Background: Many tRNA synthetases were found in the nucleus of eukaryotic cells where translation does not occur.
Results: Nuclear localization of human TyrRS is dependent on a dual-function motif also used for tRNA binding.
Conclusion: The cognate tRNA controls nuclear localization of human TyrRS.
Significance: The tRNA-controlled regulation coordinates nuclear import of a human tRNA synthetase with the demands of protein synthesis in the cytoplasm.
Keywords: Aminoacyl tRNA Synthetase, Cell Fractionation, Intracellular Trafficking, Protein Structure, Protein-Nucleic Acid Interaction
Abstract
Aminoacyl-tRNA synthetases, essential components of the cytoplasmic translation apparatus, also have nuclear functions that continue to be elucidated. However, little is known about how the distribution between cytoplasmic and nuclear compartments is controlled. Using a combination of methods, here we showed that human tyrosyl-tRNA synthetase (TyrRS) distributes to the nucleus and that the nuclear import of human TyrRS is regulated by its cognate tRNATyr. We identified a hexapeptide motif in the anticodon recognition domain that is critical for nuclear import of the synthetase. Remarkably, this nuclear localization signal (NLS) sequence motif is also important for interacting with tRNATyr. As a consequence, mutational alteration of the hexapeptide simultaneously attenuated aminoacylation and nuclear localization. Because the NLS is sterically blocked when the cognate tRNA is bound to TyrRS, we hypothesized that the nuclear distribution of TyrRS is regulated by tRNATyr. This expectation was confirmed by RNAi knockdown of tRNATyr expression, which led to robust nuclear import of TyrRS. Further bioinformatics analysis showed that to have nuclear import of TyrRS directly controlled by tRNATyr in higher organisms, the NLS of lower eukaryotes was abandoned, whereas the new NLS was evolved from an anticodon-binding hexapeptide motif. Thus, higher organisms developed a strategy to make tRNA a regulator of the nuclear trafficking of its cognate synthetase. The design in principle should coordinate nuclear import of a tRNA synthetase with the demands of protein synthesis in the cytoplasm.
Introduction
Aminoacyl-tRNA synthetases catalyze a two-step aminoacylation reaction to link amino acids with their cognate tRNAs to provide amino acid building blocks for cytoplasmic ribosomal protein synthesis (1, 2). Interestingly, many tRNA synthetases were also found in the nucleus of eukaryotic cells where translation does not occur (3, 4). At least in some organisms, the nuclear distributions of tRNA synthetase are involved with an aminoacylation-dependent quality control mechanism to ensure nuclear export of mature and functional tRNAs (5–7). In addition to tRNA proofreading, nuclear functions of tRNA synthetases include regulation of transcription and ribosomal RNA synthesis (8, 9). However, little is known about how the distribution of synthetases between cytoplasmic and nuclear compartments is controlled.
Tyrosyl-tRNA synthetase (TyrRS)2 is a class I tRNA synthetase with a Rossmann fold-based catalytic domain that contains the active site where the two-step aminoacylation reaction occurs. In the first step, tyrosine and ATP generate enzyme-bound Tyr-AMP and release PPi. In the second step, the tyrosyl moiety of Tyr-AMP is transferred to the 3′-end of tRNATyr to generate Tyr-tRNATyr. To address the question of how the distribution of a tRNA synthetase is controlled, we focused on human TyrRS because it is one of the most characterized tRNA synthetases, with crystal structures being solved from 12 different organisms including human (Protein Data Bank (PDB) database). Human TyrRS is also of broad interest because it prominently exemplifies the multifunctionality of tRNA synthetases (10) and is associated with heritable disease (11). Lastly, nuclear localization of TyrRS was previously demonstrated in Saccharomyces cerevisiae (12).
Because the nuclear localization signal (NLS) identified in yeast TyrRS is not found in the human enzyme (12), we started our investigation by detecting nuclear localization of TyrRS in human cells. Through a combination of methods, we identified a distinct NLS sequence for human TyrRS and showed that this sequence was adapted from a key motif used for tRNA anticodon binding. This scenario effectively coordinates nuclear trafficking of the synthetase with its role for protein synthesis in the cytoplasm.
