surface of APCs, resulting in the presentation of a select number of epitopes and induction of a small repertoire of autoreactive CD4 T cells. Individuals heterozygous for the high risk-associated HLA-DQ2/8 haplotype can express multiple HLA-DQ dimers on APCs, consequently resulting in the presentation of a diversity of auto-antigen-derived epitopes and induction of a broader repertoire of autoreactive CD4 T cells. This may underlie the association of HLA-DQ2/8 with development of T1D.
In this respect it is important to note that it has been shown that CD4 T cells can cross-react with the same peptide bound to either HLA-DQ2cis or HLA-DQ2trans as well (33). In agreement with this, we observed that DQ2cis and DQ2trans share peptide binding properties (Table 2). Thus, it is likely that identical peptides can bind to both dimers. This may in turn allow T cells to cross-react between DQ2cis and DQ2trans also. Thus, a combination of a larger autoantigen-derived peptide repertoire together with extensive cross-reactivity between HLA-DQ cis- and transdimers might underlie the strong association between the development of T1D and the presence of HLA-DQ2/8 heterozygosity.
Several CD4 T cell epitopes derived from beta cell proteins have been described, but their relative importance in human disease pathogenesis is unclear. For a better understanding of the involvement of autoantigen-derived peptides in the pathogenesis of T1D, detailed knowledge about the involvement of the HLA-DQ molecules involved is crucial. This study identifies an autoantigen-derived peptide repertoire that is partly unique for HLA-DQ8trans and partly shared with HLA-DQ8cis. Obviously, the significance of the identified autoantigen-derived peptides will have to be tested in functional experiments in which patient-derived CD4 T cells are tested for reactivity with the identified DQ8trans-specific candidate epitopes, a study that falls beyond the scope of the present work. Nevertheless, such studies might reveal which peptides are capable of eliciting pathogenic CD4 T cell responses in the context of the disease-associated HLA-DQ molecules, which would be a crucial step toward the development of preventive or curative measures.
Supplementary Material
Acknowledgment
The Silicon Graphics Fuel instrument and the accompanying software were obtained via the Epirus Regional Development Programme to the Epirus Institute of Technology and of the 3rd Community Support Framework of the European Union.
This work was supported by Dutch Diabetes Foundation Grant 2007.00.038, by the Celiac Disease Consortium, an Innovative Cluster approved by The Netherlands Genomics Initiative partially funded by the Dutch government (BSIK03009), by the European Union 7th Framework Programme (FP7/2007-2013) under Grant Agreement 241447 (Natural Immunomodulators as Novel Immunotherapies for Type 1 Diabetes; NAIMIT), and by a VICI grant of The Netherlands Organization for Scientific Research (VICI 918.86.611).
- T1D
- type 1 diabetes
- GAD
- glutamic acid decarboxylase
- IA-2
- insulinoma-associated 2
- APC
- antigen-processing cell
- CD
- celiac disease.
REFERENCES
- 1. Atkinson M. A., Maclaren N. K. (1994) The pathogenesis of insulin-dependent diabetes mellitus. N. Engl. J. Med. 331, 1428–1436 [DOI] [PubMed] [Google Scholar]
- 2. Bluestone J. A., Herold K., Eisenbarth G. (2010) Genetics, pathogenesis, and clinical interventions in type 1 diabetes. Nature 464, 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Belle T. L., Coppieters K. T., von Herrath M. G. (2011) Type 1 diabetes. Etiology, immunology, and therapeutic strategies. Physiol. Rev. 91, 79–118 [DOI] [PubMed] [Google Scholar]
- 4. Kent S. C., Chen Y., Bregoli L., Clemmings S. M., Kenyon N. S., Ricordi C., Hering B. J., Hafler D. A. (2005) Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435, 224–228 [DOI] [PubMed] [Google Scholar]
