Background: The class I cytokine IL-21 exerts pleiotropic effects on innate and adaptive immunity.
Results: We obtained the crystal structure of the partially glycosylated IL-21 receptor (IL-21R) bound to IL-21.
Conclusion: A sugar chain is an integral part of IL-21R.
Significance: This structure offers an insight into the putative role of the class I cytokine receptor signature motif.
Keywords: Carbohydrate Glycoprotein, Carbohydrate Structure, Crystallography, Immunology, Interleukin, Protein Structure, Receptor Structure-Function
Abstract
IL-21 is a class I cytokine that exerts pleiotropic effects on both innate and adaptive immune responses. It signals through a heterodimeric receptor complex consisting of the IL-21 receptor (IL-21R) and the common γ-chain. A hallmark of the class I cytokine receptors is the class I cytokine receptor signature motif (WSXWS). The exact role of this motif has not been determined yet; however, it has been implicated in diverse functions, including ligand binding, receptor internalization, proper folding, and export, as well as signal transduction. Furthermore, the WXXW motif is known to be a consensus sequence for C-mannosylation. Here, we present the crystal structure of IL-21 bound to IL-21R and reveal that the WSXWS motif of IL-21R is C-mannosylated at the first tryptophan. We furthermore demonstrate that a sugar chain bridges the two fibronectin domains that constitute the extracellular domain of IL-21R and anchors at the WSXWS motif through an extensive hydrogen bonding network, including mannosylation. The glycan thus transforms the V-shaped receptor into an A-frame. This finding offers a novel structural explanation of the role of the class I cytokine signature motif.
Introduction
IL-21 is a class I cytokine with a four-helix bundle structure arranged in an up-up-down-down topology typical for the class I cytokines (1). It exerts pleiotropic effects on both innate and adaptive immune responses. IL-21 is secreted by activated CD4+ T cells, in particular TH17 and T follicular helper cells, as well as natural killer cells (2). Not only do both TH17 and T follicular helper cells produce IL-21, but this cytokine also plays an important role in promoting the development of TH17 and T follicular helper cells by a feed-forward mechanism (3–8). Furthermore, IL-21 cooperates with other cytokines to increase the cytotoxicity of CD8+ T cells and promotes proliferation of CD8+ cells in the presence of antigens (9). IL-21 also influences antibody production by B cells (10). Recent studies demonstrated that IL-21 produced by CD4+ cells is critical for the ability of CD8+ T cells to control viral infection (11–13). The ability of IL-21 to augment immunity has spurred substantial interest in the therapeutic use of IL-21, and it is currently being evaluated in a number of clinical trials against, for example, metastatic melanoma and renal cancer (14).
IL-21 signals through a heterodimeric receptor complex consisting of the private chain IL-21 receptor (IL-21R)2 and the common γ-chain (γC), the latter being shared by IL-2, IL-4, IL-7, IL-9, and IL-15 (15). Upon binding of IL-21 to the receptor complex and subsequent receptor activation, signaling occurs through the Jak-STAT signaling pathway (16). The IL-21R chain binds IL-21 with high affinity and provides the majority of the binding energy (17). However, interaction with γC is required for signaling (16), and IL-21 mutants that bind IL-21R but fail to interact properly with γC act as potent antagonists of IL-21 signaling (1).
Several structures of γC class cytokines bound to one or more of their receptor chains have been solved (18–22). The receptors generally adopt very similar structures, with two type III fibronectin domains separated by a short linker. Each receptor chain adopts a V-shape, with the ligand binding at the tip of the V. The fibronectin domains each contain seven β-strands forming a sandwich-like structure. Binding of the ligand to the receptor engages residues present within the loops of both fibronectin domains.
Receptors of the γC cytokines are known to be universally N-glycosylated (20). This glycosylation is not required, however, for formation of the ligand-receptor complexes, as structures using receptors purified from Escherichia coli have been solved (20, 22). Still, the binding affinity between IL-7 and IL-7R has been demonstrated to be increased when IL-7R is glycosylated (20). Whether this is also the case for other members of the family remains to be determined.
The class I cytokine receptor family is characterized by the presence of the so-called class I cytokine receptor signature motif in the second fibronectin domain (24). The consensus sequence of the motif is WSXWS (present in 28 of 36 investigated sequences (25)). Several possible functional roles of the WSXWS sequence have been suggested, including involvement in ligand binding, receptor internalization, proper folding, export, and signal transduction (25–28). Although numerous studies have addressed the functional role of this highly conserved receptor motif, no clear picture has emerged. However, the first structures of class I cytokine receptors have clearly demonstrated that the motif is not directly involved in ligand binding (18–22, 29). Furthermore, despite the highly conserved nature of this motif, considerable sequence diversity is tolerated as demonstrated by several mutagenesis studies (25–27). The most extreme example is the human growth hormone receptor, in which the motif has the sequence YTEFS rather than WSXWS (30).
