Background: The oxygen-dependent asparaginyl hydroxylase FIH has two classes of substrate, HIF and ARD proteins.
Results: Substrate recognition by FIH is mediated by structural context and specific amino acids proximal and distal to the target asparagine.
Conclusion: Differences in sequence and structure explain distinct kinetic properties of HIF and ARD substrates.
Significance: These data demonstrate how FIH substrate selection can be achieved in vivo.
Keywords: Gene Regulation, Hydroxylase, Hypoxia, Hypoxia-inducible Factor (HIF), Protein Chemical Modification, Ankyrin Repeats, Factor Inhibiting HIF (FIH)
Abstract
Factor Inhibiting HIF (FIH) catalyzes the β-hydroxylation of asparagine residues in HIF-α transcription factors as well as ankyrin repeat domain (ARD) proteins such as Notch and Gankyrin. Although FIH-mediated hydroxylation of HIF-α is well characterized, ARDs were only recently identified as substrates, and less is known about their recognition and hydroxylation by FIH. We investigated the molecular determinants of FIH substrate recognition, with a focus on differences between HIF and ARD substrates. We show that for ARD proteins, structural context is an important determinant of FIH-recognition, but analyses of chimeric substrate proteins indicate that the ankyrin fold alone is not sufficient to explain the distinct substrate properties of the ARDs compared with HIF. For both substrates the kinetic parameters of hydroxylation are influenced by the amino acids proximal to the target asparagine. Although FIH tolerates a variety of chemically disparate residues proximal to the asparagine, we demonstrate that certain combinations of amino acids are not permissive to hydroxylation. Finally, we characterize a conserved RLL motif in HIF and demonstrate that it mediates a high affinity interaction with FIH in the presence of cell lysate or macromolecular crowding agents. Collectively, our data highlight the importance of residues proximal to the asparagine in determining hydroxylation, and identify additional substrate-specific elements that contribute to distinct properties of HIF and ARD proteins as substrates for FIH. These distinct features are likely to influence FIH substrate choice in vivo and, therefore, have important consequences for HIF regulation.
Introduction
Factor Inhibiting HIF (FIH)4 is a 2-oxoglutarate and Fe(II)-dependent dioxygenase that catalyzes post-translational hydroxylation of protein substrates (1, 2). As its name suggests, FIH was first identified as a negative regulator of the hypoxia-inducible factors (HIFs), key mediators of the transcriptional response to hypoxia (3). FIH catalyzes the hydroxylation of a single conserved asparagine (Asn) residue in the C-terminal transactivation domain (CAD) of HIF-α, a modification that blocks the recruitment of p300/CBP (CREB-binding protein)coactivators and renders the CAD transcriptionally inactive in normoxia (1, 2). The hydroxylase activity of FIH is regulated by the availability of oxygen; thus, it can function as a direct oxygen sensor and confer oxygen sensitivity to the HIF pathway (1, 4).
In addition to HIF-α, intracellular proteins containing ankyrin repeat domains (ARDs) are common targets for hydroxylation by FIH (for review, see Ref. 5). ARDs contain tandem repeats of an ankyrin structural motif made up of ∼33 amino acids arranged in a helix-turn-helix conformation, with a β-hairpin-like loop that connects adjacent repeats (6). The Asn residues targeted by FIH are located within the β-hairpin loop between repeats and are semiconserved in the ankyrin repeat consensus sequence (7). Despite the identification of numerous ARD substrates for FIH (8–14), the functional significance of ARD hydroxylation remains unclear. Several studies have demonstrated that the hydroxy-asparagine stabilizes localized regions of the ankyrin fold (14–16). However, this is not the case for all ARD substrates of FIH (17). The ability of ARD proteins to compete with HIF for hydroxylation by FIH has led to a model in which ARD proteins act in concert to regulate the activity of the HIF CAD through sequestration of FIH (10). In this scenario the recognition of each substrate and their relative affinity for FIH is an important determinant of FIH sequestration and consequently HIF regulation.
Proteomic analyses suggest that ARD hydroxylation by FIH is widespread (9) and, with more than 300 ARD proteins in the human proteome, a key issue is how FIH achieves specificity. FIH clearly displays strong preferences for specific target residues within protein substrates. However, its substrate consensus sequence is largely degenerate, and no single amino acid is completely conserved in all substrates (Fig. 1). Recent studies have shown that FIH can catalyze β-hydroxylation of aspartyl and histidinyl residues in ARDs (13, 14), indicating that even the Asn itself is not an absolute requirement. Interestingly, FIH is able to bind ARDs that are not substrates (15, 18), suggesting that distinct residues may mediate binding versus catalysis.
FIGURE 1.
The degenerate substrate consensus sequence for FIH. Sequence alignment of FIH-hydroxylation sites from selected human (h) and mouse (m) substrates is shown. The # symbol indicates the hydroxylated residue, and the numbers to the right specify the position in the amino acid sequence. Highlighted in gray are key residues that form the consensus sequence for hydroxylation by FIH (5). The consensus sequence for ankyrin repeats is included for comparison (7) and shows that key residues required for hydroxylation by FIH are conserved in ankyrin repeats. The φ symbol represents hydrophobic amino acids.
The importance of tertiary structure in substrate recognition by FIH is also unclear. Crystallographic analyses of ankyrin and HIF substrate peptides bound to FIH reveal very similar extended conformations (10, 19). However, the characteristic fold of a complete ARD is structurally distinct from the HIF CAD, which is intrinsically disordered in solution (20, 21). Such a difference suggests first, that considerable structural rearrangement of both HIF and ARD is required for hydroxylation, and second, that a fully folded ARD may be recognized by FIH in a manner quite different from peptide fragments or, indeed, the HIF CAD. In support of this, kinetic analyses indicate that FIH has a higher affinity for the ARD of Notch1 compared with the HIF CAD, with Km values of 0.2 and 50 μm, respectively (18). These values likely reflect differences in both secondary and tertiary structure as well as differences in the amino acid sequence proximal to the target Asn; however, the relative contribution of each of these factors remains to be determined.