- 5. Miller G. G., Pollack M. S., Nell L. J., Thomas J. W. (1987) Insulin-specific human T cells. Epitope specificity, major histocompatibility complex restriction, and alloreactivity to a diabetes-associated haplotype. J. Immunol. 139, 3622–3629 [PubMed] [Google Scholar]
- 6. Pinkse G. G., Tysma O. H., Bergen C. A., Kester M. G., Ossendorp F., van Veelen P. A., Keymeulen B., Pipeleers D., Drijfhout J. W., Roep B. O. (2005) Autoreactive CD8 T cells associated with beta cell destruction in type 1 diabetes. Proc. Natl. Acad. Sci. U.S.A. 102, 18425–18430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schloot N. C., Willemen S., Duinkerken G., de Vries R. R., Roep B. O. (1998) Cloned T cells from a recent onset IDDM patient reactive with insulin B-chain. J. Autoimmun. 11, 169–175 [DOI] [PubMed] [Google Scholar]
- 8. Kelemen K., Gottlieb P. A., Putnam A. L., Davidson H. W., Wegmann D. R., Hutton J. C. (2004) HLA-DQ8-associated T cell responses to the diabetes autoantigen phogrin (IA-2 beta) in human prediabetes. J. Immunol. 172, 3955–3962 [DOI] [PubMed] [Google Scholar]
- 9. Harfouch-Hammoud E., Walk T., Otto H., Jung G., Bach J. F., van Endert P. M., Caillat-Zucman S. (1999) Identification of peptides from autoantigens GAD65 and IA-2 that bind to HLA class II molecules predisposing to or protecting from type 1 diabetes. Diabetes 48, 1937–1947 [DOI] [PubMed] [Google Scholar]
- 10. Oling V., Geubtner K., Ilonen J., Reijonen H. (2010) A low antigen dose selectively promotes expansion of high avidity autoreactive T cells with distinct phenotypic characteristics. A study of human autoreactive CD4+T cells specific for GAD65. Autoimmunity 43, 573–582 [DOI] [PubMed] [Google Scholar]
- 11. Tree T. I., Duinkerken G., Willemen S., de Vries R. R., Roep B. O. (2004) HLA-DQ-regulated T cell responses to islet cell autoantigens insulin and GAD65. Diabetes 53, 1692–1699 [DOI] [PubMed] [Google Scholar]
- 12. Concannon P., Rich S. S., Nepom G. T. (2009) Genetics of type 1A diabetes. N. Engl. J. Med. 360, 1646–1654 [DOI] [PubMed] [Google Scholar]
- 13. Koeleman B. P., Lie B. A., Undlien D. E., Dudbridge F., Thorsby E., de Vries R. R., Cucca F., Roep B. O., Giphart M. J., Todd J. A. (2004) Genotype effects and epistasis in type 1 diabetes and HLA-DQ transdimer associations with disease. Genes Immun. 5, 381–388 [DOI] [PubMed] [Google Scholar]
- 14. Pociot F., McDermott M. F. (2002) Genetics of type 1 diabetes mellitus. Genes Immun. 3, 235–249 [DOI] [PubMed] [Google Scholar]
- 15. Nepom G. T., Kwok W. W. (1998) Molecular basis for HLA-DQ associations with IDDM. Diabetes 47, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 16. Moustakas A. K., Papadopoulos G. K. (2002) Molecular properties of HLA-DQ alleles conferring susceptibility to or protection from insulin-dependent diabetes mellitus. keys to the fate of islet beta cells. Am. J. Med. Genet. 115, 37–47 [DOI] [PubMed] [Google Scholar]
- 17. Erlich H., Valdes A. M., Noble J., Carlson J. A., Varney M., Concannon P., Mychaleckyj J. C., Todd J. A., Bonella P., Fear A. L., Lavant E., Louey A., Moonsamy P. (2008) HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk. Analysis of the type 1 diabetes genetics consortium families. Diabetes 57, 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carlotti F., Bazuine M., Kekarainen T., Seppen J., Pognonec P., Maassen J. A., Hoeben R. C. (2004) Lentiviral vectors efficiently transduce quiescent mature 3T3-L1 adipocytes. Mol. Ther. 9, 209–217 [DOI] [PubMed] [Google Scholar]
- 19. Eerligh P., van Lummel M., Zaldumbide A., Moustakas A. K., Duinkerken G., Bondinas G., Koeleman B. P., Papadopoulos G. K., Roep B. O. (2011) Functional consequences of HLA-DQ8 homozygosity versus heterozygosity for islet autoimmunity in type 1 diabetes. Genes Immun. 12, 415–427 [DOI] [PubMed] [Google Scholar]
- 20. van de Wal Y., Kooy Y. M., Drijfhout J. W., Amons R., Papadopoulos G. K., Koning F. (1997) Unique peptide binding characteristics of the disease-associated DQ(alpha 1*0501, beta 1*0201) vs the non-disease-associated DQ(alpha 1*0201, beta 1*0202) molecule. Immunogenetics 46, 484–492 [DOI] [PubMed] [Google Scholar]