Interestingly, WXXW is known to be a consensus sequence for C-mannosylation, where a mannose is attached to the first tryptophan (31), and this kind of modification has indeed been found in the WSXWS motif of the class I cytokine receptors IL-12B and EPOR (32, 33). The structures of class I cytokine receptors solved so far have not included this modification, possibly because the proteins have been produced in either insect cells or E. coli, where this modification is not made. The mechanism and potential function of this modification are currently unknown.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
IL-21 was expressed and purified from E. coli as described previously (34). The extracellular domain (ECD) of IL-21R was expressed in HEK293 cells and purified as described (1).
Complex Formation
The complex of IL-21 and IL-21R was formed by mixing IL-21 and IL-21R at room temperature at a ratio of 1.5:1. Elevated levels of IL-21 were used, as IL-21 is most readily available. Furthermore, as IL-21 (15–16 kDa) is much smaller than IL-21R (28 kDa), it is more easily separated from the complex (43 kDa) after gel filtration. The complex was loaded onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) and eluted with PBS (10 mm phosphate and 150 mm NaCl, pH 7.4). The fractions containing the complex were concentrated to 5 mg/ml using an Amicon Ultra-4 centrifugal filter device with a 10,000 Mr cutoff.
Crystallization
All crystals were grown at 18 °C as sitting drops with a reservoir solution containing 500 μl of 1.8–1.9 m diammonium sulfate and 0.1 m sodium acetate at pH 5.5. 1 μl of reservoir solution and 1 μl of protein solution were mixed in the pedestal. Large single crystals appeared after 10–14 days. These were flash-frozen using a cryosolution containing 3.0 m diammonium sulfate and 0.1 m sodium acetate at pH 5.5. Tantalum bromide derivatives were obtained by adding 0.1 μl of a 2 mm Ta6Br122+ solution. This was left for 2 h, at which point the crystals had turned green, indicating uptake of Ta6Br122+. The crystals were flash-cooled using a cryosolution containing 3.0 m diammonium sulfate and 0.1 m sodium acetate at pH 5.5. Crystals of selenomethionine IL-21 in complex with IL-21R were produced in the same way as for the wild type.
Data Collection and Processing
All data were collected at the Swiss Light Source at the PXIII (X06DA) beamline at 77 K (196 °C). For the native data set, 180 frames were collected with an oscillation of 1º at a wavelength of 0.98 Å. For the selenomethionine IL-21 data set, 720 frames were collected with an oscillation of 1º at a wavelength of 0.98 Å, and for the tantalum bromide data set, 1080 frames were collected with an oscillation of 1º at a wavelength of 0.98 Å. All data sets were indexed and scaled using XDS (35). All data sets belong to space group P212121, with cell dimensions of 83 × 152 × 365 Å. See Table 1 for details.
TABLE 1.
Data collection and refinement statistics for the IL-21R·IL-21 complex
SLS, Swiss Light Source; FOM, figure of merit; r.m.s.d., root mean square deviation.
| Native | Ta6Br2 | SemetIL21 | |
|---|---|---|---|
| Data collection | |||
| Beamline | SLS X06DA | SLS X06DA | SLS X06DA |
| Space group | P212121 | P212121 | P212121 |
| Cell dimensions | a = 82.2, b = 151.1, c = 364.8 Å; α = 90.0°, β = 90.0°, γ = 90.0° | a = 82.3, b = 151.2, c = 364.6 Å; α = 90.0°, β = 90.0°, γ = 90.0° | a = 82.0, b = 151.0, c = 365.6 Å; α = 90.0°, β = 90.0°, γ = 90.0° |
| Resolution (Å) | 94.7-2.8 (2.9-2.8) | 72.2-6 (8-6) | 50-8 (10-8) |
| Rmerge | 18.7 (71.9) | 2.75 (3.8) | 3.6 (4.4) |
| I/σI | 30.1 (2.08) | 52.1 (32.0) | 50.0 (43.5) |
| Completeness (%) | 99.7 (82.2) | 99.0 (97.6) | 97.6 (97.6) |
| Redundancy | 5.2 (4.4) | 16.0 (15.8) | 13.9 (15.8) |
| Initial FOM | 0.46 | ||
| FOM after density modification | 0.69 | ||
| Refinement | |||
| Resolution (Å) | 80.2-2.8 | ||
| No. reflections | 112,598 | ||
| Rwork/Rfree | 0.25/0.27 (0.37/0.38) | ||
| No. atoms | 21,835 | ||
| Protein | 20,916 | ||
| Sugar | 711 | ||
| Ligand/ion | 204 | ||
| B-factors | |||
| Wilson B-factor | 53.7 | ||
| Overall B-factor | 69.5 | ||
| Protein | 68.0 | ||
| Sugar | 101.1 | ||
| Ligand/ion | 105.6 | ||
| r.m.s.d. | |||
| Bond lengths (Å) | 0.018 | ||
| Bond angles | 1.352° | ||
Phasing