EXPERIMENTAL PROCEDURES
Confocal Immunofluorescence Microscopy
To detect the subcellular localization of TyrRS, HeLa cells were seeded on glass coverslips, grown for 24 h to 80% confluence, and then fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. After permeabilization with 0.5% Triton X-100 in PBS for 2 min and being blocked with 1% bovine serum albumin for 1 h, cells were first stained with rabbit anti-TyrRS antibody (custom-made, 1:10,000 diluted) for 3 h at room temperature followed by three washes with PBS. Cells were then incubated for 30 min with goat anti-rabbit IgG conjugated to the fluorescent Alexa Fluor 488 dye (1:3000 diluted) followed by three washes with PBS. Lastly, cells were stained with DAPI (1 μg/ml) for 15 min followed by three washes with PBS. The cells were viewed with a Bio-Rad (Zeiss) Radiance 2100 Rainbow laser scanning confocal microscope.
Cell Fractionation
Human TyrRS or mutants were cloned into pcDNA6 vector and transfected into near-confluent adherent HeLa cells. Cells were harvested after incubating overnight at 37 °C. The pooled cells were pelleted at 500 × g for 5 min and resuspended with PBS after removing the medium. The cytoplasmic and nuclear fractions were separated using the NE-PER® nuclear and cytoplasmic extraction kit (Thermo Scientific). TyrRS was detected by Western blot analysis using the custom-made anti-TyrRS antibody.
Mutagenesis
Site-directed mutagenesis of the human TyrRS gene was performed using the method of single mutagenic oligonucleotides and DpnI digestion on the template DNA as described (13).
Protein Expression and Purification
Human TyrRS or mutants were cloned into a bacterial expression vector pET-20b(+) with a C-terminal His6 tag and transformed into Escherichia coli BL21(DE3) cells. The transformed cells were grown in the presence of ampicillin at room temperature and induced with 1 mm isopropyl-1-thio-β-d-galactopyranoside to express the recombinant protein for 5–6 h. The harvested cells were lysed by ultrasonication, and the supernatants of the cell lysates were applied to a nickel-nitrilotriacetic acid (Novagen) column for purification. The purified proteins were stored in 10 mm HEPES, pH 7.5, 10 mm MgCl2, 100 mm KCl, and 2 mm DTT.
Aminoacylation and PPi-ATP Exchange Assays
The assays were performed as described (14). The aminoacylation assay was performed with 100 nm WT or mutant TyrRS proteins in 50 mm HEPES (pH 7.5), 20 mm KCl, 1 μm l-[3H]tyrosine, 19 μm of cold l-tyrosine, and 1 mg/ml total yeast tRNA. The PPi-ATP exchange assay was performed with 1 μm enzyme in 50 mm HEPES (pH 7.5), 20 mm KCl, 0.5 mm tyrosine, 1 mm total NaPPi with a 1:20 ratio of [32P]NaPPi to cold NaPPi. The concentration of the proteins was determined by the Bradford method.
Filter Binding Assay
Human tRNATyr transcript was labeled at the 5′-end with [γ-32P]ATP using the KinaseMaxTM kit (Ambion) and incubated with a concentration gradient of TyrRS (WT or mutant) from 125 nm to 4 μm in 10 mm HEPES, pH 7.5, 4 mm KCl, 0.02 mg/ml BSA, and 0.2 mm DTT for 30 min. The protein-tRNA mixtures were then spotted onto nitrocellulose filters, which were subsequently washed with 60 mm HEPES, pH 7.5, and 10 mm MgCl2 to remove the unbound tRNA. The amount of bound tRNATyr was quantified by using a scintillation counter and plotted to determine the binding constant.