- 21. Moustakas A. K., van de Wal Y., Routsias J., Kooy Y. M., van Veelen P., Drijfhout J. W., Koning F., Papadopoulos G. K. (2000) Structure of celiac disease-associated HLA-DQ8 and non-associated HLA-DQ9 alleles in complex with two disease-specific epitopes. Int. Immunol. 12, 1157–1166 [DOI] [PubMed] [Google Scholar]
- 22. Fremont D. H., Monnaie D., Nelson C. A., Hendrickson W. A., Unanue E. R. (1998) Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity 8, 305–317 [DOI] [PubMed] [Google Scholar]
- 23. Bondinas G. P., Moustakas A. K., Papadopoulos G. K. (2007) The spectrum of HLA-DQ and HLA-DR alleles, 2006. A listing correlating sequence and structure with function. Immunogenetics 59, 539–553 [DOI] [PubMed] [Google Scholar]
- 24. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 25. Stepniak D., Wiesner M., de Ru A. H., Moustakas A. K., Drijfhout J. W., Papadopoulos G. K., van Veelen P. A., Koning F. (2008) Large scale characterization of natural ligands explains the unique gluten binding properties of HLA-DQ2. J. Immunol. 180, 3268–3278 [DOI] [PubMed] [Google Scholar]
- 26. van de Wal Y., Kooy Y. M., Drijfhout J. W., Amons R., Koning F. (1996) Peptide binding characteristics of the coeliac disease-associated DQ(alpha1*0501, beta1*0201) molecule. Immunogenetics 44, 246–253 [DOI] [PubMed] [Google Scholar]
- 27. Khalil I., Deschamps I., Lepage V., al-Daccak R., Degos L., Hors J. (1992) Dose effect of cis- and trans-encoded HLA-DQ alpha beta heterodimers in IDDM susceptibility. Diabetes 41, 378–384 [DOI] [PubMed] [Google Scholar]
- 28. Godkin A., Friede T., Davenport M., Stevanovic S., Willis A., Jewell D., Hill A., Rammensee H. G. (1997) Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin-dependent diabetes susceptibility allele HLA-DQ8 (DQ 3.2). Int. Immunol. 9, 905–911 [DOI] [PubMed] [Google Scholar]
- 29. Vartdal F., Johansen B. H., Friede T., Thorpe C. J., Stevanović S., Eriksen J. E., Sletten K., Thorsby E., Rammensee H. G., Sollid L. M. (1996) The peptide binding motif of the disease associated HLA-DQ (alpha 1* 0501, beta 1* 0201) molecule. Eur. J. Immunol. 26, 2764–2772 [DOI] [PubMed] [Google Scholar]
- 30. Kooy-Winkelaar Y., van Lummel M., Moustakas A. K., Schweizer J., Mearin M. L., Mulder C. J., Roep B. O., Drijfhout J. W., Papadopoulos G. K., van Bergen J., Koning F. (2011) Gluten-specific T cells cross-react between HLA-DQ8 and the HLA-DQ2α/DQ8β transdimer. J. Immunol. 187, 5123–5129 [DOI] [PubMed] [Google Scholar]
- 31. Vader W., Kooy Y., Van Veelen P., De Ru A., Harris D., Benckhuijsen W., Peña S., Mearin L., Drijfhout J. W., Koning F. (2002) The gluten response in children with celiac disease is directed toward multiple gliadin and glutenin peptides. Gastroenterology 122, 1729–1737 [DOI] [PubMed] [Google Scholar]
- 32. van de Wal Y., Kooy Y. M., van Veelen P. A., Peña S. A., Mearin L. M., Molberg O., Lundin K. E., Sollid L. M., Mutis T., Benckhuijsen W. E., Drijfhout J. W., Koning F. (1998) Small intestinal T cells of celiac disease patients recognize a natural pepsin fragment of gliadin. Proc. Natl. Acad. Sci. U.S.A. 95, 10050–10054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tollefsen S., Arentz-Hansen H., Fleckenstein B., Molberg O., Ráki M., Kwok W. W., Jung G., Lundin K. E., Sollid L. M. (2006) HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Invest. 116, 2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim C. Y., Quarsten H., Bergseng E., Khosla C., Sollid L. M. (2004) Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl. Acad. Sci. U.S.A. 101, 4175–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee K. H., Wucherpfennig K. W., Wiley D. C. (2001) Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat. Immunol. 2, 501–507 [DOI] [PubMed] [Google Scholar]
- 36. Di Lorenzo T. P., Peakman M., Roep B. O. (2007) Translational mini-review series on type 1 diabetes. Systematic analysis of T cell epitopes in autoimmune diabetes. Clin. Exp. Immunol. 148, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.