Sites for the Ta6Br122+ derivative were found using SHELXE and SHELXD (36), and phases were calculated using Phenix (37). Phasing was done at 6 Å, where Ta6Br2 scattering approximates a single heavy atom. 25 initial sites were found in SHELXE using both anomalous and isomorphous data. Of these, 16 highest scoring sites were chosen for refinement in Phenix. Upon manual inspection, 15 of the initial 16 sites were selected for phasing. The phases from the Ta6Br122+ data set were used to find sites in the selenomethionine IL-21 data set using anomalous difference Fourier maps. This gave 50 initial hits. Manual inspection of these sites revealed that 20 were positioned as three sites with 8-fold symmetry, with four sites missing. Human IL-21 contains four methionines, but from the available NMR structure (34), we knew that two of these (Met10 and Met118) are close in the structure. This makes it likely that they would be seen as a single site at 6 Å. Initial phases were calculated from the Ta6Br122+ derivative, followed by density modification in Resolve. Using the selenomethionine IL-21 sites, eight copies of the known NMR structure of IL-21 were placed in the map. From this, initial NCS operators were calculated, and density modification was repeated using this 8-fold NCS. A polyalanine model of IL-2RB was created and divided into its two constituent fibronectin domains. These were placed in the regions of the map containing density for IL-21R. Using Resolve (38), one NCS group with eight operators was made for IL-21, as well as for each of the two fibronectin domains of IL-2RB. Upon refinement of the NCS operators, a final round of density modification and phase extension to 3.5 Å was performed in Resolve. The resulting map was of excellent quality at the resolution, and the IL-21R structure was built de novo by repeated cycles of building and refinement in Coot (39) and Phenix, respectively. The final model contains eight molecules of IL-21R and IL-21, forming the IL-21R·IL-21 complex refined to a resolution of 2.8 Å. Evaluation of the Ramachandran plot in Coot showed 94.25% and 5.75% in preferred and allowed regions, respectively.
RESULTS
Overall Structure
We have determined the structure of IL-21 in complex with the ECD of its private receptor chain IL-21R at a resolution of 2.8 Å (Fig. 1A and Table 1). The unit cell contains eight IL-21R·IL-21 complexes. The core structures of the eight IL-21R·IL-21 complexes are highly similar. The main difference observed is in the flexible loop connecting helices C and D in IL-21. The interaction surface between IL-21R and IL-21 is identical, however, in the eight structures. The following description represents molecules A (IL-21R) and B (IL-21) in the Protein Data Bank file (code 3TGX).
FIGURE 1.
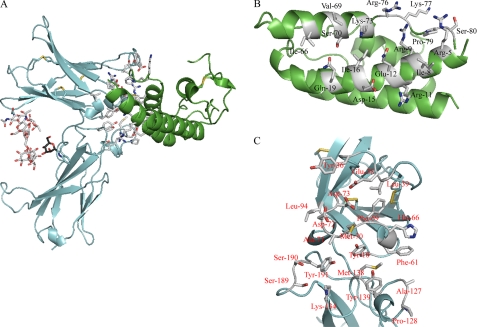
Residues in IL-21 and IL-21R involved in binding. A, structure of the IL-21R·IL-21 complex, with residues involved in the interaction between IL-21 and IL-21R shown in white. IL-21 is shown in green, and IL-21R is shown in cyan. The sugar chain originating from Asn54 is shown in white, and the mannose on Trp195 is shown in black. Distance criteria for contact assignments are ≤4.5 for van der Waals contacts and ≤3.5 for hydrogen bonds and polar interactions. B, residues in IL-21 involved in binding IL-21R. C, residues in IL-21R involved in binding IL-21.
IL-21R·IL-21 Structure
IL-21R contains two fibronectin III domains that form a V-shaped structure, with the binding site for the cytokine positioned at the tip of the V (Fig. 1B). The two fibronectin domains are connected by a short linker and bent at an angel of ∼90°. The D1 domain contains three disulfide bridges: one connecting the N terminus to β-strand 7 (Cys1–Cys90), one connecting β-strands 1 and 2 (Cys6–Cys16), and one connecting β-strands 4 and 5 (Cys46–Cys62). The membrane-proximal domain (D2) does not contain any disulfides but includes a WSXWS motif in the F′G′ loop, which is characteristic for class I cytokine receptors. IL-21 exhibits the typical type I cytokine structure composed of a four-helix bundle with an up-up-down-down topology. The finished structure consists of residues 1–208 of IL-21R and residues 2–81 and 89–123 of IL-21, with small variations in the number of built residues between the eight NCS-related molecules. The missing part of IL-21 is the loop connecting helices C and D. This part of the loop is close to neither the IL-21R-binding site nor the predicted γC-binding site.