RNA Interference (RNAi) Experiment to Knock Down tRNATyr
HeLa cells were infected with lentiviral particles harboring shRNA targeting the anticodon loop of human tRNATyr(GTA) or a nonspecific control shRNA. The sequence of the shRNA for tRNATyr(GTA) is: 5′-AGGACTGTAGATCCTTAGGTCCTCGAGGACCTAAGGATCTACAGTCCTTTTTT-3′ (the underlining indicates the RNAi targeting sequence), and the nonspecific control shRNA sequence is from RNAi vector pSilencer 2.1 (Ambion): 5′-ACTACCGTTGTTATAGGTGTTCAAGAGACACCTATAACAACGGTAGTTTTTT-3′. Twenty-four h after virus infection, the HeLa cells were harvested to assess tRNATyr (GTA) expression by Northern blot analysis of total RNA extracted using TRIzol reagent (Invitrogen). The probe used for detecting human tRNATyr(GTA) is: 5′-CGAACCAGCGACCTAAGGATCTACAGTCCTCCGCTCT-3′. The harvested cells were also subjected to cell fractionation analysis as described above.
RESULTS
Nuclear Localization of Endogenous Human TyrRS
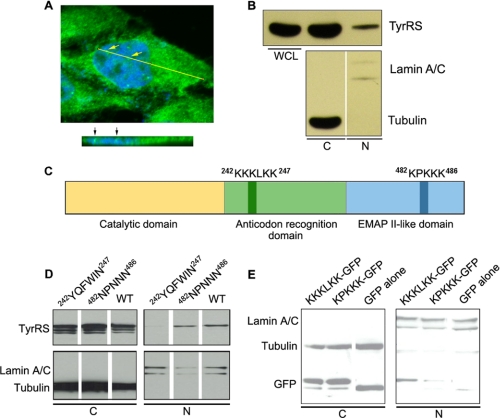
The distribution of human TyrRS in HeLa cells was investigated by immunofluorescence microscopy. Endogenous TyrRS (as detected by α-TyrRS antibodies) was clearly seen in the nucleus (Fig. 1A). To confirm the nuclear localization of TyrRS, HeLa cells were fractionated to separate nuclear and cytoplasmic components. Although the majority of TyrRS was found in the cytoplasmic fraction as expected, a small amount (less than 5%) was also detected in the nuclear fraction (Fig. 1B).
FIGURE 1.
Identification of NLS in human TyrRS. A, confocal immunofluorescence microscopy showing the nuclear distribution of TyrRS in HeLa cells (green, TyrRS; blue, DAPI). The cross section at the yellow line is shown at the bottom. B, HeLa cell fractionation analysis confirming the nuclear distribution of TyrRS. Lamin A/C and tubulin were used as nuclear (N) and cytoplasmic (C) markers, respectively. WCL, whole cell lysates. C, domain structure of human TyrRS and the locations of two potential NLSs. EMAP II, endothelial monocyte-activating protein II. D, mutagenesis and cell fractionation analyses suggesting that KKKLKK is the authentic NLS that directs the nuclear localization of human TyrRS. E, cell fractionation analysis showing that KKKLKK can facilitate the nuclear localization of GFP as a passenger protein.
Identification of NLS
We next attempted to identify the NLS of human TyrRS. The classical NLS is characterized by one or two stretches of basic residues that are surface-exposed and recognized by the acidic binding surfaces of importins (15). Based on an analysis of the primary sequence and of our previously reported high-resolution crystal structures of human TyrRS (16, 17), two potential NLSs, 242KKKLKK247 in the anticodon recognition domain and 482KPKKK486 in the C-terminal endothelial monocyte-activating protein II-like domain, were identified (Fig. 1C).