In this study we present an extensive biochemical characterization of the molecular determinants of substrate recognition by FIH, with a specific focus on key differences between ARD and HIF substrates. We show that the distinct kinetic properties of these substrates are largely determined by the amino acid sequence proximal to the target Asn, although the structural context of ARD substrates is an important determinant of binding and may, therefore, influence substrate choice in vivo. We also provide novel insights into the FIH-HIF interaction and characterize an RLL motif that mediates a high affinity interaction with FIH in the presence of cell lysate or macromolecular crowding agents. Collectively, we reveal that FIH substrate interactions are more complex than currently appreciated from available structures and make significant contributions to FIH specificity with regard to the HIF and ARD substrates and HIF regulation.
EXPERIMENTAL PROCEDURES
Plasmids are described in supplemental Methods.
Protein Expression and Purification
MBP-FIH and all thioredoxin (Trx)-His6 (6H)-tagged proteins were expressed in BL21 (DE3) Escherichia coli (E. coli) and purified by amylose and nickel (Ni2+) affinity chromatography, respectively (22). 6H-tagged proteins for circular dichroism analysis were subjected to an additional wash before elution from the Ni2+ resin in 150 mm NaCl, 20 mm Tris, pH 8.0, 50 mm imidazole and after elution were dialyzed overnight into 5 mm sodium phosphate, pH 8.0. GST-hHIF-1α proteins were induced with 0.5 mm IPTG at 37 °C for 5 h, and purified by standard glutathione-affinity chromatography methods. All proteins were buffer-exchanged using a Sephadex PD-10 column (GE Healthcare) and stored at 4 °C in 150 mm NaCl, 20 mm Tris, pH 8.0. To generate untagged FIH, affinity-purified Trx-6H-TEV-hFIH was cleaved with 6H-tagged Ac-TEV protease (Invitrogen) according to the manufacturer's directions, after which the Trx-6H tag and Ac-TEV protease were removed by standard Ni2+-affinity chromatography. Protein concentrations were determined using calculated extinction coefficients and measured absorbance at 280 nm, and purity (generally ≥90%) was assessed by SDS-PAGE with Coomassie staining.
Peptides
mNotch1 Site1 short (1930–1949) and long (1930–1963) peptides have been described previously (18).
Antibodies
Anti-FIH immunoblots were performed as described (22) using polyclonal antibodies from Santa Cruz (sc26219) at 1:500 or Novus (NB100–428) at 1:1000. HRP-conjugated secondary antibodies were obtained from Thermo Scientific.
Reporter Assays
Dual luciferase reporter assays were performed as described previously (23) using HEK-293T cells, wild type or FIH −/− mouse embryonic fibroblast (MEF) cells (24).
CO2 Capture Assays
Assays were performed as described previously (22) with a saturating amount of FIH and cofactors at final concentrations of 40 μm [1-14C]2-oxoglutarate, 1 mm ascorbate, and 100 μm FeSO4. For kinetic experiments, the concentration of FIH was optimized to ensure that activity was linear over the assay time and that less than 40% of substrate was consumed. The data were converted to Vo (nmol 2-oxoglutarate turnover/mg FIH/min), and Graphpad PRISM software was employed to fit data to a hyperbolic curve and estimate kinetic constants (apparent Km and Vmax).
In Vitro Pulldown Assays
Recombinant Trx-6H- or GST-tagged proteins were purified by Ni2+- or glutathione affinity chromatography, respectively, retained on the resin, and normalized for protein concentration (as assessed by SDS/PAGE and Coomassie staining) by adding unbound resin. Each pulldown contained 20 μl of bait resin (with equivalent molar amounts of bound protein) together with purified untagged FIH and was performed as previously described (22) in 300 μl of whole cell extract buffer (WCEB) alone, WCEB with 5% bovine serum albumin (to match protein concentration of lysate), or 300 μl of lysate from FIH−/− MEFs in WCEB. 30 μl of loading buffer was used to elute interacting proteins (and bait proteins), and 15 μl of each eluent was analyzed by SDS/PAGE and Western blotting as described (22). For pulldown assays utilizing crowding agents, whole cell extract buffer was supplemented with 26% BSA or 14% PEG (MW 20 000 g/mol), and an additional washing step was employed (4 washes in total).
Fluorescence Polarization
Fluorescence polarization (FP) competition binding assays were performed as described previously (18). Graphpad PRISM software was used to perform nonlinear regression analysis of the data and determine Ki values for the competing substrate proteins.
Molecular Modeling
The x-ray crystal structure of Notch1 (1936–1945) complexed with FIH (10) was used as the basis for modeling Notch-1 (1936–1951). The backbone conformation of residues 1946–1951 was modeled using the corresponding residues from the crystal structure of HIF-1α CAD in complex with FIH (PDB ID 1H2K) (19), and Notch-1 side chains were constructed using SCWRL4 (25) and combined with those from the crystal structure. Notch4 and its mutants as well as mutations of Notch1 were modeled by side-chain replacement on this backbone conformation using SCWRL4. Missing heavy atoms from the FIH chain were added using the DockPrep utility of UCSF Chimera (26). Hydrogen atom coordinates were calculated, and peptide force field parameters were prepared using XPLOR-NIH v2.25 (27) and the CHARMM force field (28). Force field parameters for Fe(II) and 2-oxoglutarate were used as described previously (23). Molecular dynamics were carried out using NAMD v2.6 (29). Initially, 30,000 energy minimization steps were performed on reconstructed residues and hydrogen atoms. The structure was solvated using SOLVATE v1.3 as employed in VMD v1.8.7b5 (30), and an 18 Å radius around the hydroxylated Asn residue was used in 1-ns molecular dynamics simulations with a time step of 1 fs. Simulations were carried out at 300 K with a dielectric constant of 1. A switch function on van der Waals forces was applied to interactions exceeding 8 Å, and truncation occurred at 12 Å. Reassignment of the non-bonded interactions was done every 20 time steps.