IL-21R·IL-21 Interface
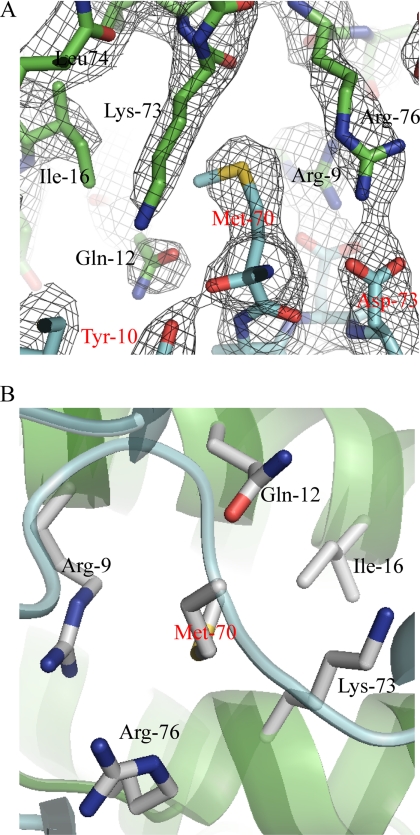
The interaction between IL-21 and IL-21R is mediated by residues present in helices A and C and by a small part of the CD loop immediately following helix C of IL-21 (Fig. 1, A and B, and supplemental Table S1). The total binding surface contributed by both IL-21 and IL-21R is 990 Å2 (calculated using PDBe PISA (40)) and includes extensive sets of polar and apolar interactions (supplemental Table S1). Ten different residues of IL-21 participate in polar interactions with 11 residues of IL-21R. There are 14 residues of IL-21 forming van der Waals contacts with 16 residues of IL-21R. In IL-21, Arg5, Arg9, and Gln12 of helix A and Arg76 and Lys73 of helix C are the major contributors to the binding surface. All five residues form extensive polar and van der Waals interactions with residues of IL-21R (supplemental Table S1). Arg9, Gln12, Arg76, and Lys73 along with Ile16 form a pocket for Met70 of IL-21R, the main contributor to the binding surface in IL-21R. It is interesting to note that the large hydrophobic side chain of Met70 fits into a pocket of mainly hydrophilic residues. Met70 seems to play a crucial role in positioning Arg9, Gln12, Arg76, and Lys73 correctly in relation to their other interaction partners. Thus, Met70 positions Arg9 and Arg76 of IL-21 for interaction with Asp72 and Asp73 of IL-21R, respectively (Fig. 2, A and B). The IL-21-binding residues of IL-21R are located in the loops connecting the β-strands. The AB, CD, EF, B′C′, and F′G′ loops and the linker all contain residues involved in binding. In IL-21R, Tyr36 in the CD loop, Met70 and Asp72 in the EF loop, and Tyr129 in the B′C′ loop contribute the most to the binding surface. The most important loop is the EF loop, which supplies 7 of the 20 amino acids of IL-21R that are involved in binding IL-21 (Fig. 1C and supplemental Table S1).
FIGURE 2.
IL-21-binding pocket for Met70 of IL-21R. A, 2Fo − Fc electron density map showing part of the binding interface between IL-21 and IL-21R centered at Met70 (color code as described for Fig. 1). B, binding pocket in IL-21 for Met70 of IL-21R viewed from IL-21R.
Comparison of Free and Receptor-bound IL-21
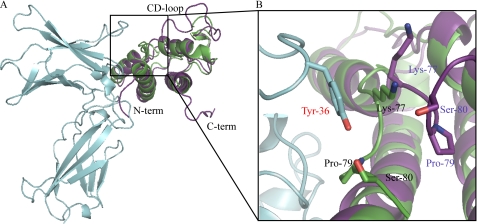
The structure of free IL-21 has been previously solved by NMR (34). In contrast to IL-21R-bound IL-21, the structure of the free form contains the N- and C-terminal parts, as well as the CD loop. These are highly disordered, however, explaining why they are absent in the IL-21R·IL-21 structure determined by crystallography. This difference is thus a result of the differences between the techniques of NMR and x-ray crystallography rather than a consequence of ligand binding. In solution, IL-21 exists in equilibrium between two distinct structures, in which helix C is either partially unfolded or formed. The crystal structure reported here indicates that helix C of IL-21 is stabilized upon binding to the receptor. The root mean square deviation using 106 backbone Cα atoms of IL-21 is 1.6 Å (calculated using SSM superimpose in Coot (41)), with the main differences seen in the first part of the CD loop and in the beginning of helix A. In the receptor-bound structure, the N-terminal part of helix A is slightly elongated compared with the free form. This is likely a result of receptor binding, as Arg5 of IL-21 is present in this region and interacts with IL-21R (supplemental Table S1). The most noticeable effect of IL-21R binding is observed in the first part of the CD loop of IL-21 (Fig. 3). The CD loop is positioned toward the receptor, allowing Lys77, Pro79, and Ser80 to form a binding pocket for Tyr36 of the receptor. Previously, a homology model of the IL-21R γC·IL-21 complex was built using the NMR model of IL-21 and the structures of IL-2 in complex with IL-2RA, IL-2RB, and γC, as well as IL-4 in complex with IL-4RA and γC (34). The position of the IL-21R chain predicted in this work is in good agreement with the IL-21R·IL-21 crystal structure presented here.