To determine which motif is the true NLS, we cloned wild type (WT) TyrRS and created two mutant forms: 242YQFWIN247 TyrRS to replace 242KKKLKK247 with the corresponding motif in E. coli TyrRS and 482NPNNN486 TyrRS to replace 482KPKKK486. (TyrRS from bacteria, archaea, and lower eukaryotes does not have endothelial monocyte-activating protein II-like domain.) Each clone was transfected into HeLa cells and analyzed by cell fractionation. Consistent with the previous results with endogenous TyrRS, overexpressed WT TyrRS also distributed to the nucleus (Fig. 1D). In contrast to WT and 482NPNNN486 TyrRS, 242YQFWIN247 TyrRS did not enter the nucleus (Fig. 1D), suggesting that 242KKKLKK247 is the nuclear targeting signal for human TyrRS. To confirm this result, we fused the KKKLKK and, separately, the KPKKK motif to the N terminus of GFP that was chosen as a passenger protein. Cell fractionation analysis showed that KKKLKK, but not KPKKK, facilitated nuclear localization of GTP (Fig. 1E). Thus, the 242KKKLKK247 motif is necessary and sufficient to facilitate nuclear import of human TyrRS.
NLS Is Positioned for tRNA Interaction
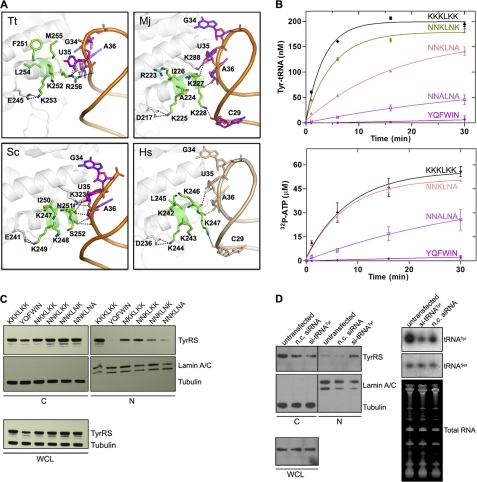
The hexapeptide NLS motif 242KKKLKK247 is located on an α-helix in the anticodon recognition domain of human TyrRS (Fig. 2A). According to the crystal structures (18–20), the corresponding motif in Thermus thermophilus 251FKKLMR256 is responsible for anticodon recognition (Fig. 2A); Arg-256, the last residue of the motif, makes base-specific and backbone interactions with U35 of the GUA anticodon. In Methanococcus jannaschii and S. cerevisiae, the corresponding motifs (223RAKIKK228 and 247KKKINS252) maintain strong interactions, through the last two residues of the hexapeptide motif, to the backbone atoms of the GUA anticodon (Fig. 2A); however, base-specific recognition of U35 was taken over by another helix adjacent to the hexapeptide motif. Nevertheless, in all above cases, this hexapeptide motif is involved in tRNA binding.
FIGURE 2.
KKKLKK is a tRNATyr-binding element. A, the interaction of tRNATyr with the KKKLKK motif in human and with the KKKLKK-corresponding motif in other organisms (Tt, T. thermophilus (PDB 1H3E); Sc, S. cerevisiae (PDB 2DLC); Mj, M. jannaschii (PDB 1J1U); Hs, Homo sapiens (modeled; PDB 1N3L for protein alone)). All structures were drawn in PyMOL (22). B, aminoacylation (left) and PPi-ATP exchange (right) assays dissecting the role of the NLS motif in aminoacylation and tRNA binding. Error bars indicate S.D. C, cell fractionation analysis showing a progressively diminished nuclear (N) localization with progressive replacement of lysine residues within the NLS. C, cytoplasmic. WCL, whole cell lysates. D, cell fractionation analysis showing a significant increase of TyrRS nuclear distribution when the cellular level of tRNATyr was knocked down using an siRNA directed against tRNATyr. In contrast, use of a nonspecific control siRNA (n.c. siRNA) had no effect on nuclear import of TyrRS.