Circular Dichroism
CD spectra were recorded on a JASCO J-815 CD spectrometer. Measurements were carried out at 20 °C in a 0.1-cm path length quartz cuvette, with protein concentrations ranging from 0.05 to 0.4 mg/ml in 5 mm sodium phosphate, pH 8.0. Spectra were recorded in the wavelength range of 300 to 185 nm at 0.2-nm intervals, and each spectrum was an average of 5 scans. Spectra were base-line-corrected by subtraction of the spectrum for buffer alone and smoothed using the “means-movement” smoothing method in the Spectra Manager software. Data were expressed as the mean residue ellipticity ([θ], deg cm2 dmol−1) and in some cases normalized to the ellipticity measured at 207 nm to minimize interference from small differences in protein concentration (31). The experimental data in the 190–260 nm range were analyzed using DICHROWEB (32), and the CDSSTR deconvolution method was used to estimate the secondary structural content using reference set 7 (33). For each protein, a minimum of three scans was performed from at least two independent protein preparations. Thermal denaturation experiments were performed at protein concentrations of 0.2 mg/ml in 5 mm sodium phosphate buffer, pH 8.0. The ellipticity at 220 nm (θ220) was monitored continuously as the temperature increased from 4 to 90 °C at a ramp rate of 1 °C/min. Data were base-line-corrected and expressed as a percentage of the θ220 value measured at 4 °C. Graphpad PRISM software was employed to fit data to a sigmoidal curve and estimate apparent Tm values, as denaturation was irreversible.
RESULTS
Folded ARD Can Bind FIH
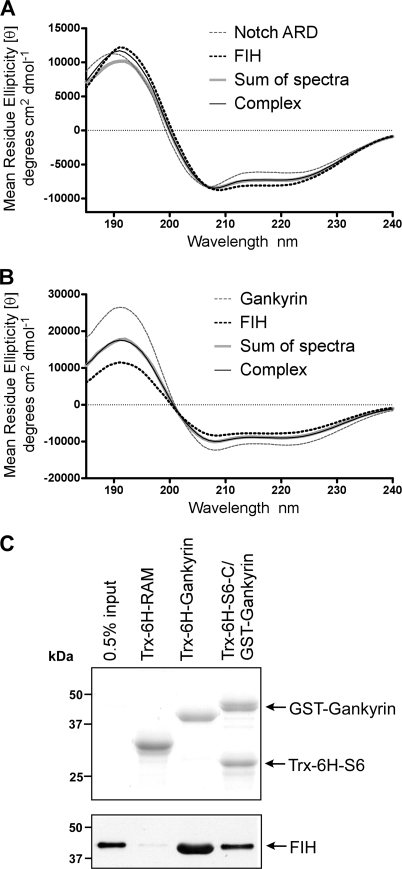
Gankyrin and Notch1 were used as representative ARD substrates to investigate the importance of secondary and tertiary structure in ARD recognition by FIH. Although both proteins contain seven ankyrin repeats, the ARD of Notch1 contains two Asn residues (Asn-1945/2012) that are targeted for hydroxylation by FIH (10, 34), whereas a single Asn residue in Gankyrin (Asn-100) is hydroxylated (Ref. 8; supplemental Fig. S1). Previous crystallographic studies with Notch peptides in complex with FIH suggest that the ARD must unfold to some extent to enable hydroxylation (10). However, it is not clear whether FIH can bind a folded ARD (regardless of hydroxylation) and likewise whether a structural rearrangement would involve a global unfolding of the ARD or localized unfolding of the repeats adjacent to the hydroxylation site. To determine the type and extent of the conformational changes that occur upon complex formation, far-UV circular dichroism (CD) was used to monitor major changes in the secondary structure of the Notch1 ARD and FIH upon complex formation (35).
The individual spectra for purified FIH and the Notch1 ARD are typical of structured proteins with a combination of α-helical and β-sheet/β-turn content (Fig. 2A, supplemental Table S1), consistent with the published structures for these proteins (19, 36–38). Although there are two binding sites for FIH in Notch1 ARD, Site 1 and Site 2 have different affinities with Km values of 0.7 and 13 μm, respectively (18). Therefore, FIH and the Notch1 ARD were combined in equimolar amounts (5.5 μm), and the spectrum of the resultant complex was obtained. Note that at this concentration the binding kinetics are consistent with a single high affinity interaction with Site 1 (18). The spectrum of the FIH-Notch complex overlays very well with the sum of the individual spectra for Notch and FIH, with a slight deviation in ellipticity at lower wavelengths (Fig. 2A). Similar results were obtained with a 2-fold molar excess of either FIH or Notch1 ARD (data not shown) and also for FIH and Gankyrin (Fig. 2B). The similarity between the sum of the individual spectra and the spectra of the complexes indicates that only minor structural changes occur to the ARD proteins or FIH upon binding.
FIGURE 2.
The importance of ankyrin structure in FIH recognition. A and B, shown are far-UV CD spectra of 6H mouse FIH in complex with 6H mouse Notch1 ARD (A) or 6H human Gankyrin (B). In both cases, the “complex” spectrum was measured for a 1:1 molar mixture of FIH and substrate and is superimposed for comparison with the sum of the spectra of the isolated components. The individual spectra for 6H-FIH, 6H-Notch, and 6H-Gankyrin are also presented. The concentration of each protein was 5.5 μm in 5 mm sodium phosphate, pH 8.0. Data are representative of three independent experiments. C, affinity pulldown assays in which Trx-6H-tagged bait proteins were retained on Ni2+-resin after purification and incubated with recombinant FIH. Captured complexes were analyzed by SDS-PAGE and immunoblotting with an anti-FIH antibody (Santa Cruz; lower panel) or Coomassie staining (upper panel) to visualize bait proteins. A representative pulldown assay of >3 independent experiments is shown. For the Gankyrin-S6 complex, GST-Gankyrin was co-purified with Trx-6H-S6-C. The RAM domain of Notch, which does not interact with FIH and is not hydroxylated (34), was used as a negative control. The input lane contains 0.5% of the total amount of FIH incubated with the bait resin.