FIGURE 3.
Comparison of IL-21 in solution and IL-21 bound to IL-21R. The structure of IL-21 in free form solved by NMR (Protein Data Bank code 2OQP; purple) was superimposed onto the IL-21R·IL-21 crystal structure (color code as described for in Fig. 1) using SSM superimpose in Coot. The cutout shows an enlargement of the first part of the CD loop of IL-21. Residues of IL-21 forming a pocket for Tyr36 of the receptor are labeled and color-coded as described for the respective structures.
Sugar Chain Attached to Asn54 Is Essential for Proper Expression of ECD of IL-21R
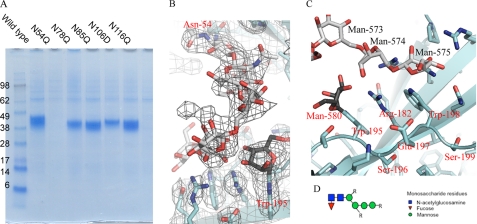
The ECD of IL-21R contains five potential N-linked glycosylation sites (Asn54, Asn78, Asn85, Asn106, and Asn116), and the purified ECD of IL-21R recombinantly expressed in HEK293 cells includes ∼10-kDa glycans. To establish which sites might be required for receptor integrity, all potential Asn-linked glycosylation sites were individually mutated to Gln, and the effect was evaluated by expression in HEK293 cells. Only the Asn54-to-Gln mutation had a profound effect, as it led to an almost complete loss of secreted protein (Fig. 4A). To avoid interference from nonessential sugar residues in both crystallization and biochemical tests, the glycan-minimized ECD of IL-21R (N78Q, N85Q, N106D, and N116Q) was expressed in HEK293 cells. This was the protein used for crystallization.
FIGURE 4.
Carbohydrate chain attached to Asn54 anchors at class I cytokine receptor signature motif (WSXWS). A, expression followed by His-tagged purification of the ECD of IL-21R, the wild type, or variants with mutations of the five predicted N-linked glycosylation sites as indicated. B, 2Fo − Fc electron density map (at 1.5σ) around the sugar chain in IL-21R. The color code is as described for Fig. 1. C, mannosylated Trp195 and Arg182 form multiple hydrogen bonds with the carbohydrate chain originating at Asn54. Arg182 is sandwiched between the two tryptophans of the WSXWS motif, forming the W-R-W zipper described in the text. D, the glycan structure is heterogeneous. R indicates that extra sugar residues might exist at this position, which cannot be built in the electron density. The core structure depicted here is found in the large majority of the glycans. The full glycan structure was determined by mass spectrometry (supplemental Table S2).
Two Domains of IL-21R Are Bridged by Sugar Chain That Could Potentially Stabilize Receptor
To our surprise, we found that Asn54 is connected to the highly conserved WSXWS motif by an N-linked glycan bridge, which forms a cross-bar, turning the flexible V-shape, formed by the two fibronectin domains, into a seemingly rigidified A-frame (Figs. 1A and 4, B and C). Digestion with peptide:N-glucosidase F to deglycosylate the protein failed (data not shown), suggesting that the sugar-based cross-link is an integral part of IL-21R and therefore might be protected from digestion. The composition of the sugar chain attached to Asn54 was investigated by LC-MS, revealing that the glycosylation is rather heterogeneous (supplemental Table S2). However, a common core consisting of two N-acetylglucosamines and four hexoses, most likely mannose, was observed (Fig. 4D and supplemental Table S2). In most of the sugar chains, a fucose is also present. The IL-21R structure presented here has been built to represent this most prevalent glycan chain, i.e. with two N-acetylglucosamines, four mannoses, and one fucose residue (Figs. 1A and 4B). The electron density and the LC-MS analysis indicate that additional sugar residues are present in the receptor chain (Fig. 4, B and D, and supplemental Table S2). However, these cannot be placed with confidence due to the heterogeneity of the glycosylation and the possible flexibility of the sugars, which do not engage in the hydrogen bonding network described above. The position of the additional glycosylation is indicated in Fig. 4D.
Mannosylation at First Tryptophan in WSXWS Motif Forms Extensive Hydrogen Bonding Network with Sugar Chain Originating from Asn54
The hydrogen bonding network between the sugar chain and the WSXWS motif offers a unique insight into how glycans can be integrated into protein structures (Fig. 4C and Table 2). Arg182 is held tightly in place between the two Trp residues, with its charged headgroup exposed, allowing it to form hydrogen bonds with the sugar chain. Furthermore, an additional density was observed around Trp195, suggesting that this residue was modified. Indeed, protease digestion followed by LC-MS analysis confirmed that Trp195 is C-mannosylated at C1 of the indole ring (data not shown). Thus, the carbohydrate chain originating from Asn54 forms multiple hydrogen bonds not only with Arg182 but also with mannosylated Trp195 (Fig. 4C and Table 2).