Mutagenic Analysis Confirms Dual Role of NLS
In the absence of a structure for the human TyrRS-tRNATyr complex, to confirm the role of KKKLKK in tRNA binding, we created a series of mutations of the hexapeptide motif. These mutations progressively replaced charged lysines with neutral residues (asparagine or alanine) to give 242NKKLKK247, 242NNKLKK247, 242NNKLNK247, 242NNKLNA247, and 242NNALNA247 TyrRSs (the underlining indicates mutated residue(s)). The mutant enzymes were expressed in E. coli, purified, and used in three different assays. First, tyrosine-dependent PPi-ATP exchange was used to monitor adenylate synthesis in the absence of tRNATyr. Second, the overall aminoacylation activity was determined using tRNATyr as the acceptor. Lastly, a nitrocellulose filter binding assay was used to determine the binding constant with tRNATyr. Progressive replacement of Lys-242, Lys-243, and Lys-246 with Asn only modestly affected the aminoacylation activity, whereas the additional K247A mutation (to give NNKLNA) decreased the initial rate of aminoacylation to less than 50% of the WT protein (Fig. 2B). However, the NNKLNA mutant had WT-like activity in the PPi-ATP exchange reaction (Fig. 2B), suggesting that the side chain of Lys-247 is important for tRNA binding. Consistently, the filter binding assay indicated that although the Kd of NNKLNK TyrRS for tRNATyr (123 nm) is similar to that of WT TyrRS (119 nm), the Kd of NNKLNA TyrRS has increased to 166 nm (Table 1), suggesting a weakened interaction with the cognate tRNA as a result of K247A mutation. Possibly, the positively charged side chain of Lys-247 binds to a negatively charged phosphate of the anticodon stem loop, in a way similar to that of Lys-228 of M. jannaschii TyrRS (Fig. 2A).
TABLE 1.
Dissociation constants of human TyrRS and its mutants with tRNATyr
| 242KKKLKK247 (WT) | NNKLNK | NNKLNA | NNALNA | YQFWIN | |
|---|---|---|---|---|---|
| Kd (nm) | 119 ± 24 | 123 ± 20 | 166 ± 21 | 190 ± 52 | 562 ± 84 |
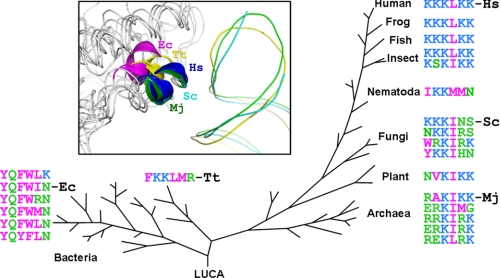
Additional replacement of Lys-244 with Ala (to give NNALNA) further decreased the synthetase-tRNA interaction (Kd = 190 nm; Table 1) and affected both aminoacylation and adenylate synthesis activities (Fig. 2B). It is worth noting that Lys-244 forms a conserved salt bridge interaction with Asp-236 and is the most conserved residue of the hexapeptide motif during evolution (Fig. 3). Most likely, Lys-244 plays a structural role to ensure the correct positioning of the hexapeptide, which, in some way, also affects the active site. Consistently, replacement of the entire hexapeptide from human TyrRS with its counterpart from E. coli TyrRS (YQFWIN) dramatically decreased the interaction with tRNATyr (Kd = 562 nm; Table 1) and completely abolished aminoacylation as well as adenylate synthesis activities (Fig. 2B). Therefore, our mutagenesis and enzymatic analyses suggested a close communication between the active site of the catalytic domain and the site of the NLS motif. However, with regard to tRNA binding, the role of Lys-244 should be indirect.
FIGURE 3.
Evolution of KKKLKK-corresponding motif. The hydrophobic and hydrophilic residues are colored in magenta and green, respectively, while the lysines are colored in blue. Inset, superposition of E. coli (Ec; PDB 2YXN), T. thermophilus (Tt; PDB 1H3E), M. jannaschii (Mj; PDB 1J1U), S. cerevisiae (Sc; PDB 2DLC), and H. sapiens (Hs; PDB 1N3L) TyrRS structures to show the outward conformational shift of the KKKLKK-corresponding motif during evolution.
As for nuclear targeting, each additional replacement of Lys-242, Lys-243, Lys-246, and Lys-247 within the NLS progressively diminished nuclear location of TyrRS (Fig. 2C). This result is consistent with the idea that the charge-charge interactions of TyrRS with importins are dispersed over several contiguous residues within the synthetase.