To further investigate the involvement of changes to the tertiary structure in FIH-ARD binding, we exploited the high affinity interaction between Gankyrin and the S6 ATPase of the 26 S proteasome (39). The C-terminal domain of S6 (S6-C) binds to Gankyrin across the concave surface formed by the β-hairpin turns of all seven ankyrin repeats, forming a stable complex and thus constraining Gankyrin in a rigid ankyrin fold (40). We co-purified Trx-6H-S6-C bound to GST-tagged Gankyrin via Ni2+ affinity chromatography. This allowed us to select for the 1:1 S6-C-Gankyrin complexes, as Trx-6H-S6-C is insoluble in the absence of Gankyrin, and unbound GST-Gankyrin is not purified because it lacks a 6H tag. In vitro affinity pulldown assays were performed using Trx-6H-tagged bait proteins bound to Ni2+-resin (Fig. 2C, lower panel) to capture recombinant FIH. Trx-6H-Gankyrin and the RAM (RBP-Jκ-associated molecule) domain of Notch1, which we have previously shown does not interact with FIH (34), were also included. FIH was captured by both the S6-C-Gankyrin complex and Trx-6H-Gankyrin but not the RAM domain.
Together with data from the CD experiments, these results demonstrate that FIH can indeed bind to a fully folded and structurally constrained ARD. However, previous studies suggest that a constrained ARD fold is not favorable for hydroxylation by FIH (16). Thus, our data support a model in which a structurally constrained ARD can bind to FIH but must unfold to some extent to enable hydroxylation.
Structural Determinants of Recognition by FIH Differ for ARD and HIF Substrates
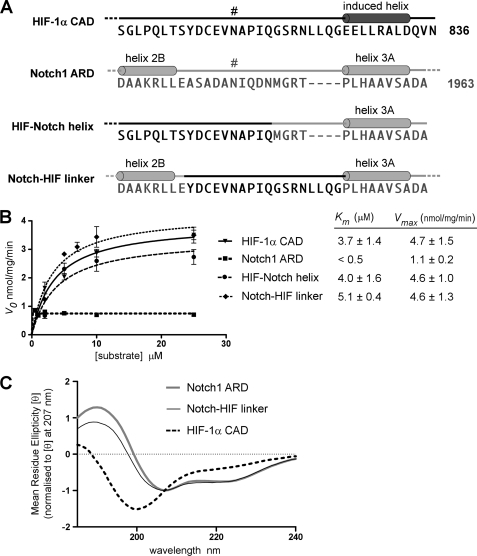
Substrate recognition by FIH is likely to be influenced by a combination of primary, secondary, and tertiary structure and may differ between classes of substrate. We have previously reported that hydroxylation of Asn-1945 in Notch1 (site 1) is influenced by the length of the peptide substrate, and kinetic analyses point to a short region proximal to the Asn as a key determinant of binding affinity (18). Specifically, FIH has a considerably higher affinity (>100 fold) for a Notch1 peptide that includes 15 residues (1950–1963) C-terminal to the hydroxylation site compared with a peptide that terminates shortly after the hydroxylation site. This extended C-terminal region is likely to interact directly with FIH, although there is no direct evidence for this interaction, as the published crystal structure of the FIH-Notch complex involves shorter Notch peptides lacking these residues (10). Although this region forms an α-helix (helix 3A) in the context of the Notch1 ARD, secondary structure is unlikely to contribute to the affinity difference, as Notch peptides with or without the “helix 3A” residues are disordered (supplemental Fig. S2). Interestingly, the equivalent residues in HIF, relative to Asn-803, form an induced α-helix when bound directly to FIH but do not make a major contribution to the affinity of FIH for HIF (18, 19).
Thus, we sought to determine whether the relatively high affinity of FIH for ARD substrates such as Notch, compared with a lower affinity for HIF, was due in part to this C-terminal helix and whether Notch helix 3A could render the HIF CAD a higher affinity substrate for FIH. To this end, the last 18 amino acids of the HIF-1α CAD containing the induced helix were replaced with the C-terminal helix 3A sequence from Notch1 (Fig. 3A). The chimeric HIF CAD/Notch helix 3A protein (HIF-Notch helix) was analyzed by CO2 capture assay and displayed an apparent Km value comparable with that obtained for the wild type CAD, indicating that the addition of this helix does not significantly alter binding or hydroxylation of HIF (Fig. 3B). Although the affinity of FIH for Notch peptides is strongly influenced by this C-terminal helix, these data indicate that the helix itself is not sufficient to alter the affinity of the interaction with HIF. Thus, the overall affinity of FIH for HIF may be largely dictated by the strength of contacts made at the hydroxylation site itself and/or by N-terminal-positioned residues, as opposed to the more C-terminal contact site.
FIGURE 3.
Structural determinants of FIH substrate recognition. A, shown are amino acid sequences of HIF/Notch wild type and constructed chimeric proteins in the region containing the hydroxylated asparagine residues, Asn-813 in mHIF-1α and Asn-1945 (Site 1) in mNotch1 (indicated by #). The C-terminal helix that is induced in HIF-1α upon binding to FIH (10) and helices 2B and 3A of the Notch1 ARD (37) are shown. The HIF-Notch helix chimera was generated by replacing the C-terminal-induced helix (amino acids 819–836) of the mHIF-1a CAD with helix 3A from mNotch1 (amino acids 1950–1963), maintaining the spacing of Notch between the target Asn and helix 3A. The Notch-HIF linker chimera was made by replacing the β-hairpin turn between repeats 2 and 3 of mNotch1 ARD with a linker region (amino acids 808–825) from mHIF-1α containing the FIH recognition site. B, Trx-6H-tagged wild type and chimeric proteins described in A were analyzed in CO2 capture assays at concentrations ranging from 0.5 to 25.0 μm. Reactions were incubated for 22 min at 37 °C and contained 120 nm MBP-mouse FIH and saturating concentrations of cofactors. Data are expressed as Vo (nmol/min/mg FIH), and Km values were calculated using Graphpad PRISM software. A representative curve (mean of triplicates ± S.E.) is shown for each protein, and Km values are the mean of three independent experiments ± S.D. C, Far-UV CD spectra were determined for 6H-mNotch1 ARD, 6H-Notch-HIF-linker, and untagged hHIF-1α CAD. The concentration of each protein was 0.2 mg/ml in 5 mm sodium phosphate pH 8.0. Data were converted to mean residue ellipticity and normalized to the ellipticity at 207 nm (31) and are representative of three independent scans.