TABLE 2.
Hydrogen bonds connecting the Asn54N-linked sugar to the second fibronectin domain of IL-21R and Trp195-linked Man580
| N-Linked sugar/atom | IL-21R or Man580 | Atom | Distance |
|---|---|---|---|
| Å | |||
| Man574 | |||
| O2 | Man580 | O4 | 2.4 |
| O2 | Man580 | O2 | 2.1 |
| O2 | Arg186 | NH1 | 3.3 |
| O2 | Arg154 | NH1 | 3.5 |
| O2 | Arg186 | N2 | 3.2 |
| O3 | Man580 | O4 | 2.9 |
| O5 | Arg154 | NH2 | 3.3 |
| O6 | Trp198 | NE1 | 3.4 |
| Man575 | |||
| O3 | Gln140 | OE1 | 2.8 |
| O3 | Gln140 | NE2 | 2.7 |
| O3 | Arg142 | NH2 | 3.5 |
| O4 | Gln180 | OE1 | 3.3 |
| O5 | Trp198 | NE1 | 2.9 |
DISCUSSION
The complex between IL-21R and IL-21 bears considerable resemblance to the other γC-binding cytokines complexed with their corresponding receptors. The binding interactions are mediated by residues present in the loops of both of the fibronectin domains of the receptor contacting residues of helices A and C and the first part of the CD loop of IL-21. The overall structure of IL-21 bound to IL-21R is quite similar to the structure of IL-21 in solution, with the most prominent difference being a rearrangement of the first part of the CD loop, leading to formation of a pocket providing binding of Tyr36 of the receptor. Free IL-21 exists in two distinct forms in solution; one has a structured helix C and the other an unstructured region within this segment. The IL-21R·IL-21 crystal structure demonstrates that it is the IL-21 conformation that has a structured helix C that is bound by the receptor. In line with this, it was previously determined that stabilization of helix C and the first part of the CD loop of IL-21 by replacing them with the equivalent part of IL-4 leads to a ligand of significantly higher activity (34). Helix C in IL-4 is more ordered than its counterpart in IL-21. The increase in ligand potency was thus proposed to arise as a result of chimeric IL-21 presenting helix C and the CD loop more favorable for functional interaction with IL-21R.
Our structure is the first type II cytokine structure to display partial mammalian glycosylation. We observed that the highly conserved WSXWS motif is C-mannosylated at the first Trp, as would be expected from prior biochemical work (31–33, 42). Thus, for the first time, we can visualize the structural consequences of this modification. Interestingly, the structure furthermore reveals that Asn54 of the D1 domain of IL-21R is connected to the WSXWS motif in the D2 domain by an N-linked glycan bridge. The sugar chains are in solvent channels in the crystal and are thus not close to any crystal contacts. The position of the sugar is thus not an artifact of the crystal packing. We have shown that the sugar chain attached to Asn54 is required for proper cell surface expression of IL-21R, whereas the remaining sugars are not. We consider it unlikely that the general folding of the receptor is impaired by the absence of Asn54 glycosylation. In the previously published structures of class I cytokine receptor complexes, the ECDs of the receptors were produced in insect cells, which do not yield the same type of glycosylation as mammalian cells (19, 21, 29, 43, 44). As these structures were solved without the carbohydrate bridge, it cannot be essential for the folding of individual domains. Previous studies have shown, however, that even minimal glycosylation of IL-7R enhances the affinity for IL-7 (20). Whether this is also the case for other members of the family remains to be determined. Biochemical studies of EPOR have shown that mutations within the WSXWS motif impair export of EPOR to the cell surface and lead to accumulation in the Golgi (25, 45). In particular, the first Trp of the WSXWS showed no tolerance for substitution. Interestingly, EPOR has an N-terminal helix that takes up the space between the D1 and D2 domains, making contact with the WSXWS motif (23). This helix thus occupies the same position as the glycan bridge in IL-21R. It is tempting to speculate that the sugar bridge has an influence on IL-21R signaling, possible via a stabilizing effect on the ECD of IL-21R. However, we have been unable to address this important question properly, as mutations that eliminate the sugar chain originating from Asn54 also impair cell surface expression of the receptor.
Several members of the class I cytokine receptor family have potential N-linked glycosylation sites in the D1 domain facing the D2 domain (e.g. IL-2RB has Asn17, γC has Asn49, and IL-7R has Asn29) (17, 20). Most of the previous structures of type I cytokine complexes use receptors expressed in insect cells, which show less complex glycosylation patterns than mammalian cells and do not mannosylate the WSXWS sequence (19, 21, 29, 43). These structures can thus not help elucidate whether the N-linked glycan bridge observed in the IL-21R structure, is a specific feature of IL-21R or a more general feature of the class I cytokine receptor family.