Dual Role of NLS Provides tRNA-controlled Nuclear Trafficking
We surmised from these results that tRNA binding might block full access of the hexapeptide to the nuclear import machinery. If this were the case, then the level of tRNA inside the cell might regulate the nuclear trafficking of TyrRS. To test this possibility, we tried to knock down expression of tRNATyr by introduction of a specific siRNA directed against tRNATyr. The knockdown of tRNATyr in HeLa cells was confirmed by Northern blot analysis. Remarkably, knockdown of tRNATyr dramatically increased nuclear localization of the endogenous TyrRS in HeLa cells (Fig. 2D). In contrast, use of a nonspecific control siRNA, although causing some diminution in the production of RNA in general, had no effect on nuclear import of TyrRS. Thus, having an NLS and anticodon-binding element overlap makes tRNATyr a regulator of the nuclear trafficking of its cognate synthetase.
DISCUSSION
The dual-function KKKLKK hexapeptide motif first appeared in Drosophila melanogaster and has been stably maintained in evolution ever since (Fig. 3). Thus, from insects to vertebrates and to mammals, the hexapeptide motif is most likely used for tRNATyr binding as well as for targeting nuclear localization of TyrRS. For most tRNA synthetases, anticodon binding is critical for the essential aminoacylation activity, so there is strong selective pressure for maintenance of this structural element. Interestingly, the hexapeptide motif in TyrRS transitions from predominantly hydrophobic in bacteria to predominantly hydrophilic in archaea and eukaryotes (Fig. 3). The transition in sequence is also accompanied by an outward conformational shift of this hexapeptide to become more surface-exposed and presumably more accessible to the nuclear import machinery. In S. cerevisiae, although this motif contains three lysines, they were not sufficient to direct yeast TyrRS to the nucleus (12). Instead, nuclear localization is driven by a separate lysine-rich peptide that is unique to TyrRS in fungi. Mutations that altered the basic amino acids of that NLS affected the nuclear localization of the synthetase, but did not perturb aminoacylation (12). Thus, in contrast to the mammalian system investigated here, the nuclear targeting motif in yeast is segregated from key elements for aminoacylation.
To our knowledge, this is the first report of nuclear localization of a tRNA synthetase being regulated by its cognate tRNA. In Xenopus laevis, where aminoacylated tRNATyr was found in the nucleus (5), the nuclear presence of TyrRS was strongly suggested. The aminoacylated tRNA was exported more efficiently than was uncharged tRNA, so that the preference for the charged tRNA serves as a proofreading mechanism to ensure that mature and functional tRNAs are exported into the cytoplasm for protein synthesis. In a different vein, from yeast to vertebrates, retrograde trafficking of tRNA from the cytoplasm to the nucleus occurs under stress conditions such as amino acid deprivation (21). This retrograde tRNA trafficking is thought to down-regulate gene expression through general inhibition of translation under conditions that are unfavorable for protein synthesis. This kind of response would presumably deprive tRNATyr from the cytoplasm and, according to the results here, would stimulate nuclear localization of TyrRS. In this scenario, the nuclear accumulated TyrRS should not only serve for proofreading tRNATyr, but could also affect other to be determined functions in the nucleus. These functions are perhaps related to a physiological response to stress conditions that cut back protein synthesis. However, most importantly, the novel design reported here, which makes tRNA a regulator of the nuclear trafficking of its cognate synthetase, still prioritizes the demands of protein synthesis in the cytoplasm.
Acknowledgment
We thank Professor Paul Schimmel for valuable scientific insight and help on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 88278 (to X.-L. Y.). This work was also supported by the Skaggs Foundation and a fellowship from the National Foundation for Cancer Research.
This article was selected as a Paper of the Week.
- TyrRS
- tyrosyl-tRNA synthetase
- NLS
- nuclear localization signal.