Consistent with previous kinetic analyses of HIF and Notch substrates, the Km of FIH for the Notch1 ARD is considerably lower than that measured for HIF-1α (5, 18). Because the HIF CAD lacks any discernable structure before binding FIH, this difference in affinity may reflect a preference for ankyrin repeat structure in substrate recognition. Although an ARD must unfold to enable hydroxylation, an intact ARD is still recognized by FIH and is, therefore, likely to contribute to the kinetic parameters of hydroxylation. To investigate whether the lack of an ankyrin fold for HIF was responsible for its relatively low affinity, we sought to analyze hydroxylation of HIF within the structural context of an ARD. A second chimeric protein, Notch-HIF linker (Fig. 3A), was generated by inserting the hydroxylation site from mHIF-1α as a linker between repeats 2 and 3 of the Notch1 ARD in place of the β-hairpin loop containing the Site 1 Asn in Notch. To ensure that the HIF linker was the only possible site of hydroxylation by FIH, the Site 2 Asn in Notch (Asn-2012) was mutated to a Gln residue. Conservative mutation of both target Asn residues in Notch1 was found to prevent hydroxylation by FIH without disrupting the structure of the Notch ARD (supplemental Fig. S3, A and B). The activity of FIH with each of the Asn-Gln point mutants was consistent with previous analyses of Asn-Ala mutants of Notch1, with Site 1 being favored over Site 2 (10, 34). Far-UV CD spectroscopy indicated that the Notch-HIF-linker chimera was similar in structure to the Notch1 ARD (Fig. 3C). Differences in these spectra likely reflect an inability of the HIF linker to form a β-turn between repeats 2 and 3, which may in turn alter the stacking of adjacent repeats in the ARD. Nonetheless, the spectrum is clearly distinct from that of the HIF-1α CAD, which is typical of a disordered protein, as indicated by the large negative ellipticity at 198 nm. Despite its pseudo-ankyrin conformation, the Notch-HIF linker chimera displayed very similar kinetic parameters to the wild type HIF-1α CAD, with significantly higher apparent Km and Vmax values than those obtained for the Notch1 ARD (Fig. 3B).
Given that a constrained ARD is non-permissive to hydroxylation (16), our pseudo-ankyrin chimera must be capable of unfolding, after which it apparently permits, but does not significantly influence, HIF hydroxylation kinetics. Thus, in this context the secondary/tertiary structure of HIF has no major influence on the efficiency of hydroxylation by FIH. This further supports the hypothesis that the sequence of amino acids more proximal or more N-terminal to the Asn may be of particular importance in hydroxylation of HIF substrates, in contrast with ARD recognition, which is more strongly influenced by structural context and residues C-terminal to the Asn.
Amino Acids Proximal to Target Asparagine Are Critical Determinants of Hydroxylation
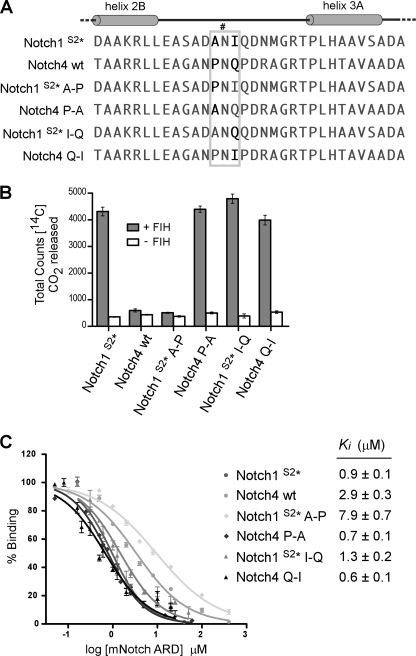
Having clarified the importance of structure in ARD recognition and hydroxylation by FIH, we sought to determine the contribution of residues proximal to the target Asn in ARD hydroxylation efficiency. We predicted that proximal residue identity could have a strong impact because the ARD of Notch4 is structurally analogous to other ARD substrates and is able to bind FIH, yet it is not hydroxylated by FIH in vitro (18, 34). This is of particular interest given the presence of an Asn residue (Asn-1656) in an equivalent position to the Asn that is hydroxylated at Site 1 in the other Notch ARD proteins. Notch4, therefore, provides a useful tool for identifying the residues that are required for hydroxylation of ARD proteins as distinct from those that are required for binding, although these may overlap.
Two obvious differences between Notch4 and Notch1 are the amino acids immediately adjacent to the Asn, with Pro (−1) and Gln (+1) in Notch4 compared with Ala and Ile in Notch1 (Fig. 4A). To investigate their contribution to binding and hydroxylation, these amino acids in Notch4 were mutated to those at the corresponding positions in Notch1 (Pro-1655A and Gln-1657I). The reciprocal mutations were made in Notch1 (A1944P and I1946Q) along with substitution of the Site 2 Asn to Gln (Notch1S2*) to prevent interference from hydroxylation at this second site (supplemental Fig. S3).
FIGURE 4.
Residues directly adjacent to the Asn in Notch influence hydroxylation and binding by FIH. A, shown is sequence alignment of hydroxylation Site 1, containing Asn-1945 (indicated by #), in mNotch1 and the corresponding region from the ARD of mNotch4, containing Asn-1656. Mutagenesis of the Notch1 and Notch4 ARDs was performed to generate Notch1S2* (N2012Q), Notch1S2* A-P (A1944P/N2012Q), Notch1S2* I-Q (I1946Q/N2012Q), Notch4 P-A (P1655A), and Notch4 Q-I (Q1657I). The gray box highlights the region in which the mutagenesis was performed. B, proteins described in A were tested as substrates for FIH in CO2 capture assays, with 50 μm concentrations of each ARD protein and a saturating amount (500 nm) of recombinant MBP-mouse FIH. Data are the mean of triplicate reactions ± S.D. and are representative of >3 independent experiments. C, affinity-purified Trx-6H-tagged Notch ARD proteins from B were assayed for their ability to compete with a fluorescently labeled Notch peptide in FP competition binding experiments. Ki values were determined using Graphpad PRISM software. Data are the average of three independent experiments ± S.E.