Acknowledgments
We thank V. Olieric (Swiss Light Source) for assistance with synchrotron data collection. Beamline access was supported in part by the Danscatt Consortium.
This work was supported by a research fellowship from the Danish Cancer Society (to O. J. H.) and by grants from the Danish Cancer Society, the Danish Council for Independent Research Natural Science, and the Novo Nordisk Foundation (to R. H.).

This article contains supplemental Tables S1–S3.
The atomic coordinates and structure factors (code 3TGX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IL-21R
- IL-21 receptor
- γC
- γ-chain
- ECD
- extracellular domain.
REFERENCES
- 1. Kang L., Bondensgaard K., Li T., Hartmann R., Hjorth S. A. (2010) Rational design of interleukin-21 antagonist through selective elimination of the γC binding epitope. J. Biol. Chem. 285, 12223–12231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spolski R., Leonard W. J. (2010) IL-21 and T follicular helper cells. Int. Immunol. 22, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnston R. J., Poholek A. C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A. L., Craft J., Crotty S. (2009) Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Korn T., Bettelli E., Gao W., Awasthi A., Jäger A., Strom T. B., Oukka M., Kuchroo V. K. (2007) IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448, 484–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nurieva R., Yang X. O., Martinez G., Zhang Y., Panopoulos A. D., Ma L., Schluns K., Tian Q., Watowich S. S., Jetten A. M., Dong C. (2007) Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 [DOI] [PubMed] [Google Scholar]
- 6. Nurieva R. I., Chung Y., Hwang D., Yang X. O., Kang H. S., Ma L., Wang Y. H., Watowich S. S., Jetten A. M., Tian Q., Dong C. (2008) Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity 29, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vogelzang A., McGuire H. M., Yu D., Sprent J., Mackay C. R., King C. (2008) A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity 29, 127–137 [DOI] [PubMed] [Google Scholar]
- 8. Nurieva R. I., Chung Y., Martinez G. J., Yang X. O., Tanaka S., Matskevitch T. D., Wang Y. H., Dong C. (2009) Bcl6 mediates the development of T follicular helper cells. Science 325, 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng R., Spolski R., Finkelstein S. E., Oh S., Kovanen P. E., Hinrichs C. S., Pise-Masison C. A., Radonovich M. F., Brady J. N., Restifo N. P., Berzofsky J. A., Leonard W. J. (2005) Synergy of IL-21 and IL-15 in regulating CD8+ T cell expansion and function. J. Exp. Med. 201, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozaki K., Spolski R., Feng C. G., Qi C. F., Cheng J., Sher A., Morse H. C., 3rd, Liu C., Schwartzberg P. L., Leonard W. J. (2002) A critical role for IL-21 in regulating immunoglobulin production. Science 298, 1630–1634 [DOI] [PubMed] [Google Scholar]
- 11. Elsaesser H., Sauer K., Brooks D. G. (2009) IL-21 is required to control chronic viral infection. Science 324, 1569–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fröhlich A., Kisielow J., Schmitz I., Freigang S., Shamshiev A. T., Weber J., Marsland B. J., Oxenius A., Kopf M. (2009) IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science 324, 1576–1580 [DOI] [PubMed] [Google Scholar]
- 13. Yi J. S., Du M., Zajac A. J. (2009) A vital role for interleukin-21 in the control of a chronic viral infection. Science 324, 1572–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hashmi M. H., Van Veldhuizen P. J. (2010) Interleukin-21: updated review of Phase I and II clinical trials in metastatic renal cell carcinoma, metastatic melanoma, and relapsed/refractory indolent non-Hodgkin lymphoma. Expert Opin. Biol. Ther. 10, 807–817 [DOI] [PubMed] [Google Scholar]
- 15. Rochman Y., Spolski R., Leonard W. J. (2009) New insights into the regulation of T cells by γC family cytokines. Nat. Rev. Immunol. 9, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asao H., Okuyama C., Kumaki S., Ishii N., Tsuchiya S., Foster D., Sugamura K. (2001) Cutting edge: the common γ-chain is an indispensable subunit of the IL-21 receptor complex. J. Immunol. 167, 1–5 [DOI] [PubMed] [Google Scholar]
- 17. Zhang J. L., Foster D., Sebald W. (2003) Human IL-21 and IL-4 bind to partially overlapping epitopes of common γ-chain. Biochem. Biophys. Res. Commun. 300, 291–296 [DOI] [PubMed] [Google Scholar]