REFERENCES
- 1. Carter C. W., Jr. (1993) Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 [DOI] [PubMed] [Google Scholar]
- 2. Giegé R. (2006) The early history of tRNA recognition by aminoacyl-tRNA synthetases. J. Biosci. 31, 477–488 [DOI] [PubMed] [Google Scholar]
- 3. Nathanson L., Deutscher M. P. (2000) Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 275, 31559–31562 [DOI] [PubMed] [Google Scholar]
- 4. Popenko V. I., Ivanova J. L., Cherny N. E., Filonenko V. V., Beresten S. F., Wolfson A. D., Kisselev L. L. (1994) Compartmentalization of certain components of the protein synthesis apparatus in mammalian cells. Eur. J. Cell Biol. 65, 60–69 [PubMed] [Google Scholar]
- 5. Lund E., Dahlberg J. E. (1998) Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282, 2082–2085 [DOI] [PubMed] [Google Scholar]
- 6. Sarkar S., Azad A. K., Hopper A. K. (1999) Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 96, 14366–14371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grosshans H., Hurt E., Simos G. (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 14, 830–840 [PMC free article] [PubMed] [Google Scholar]
- 8. Yannay-Cohen N., Carmi-Levy I., Kay G., Yang C. M., Han J. M., Kemeny D. M., Kim S., Nechushtan H., Razin E. (2009) LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol. Cell 34, 603–611 [DOI] [PubMed] [Google Scholar]
- 9. Ko Y. G., Kang Y. S., Kim E. K., Park S. G., Kim S. (2000) Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 149, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wakasugi K., Schimmel P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 [DOI] [PubMed] [Google Scholar]
- 11. Jordanova A., Irobi J., Thomas F. P., Van Dijck P., Meerschaert K., Dewil M., Dierick I., Jacobs A., De Vriendt E., Guergueltcheva V., Rao C. V., Tournev I., Gondim F. A., D'Hooghe M., Van Gerwen V., Callaerts P., Van Den Bosch L., Timmermans J. P., Robberecht W., Gettemans J., Thevelein J. M., De Jonghe P., Kremensky I., Timmerman V. (2006) Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat. Genet. 38, 197–202 [DOI] [PubMed] [Google Scholar]
- 12. Azad A. K., Stanford D. R., Sarkar S., Hopper A. K. (2001) Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell 12, 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shenoy A. R., Visweswariah S. S. (2003) Site-directed mutagenesis using a single mutagenic oligonucleotide and DpnI digestion of template DNA. Anal. Biochem. 319, 335–336 [DOI] [PubMed] [Google Scholar]
- 14. Beebe K., Waas W., Druzina Z., Guo M., Schimmel P. (2007) A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal. Biochem. 368, 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 16. Yang X. L., Skene R. J., McRee D. E., Schimmel P. (2002) Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc. Natl. Acad. Sci. U.S.A. 99, 15369–15374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X. L., Liu F. M., Skene R. J., McRee D. E., Schimmel P. (2003) Crystal structure of an EMAP-II-like cytokine released from a human tRNA synthetase. Helvetica Chimica Acta 86, 1246–1257 [Google Scholar]
- 18. Tsunoda M., Kusakabe Y., Tanaka N., Ohno S., Nakamura M., Senda T., Moriguchi T., Asai N., Sekine M., Yokogawa T., Nishikawa K., Nakamura K. T. (2007) Structural basis for recognition of cognate tRNA by tyrosyl-tRNA synthetase from three kingdoms. Nucleic Acids Res. 35, 4289–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobayashi T., Nureki O., Ishitani R., Yaremchuk A., Tukalo M., Cusack S., Sakamoto K., Yokoyama S. (2003) Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 10, 425–432 [DOI] [PubMed] [Google Scholar]
- 20. Yaremchuk A., Kriklivyi I., Tukalo M., Cusack S. (2002) Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 21, 3829–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hopper A. K., Shaheen H. H. (2008) A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol. 18, 98–104 [DOI] [PubMed] [Google Scholar]
- 22. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 0.99rc6, Schrödinger, LLC, New York [Google Scholar]