All proteins were analyzed by far-UV CD spectroscopy and gave very similar CD profiles, confirming that the point mutations did not significantly disrupt the structure of the ARD proteins (supplemental Fig. S3C). Most notably, mutation of the −1 Pro in Notch4 to Ala (Notch4 P-A) was sufficient to enable efficient hydroxylation of Notch4 in vitro, as inferred by CO2 capture assays (Fig. 4B). Likewise, insertion of a Pro in this −1 position in Notch1 (Notch1 A-P) abolished activity, suggesting that a prolyl residue may not be tolerated in this position by FIH. Notch1 hydroxylation was largely unaffected by mutation of the +1 Ile to Gln (Notch1 I-Q). However, the reciprocal mutation in Notch4 (Notch4 Q-I) also rendered it a substrate for FIH despite the presence of a Pro at the −1 position.
A quantitative FP-based binding assay was used to determine the relative affinities for each of the ARD proteins for FIH (18). The Notch1 or Notch4 proteins that were substrates for FIH clearly demonstrated a higher binding affinity than those that were not hydroxylated (Fig. 4C). These data reveal that the amino acids directly adjacent to the target Asn residue contribute to binding by FIH and are critical determinants of catalysis, such that single substitutions are capable of either preventing or enabling hydroxylation by FIH.
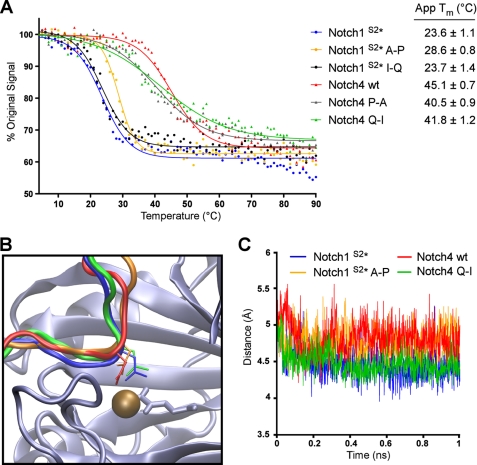
Biophysical characterization of consensus ARD proteins has shown that thermodynamic stability is a factor in determining whether an ARD protein will be hydroxylated by FIH (15), with stability of individual repeats likely to be of particular importance. Thus, we hypothesized that the point mutations in Notch1 and Notch4 may influence hydroxylation by altering the stability of the ARD. CD spectroscopy was employed to monitor temperature-induced changes in ellipticity at 220 nm and determine the apparent Tm for each protein (Fig. 5A). Comparison of the apparent Tm values with data from CO2 capture assays showed a striking correlation between the stability of an ARD and its ability to be hydroxylated by FIH. The Notch1 A-P mutation, which prevents hydroxylation by FIH, increased the thermal stability of the Notch1 ARD by 5 °C, whereas the Notch1 I-Q mutation had little influence on either stability or hydroxylation. It is important to note that it is not the absolute Tm that is important but rather the change in Tm, as the point mutations are likely to influence the thermal stability of a single repeat within the context of an ARD containing 7 repeats. So whereas the Tm for Notch1 A-P remains closer to the Tm of wild type Notch1 than Notch4, the change compared with Notch1 is likely to be significant. Likewise, the apparent Tm values for the Notch4 P-A and Q-I mutants were lower than the value determined for the wild type Notch4 protein, indicating that these mutations have a small destabilizing effect on the Notch4 ARD.
FIGURE 5.
ARD hydroxylation is influenced by thermodynamic stability and positioning of the target Asn. A, CD spectroscopy was employed to analyze the thermal denaturation of Trx-6H-tagged Notch wild type and mutant ARD proteins (described in Fig. 4A). The ellipticity at 220 nm (θ220) was monitored continuously as the temperature increased from 4 to 90 °C. Data are expressed as a percentage of the θ220 value at 4 °C, and apparent Tm values were determined using Graphpad PRISM software. A representative denaturation curve is shown for each protein, and apparent Tm values are the average of three independent experiments ± S.D. B, Notch peptide conformations during molecular dynamics simulation with FIH are shown. Schematic representation shows that peptides from the substrate proteins Notch1 wt (shown in blue) and Notch4 Q-I (green) exhibit a tighter turn around the target Asn residue, as compared with non-substrate peptides Notch1 A-P (orange) and Notch4 wt (red). C, this characteristic can be shown quantitatively throughout the 1-ns duration of molecular dynamics simulation via the measured distance between the Cα of the residue at the −1 position and the backbone N of the +1 residue.
The ability of these proteins to be hydroxylated by FIH may not, however, be entirely attributed to changes in thermodynamic stability of the ARD. Based on the crystal structure of FIH in complex with HIF and Notch peptides, the residues at the +1 and −1 positions relative to the Asn form a backbone interaction that projects the side chain of the Asn into the active site of FIH (10). Therefore, the identity of the amino acids at these positions may also dictate their ability to appropriately position the Asn for hydroxylation by FIH. To investigate this hypothesis, molecular dynamics simulation was used to examine the interaction between FIH and four of the Notch variants; two substrate proteins (Notch1 wild type and Notch4 Q-I) and two non-substrate proteins (Notch4 wild type and Notch1 A-P). A region of the Notch1 ARD surrounding Asn-1945 (comprising residues 1936–1951) was modeled in complex with FIH, and corresponding models were constructed for the Notch1 A-P mutant as well as the Notch4 wild type and Q-I mutant proteins (see “Experimental Procedures” for details).
The most notable difference between the simulations was the conformation adopted by the substrate peptide backbone, with little change observed in the conformation of FIH. Although the Notch1 wild type and Notch4 Q-I maintained a tight turn across residues at the −1 and +1 positions relative to the Asn, this was less evident for the Notch4 wild type and Notch1 A-P mutant (Fig. 5B). The extent of this difference and its maintenance throughout the duration of the simulation is demonstrated by comparing the distance across the turn. Specifically, the distance between the Cα of the residue at the −1 position and the backbone nitrogen of the +1 residue was measured every 100 fs of the simulation and plotted against time (Fig. 5C).