- 18. Bravo J., Staunton D., Heath J. K., Jones E. Y. (1998) Crystal structure of a cytokine-binding region of gp130. EMBO J. 17, 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., Heller N. M., Keegan A. D., Garcia K. C. (2008) Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McElroy C. A., Dohm J. A., Walsh S. T. (2009) Structural and biophysical studies of the human IL-7/IL-7Rα complex. Structure 17, 54–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X., Rickert M., Garcia K. C. (2005) Structure of the quaternary complex of interleukin-2 with its α, β, and γC receptors. Science 310, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 22. Hage T., Sebald W., Reinemer P. (1999) Crystal structure of the interleukin-4/receptor α-chain complex reveals a mosaic binding interface. Cell 97, 271–281 [DOI] [PubMed] [Google Scholar]
- 23. Livnah O., Stura E. A., Middleton S. A., Johnson D. L., Jolliffe L. K., Wilson I. A. (1999) Crystallographic evidence for preformed dimers of erythropoietin receptor before ligand activation. Science 283, 987–990 [DOI] [PubMed] [Google Scholar]
- 24. Tauber M. T., Porra V., Dastot F., Molinas C., Amselem S., Cholin S., Rochiccioli P., Bieth E. (1998) Heterozygous mutation in the WSXWS equivalent motif of the growth hormone receptor in a child with poor response to growth hormone therapy. Growth Horm. IGF Res. 8, 211–216 [DOI] [PubMed] [Google Scholar]
- 25. Hilton D. J., Watowich S. S., Katz L., Lodish H. F. (1996) Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J. Biol. Chem. 271, 4699–4708 [DOI] [PubMed] [Google Scholar]
- 26. Chiba T., Amanuma H., Todokoro K. (1992) Tryptophan residue of Trp-Ser-X-Trp-Ser motif in extracellular domains of erythropoietin receptor is essential for signal transduction. Biochem. Biophys. Res. Commun. 184, 485–490 [DOI] [PubMed] [Google Scholar]
- 27. Miyazaki T., Maruyama M., Yamada G., Hatakeyama M., Taniguchi T. (1991) The integrity of the conserved “WS motif” common to IL-2 and other cytokine receptors is essential for ligand binding and signal transduction. EMBO J. 10, 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quelle D. E., Quelle F. W., Wojchowski D. M. (1992) Mutations in the WSAWSE and cytosolic domains of the erythropoietin receptor affect signal transduction and ligand binding and internalization. Mol. Cell. Biol. 12, 4553–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hansen G., Hercus T. R., McClure B. J., Stomski F. C., Dottore M., Powell J., Ramshaw H., Woodcock J. M., Xu Y., Guthridge M., McKinstry W. J., Lopez A. F., Parker M. W. (2008) The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell 134, 496–507 [DOI] [PubMed] [Google Scholar]
- 30. Baumgartner J. W., Wells C. A., Chen C. M., Waters M. J. (1994) The role of the WSXWS equivalent motif in growth hormone receptor function. J. Biol. Chem. 269, 29094–29101 [PubMed] [Google Scholar]
- 31. Furmanek A., Hofsteenge J. (2000) Protein C-mannosylation: facts and questions. Acta Biochim. Pol. 47, 781–789 [PubMed] [Google Scholar]
- 32. Doucey M. A., Hess D., Blommers M. J., Hofsteenge J. (1999) Recombinant human interleukin-12 is the second example of a C-mannosylated protein. Glycobiology 9, 435–441 [DOI] [PubMed] [Google Scholar]
- 33. Furmanek A., Hess D., Rogniaux H., Hofsteenge J. (2003) The WSAWS motif is C-hexosylated in a soluble form of the erythropoietin receptor. Biochemistry 42, 8452–8458 [DOI] [PubMed] [Google Scholar]
- 34. Bondensgaard K., Breinholt J., Madsen D., Omkvist D. H., Kang L., Worsaae A., Becker P., Schiødt C. B., Hjorth S. A. (2007) The existence of multiple conformers of interleukin-21 directs engineering of a superpotent analogue. J. Biol. Chem. 282, 23326–23336 [DOI] [PubMed] [Google Scholar]
- 35. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Terwilliger T. C. (2003) Improving macromolecular atomic models at moderate resolution by automated iterative model building, statistical density modification, and refinement. Acta Crystallogr. D Biol. Crystallogr. 59, 1174–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Velankar S., Kleywegt G. J. (2011) The Protein Data Bank in Europe (PDBe): bringing structure to biology. Acta Crystallogr. D Biol. Crystallogr. 67, 324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42. Hofsteenge J., Müller D. R., de Beer T., Löffler A., Richter W. J., Vliegenthart J. F. (1994) New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry 33, 13524–13530 [DOI] [PubMed] [Google Scholar]
- 43. Chow D., He X., Snow A. L., Rose-John S., Garcia K. C. (2001) Structure of an extracellular gp130 cytokine receptor signaling complex. Science 291, 2150–2155 [DOI] [PubMed] [Google Scholar]
- 44. Stauber D. J., Debler E. W., Horton P. A., Smith K. A., Wilson I. A. (2006) Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hilton D. J., Watowich S. S., Murray P. J., Lodish H. F. (1995) Increased cell surface expression and enhanced folding in the endoplasmic reticulum of a mutant erythropoietin receptor. Proc. Natl. Acad. Sci. U.S.A. 92, 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]