It is clear from this analysis that the proteins that are hydroxylated by FIH (Notch1 wild type and Notch4 Q-I) maintain a tighter and more stable turn, with the distance remaining around 4.5 Å for the duration of the simulation compared with 4.8–5.0 Å for the non-substrate proteins (Notch4 wild type and Notch1 A-P). The turn surrounding the hydroxylated Asn has previously been noted for its importance in allowing positioning of the Asn side chain within the active site of FIH (19). In simulations of the non-substrate peptides, the Asn side chain does escape its key position in the active site (as shown in Fig. 5B), whereas the wider path of the peptide also deforms the active site to a small extent. It must be noted, however, that our simulations begin with a preformed complex and that the true biochemical consequence of a more extended backbone conformation may simply be that the peptide cannot dock appropriately into the active site at all.
The simulations also reveal that for the wild type Notch1 peptide, the turn is maintained by hydrogen bonding between the amide hydrogens of the −1 and +1 residues, with a water molecule between them (supplemental Fig. S4A). Introduction of a Pro at the −1 position removes the requisite amide hydrogen, explaining the lack of hydroxylation of the wild type Notch4 protein (supplemental Fig. S4C). One would expect the same argument to apply to the Notch4 Q-I mutant; however, the Pro and Ile residues at −1 and +1 form a close hydrophobic interaction that performs a similar function to the aforementioned hydrogen bond (supplemental Fig. S4D).
Finally, although the Notch1 A-P mutant similarly has hydrophobic Pro and Ile residues at −1 and +1, the deviation of the peptide due to the −1 Pro promotes an interaction between Notch1 Asp-1943 and FIH Lys-107. Combined with an interaction between Notch1 Gln-1947 and FIH Glu-105 and Arg-120, this restrains the whole peptide in its more extended form (supplemental Fig. S4B). Thus, similar to previous studies on the HIF CAD (23), the identity of ARD residues proximal to the Asn can impact hydroxylation efficiency by compromising the positioning of the acceptor Asn in the FIH active site. However, the structured nature of the ARD compared with the disordered HIF CAD means that mutations next to the Asn may also influence the thermal stability of the proteins and thus determine whether the unfolding required for hydroxylation can occur.
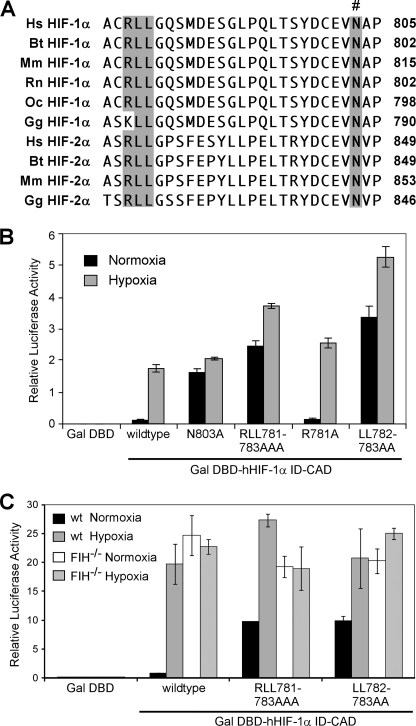
RLL Motif Is Required for FIH-mediated Normoxic Repression of HIF CAD
Given that the FIH consensus sequence (Fig. 1) underestimates the importance of the amino acids directly adjacent to the Asn, we searched the literature for more distant primary sequence motifs and identified a conserved arginine-dileucine (RLL) motif in HIF-α that is likely to be important for recognition by FIH. The RLL motif is located N-terminal to the CAD in HIF-α within the inhibitory domain (ID, Fig. 6A), a highly conserved region that is required for normoxic repression of the CAD (41, 42). Triplicate alanine mutagenesis (RLL-AAA) of this motif in both HIF-1α and HIF-2α results in constitutive CAD activity (43), suggesting that these residues are important for recognition and catalysis by FIH.
FIGURE 6.
A dileucine motif within the inhibitory domain is essential for normoxic repression of the HIF-CAD. A, alignment of part of the HIF-α ID-CAD sequence shows conservation of an arginine-dileucine (RLL) motif, highlighted in gray, between HIF-1α and HIF-2α vertebrate orthologues. The asparagine hydroxylated by FIH is indicated by #. B, shown are reporter assays in which HEK-293T cells were transiently transfected with empty pGalDBD or pGalDBD hHIF-1α ID-CAD (737–826) wild type, N803A, R781A/L783A/L783A, R781A, or L782A/L783A mutants together with p-G5E1B-Luc and pRLTK. Transfected cells were treated for 16 h with normoxia or hypoxia (0.1% O2), and firefly luciferase activity was quantified relative to renilla luciferase activity. Data are the mean of triplicates ± S.E. and are representative of three independent experiments. C, reporter gene assays in which wild type (wt) or FIH−/− MEF cells were transiently transfected with p-G5E1B-Luc, pRLTK, and GALDBD-hHIF-1α ID-CAD (737–826) wild type, R781A/L783A/L783A, or L782A/L783A mutants and treated for 16 h with normoxia or hypoxia (0.1% O2) followed by analysis of relative luciferase activity. Data are the mean of triplicates ± S.E. and are representative of three independent experiments.
To independently confirm a necessity for the RLL motif in normoxic repression of the CAD and to directly compare the contribution of its chemically distinct components (the arginine residue and dileucine motif), we performed reporter gene assays with Gal-DNA binding domain (GalDBD)-HIF ID-CAD chimeras in HEK-293T cells. As shown in Fig. 6B, the wild type ID-CAD exhibits low normoxic activity but is induced ∼17 fold in hypoxia. The N803A mutant is constitutively active, which is expected given that it cannot be hydroxylated by FIH. The R-A mutant behaved similarly to the wild type protein, showing that this non-conservative mutation is tolerated by FIH. However, both the RLL-AAA and LL-AA mutants displayed much higher normoxic activity than the wild type ID-CAD and less than 2-fold induction by hypoxia. These results imply that the RLL-AAA and LL-AA mutant HIF-α proteins resist hydroxylation by FIH and thus evade normoxic repression.
To confirm that the observed normoxic repression of the HIF-1α CAD is due to hydroxylation by FIH, similar assays were performed in wild type and FIH−/− MEFs (Fig. 6C). As expected, the wild type ID-CAD and LL-AA/RLL-AAA mutants were all constitutively active in the absence of endogenous FIH, indicating that both FIH and an intact RLL motif are required for efficient repression of the HIF CAD in normoxia. Furthermore, as the RLL-AAA and LL-AA mutants behave in essentially the same manner, and the two leucine residues constitute the essential component of this motif. Interestingly, a small but consistent hypoxic induction was observed for the RLL-AAA and LL-AA mutants in both HEK-293T cells and wild type MEFs but was abolished in the FIH−/− MEFs, suggesting that these mutations hinder, but do not completely inhibit hydroxylation by FIH.
RLL Motif Is High Affinity Binding Determinant for FIH
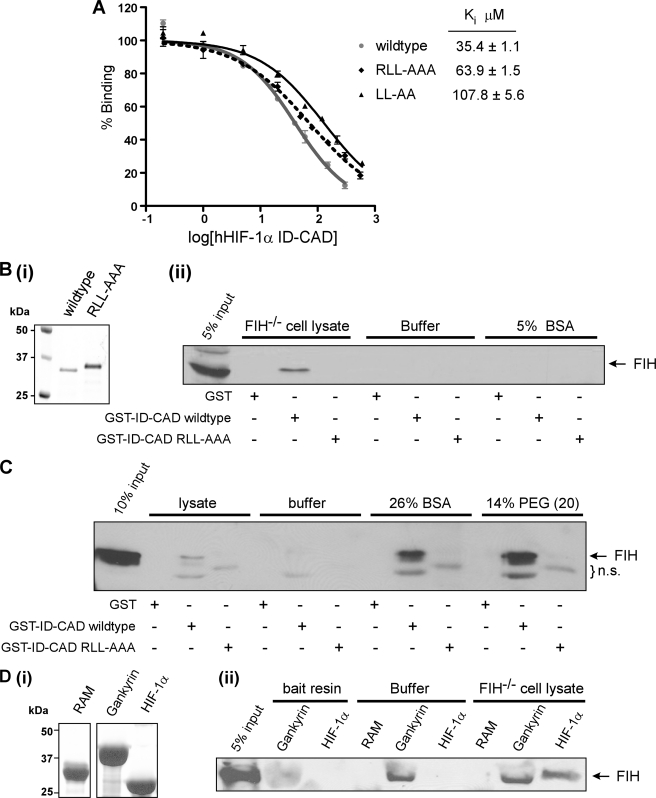
The reporter gene assay data indicate that the RLL motif is a critical determinant of recognition and hydroxylation by FIH. However, the x-ray crystal structure of the FIH-HIF complex reveals a bipartite contact site that lies entirely within the 40-amino acid CAD and fails to implicate the RLL motif in binding FIH (19). We sought to determine whether the RLL motif is capable of influencing the ability of FIH to bind HIF-1α in solution. Mutation of the RLL motif to AAA caused a 2-fold decrease in the affinity of FIH for the HIF-1α ID-CAD, as determined by FP competition binding assays, with Ki values of 35 and 64 μm obtained for the wild type and RLL-AAA proteins, respectively (Fig. 7A). This is a potentially important difference although not as great as expected given the severe effect of the RLL-AAA mutation in cell-based activity assays. Thus we hypothesized that a cellular factor might further amplify the function of the RLL motif in FIH binding.
FIGURE 7.
An intact RLL motif in the HIF ID-CAD promotes a high affinity interaction between FIH and HIF in the presence of cell-lysate or macromolecular crowding agents. A, affinity-purified Trx-6H-tagged hHIF-1α ID-CAD (737–826) wild type, R781A/L783A/L783A, or L782A/L783A mutants were assayed for their ability to compete with a fluorescently labeled Notch peptide in FP competition binding experiments. Ki values were determined using Graphpad PRISM software and are the average of three independent experiments ± S.D. B, GST and GST-hHIF-1α ID-CAD wild type or R781A/L783A/L783A mutant proteins were used as bait for in vitro affinity pulldown assays with recombinant human FIH. Pulldown assays were performed in FIH−/− cell lysate, buffer alone, or buffer normalized to the lysate protein concentration with BSA. Bait proteins were analyzed by SDS-PAGE with Coomassie staining (i), and interacting FIH was detected by SDS/PAGE and Western blotting with a Novus anti-FIH antibody (ii). C, bait proteins from B were used in pulldown assays, which were performed as described but with the provision of FIH−/− lysate, buffer alone, or buffer supplemented with 26% BSA or 14% PEG (MW 20,000 g/mol). Several nonspecific bands were detected by the antibody (n.s.). D, Trx-6H-tagged hHIF-1α ID-CAD, Gankyrin, or RAM (negative control) were used as bait to capture recombinant FIH. Bait proteins were analyzed by SDS-PAGE with Coomassie staining (i), and interacting FIH was detected by SDS-PAGE/immunoblotting (ii) with an anti-FIH antibody (Novus). Note that untagged FIH has a similar molecular weight to Trx-6H-tagged Gankyrin, and consequently, in the samples containing Trx-6H-tagged Gankyrin, the migration of FIH is altered, with a lower apparent molecular weight. All pulldown assays are representative of at least three independent experiments.
Therefore, to further explore this hypothesis, we performed affinity pulldown assays using GST-tagged HIF ID-CAD proteins as bait to capture recombinant FIH in the presence of either buffer or mammalian cell lysate. As shown in Fig. 7B, the relatively low affinity of FIH for the HIF CAD prevented detection of FIH bound to HIF under standard assay conditions (buffer alone). However, the inclusion of mammalian cell lysate, but not BSA, was sufficient to enhance the binding affinity such that a detectable level of FIH was found to interact with the HIF ID-CAD in assays performed in the presence of FIH−/− MEF cell lysate (Fig. 7B). Of particular interest, however, was that the same lysate was unable to promote an interaction between FIH and the RLL-AAA mutant ID-CAD. Collectively, these data suggest that cell lysate amplifies the function of the RLL motif as a potential high affinity binding site for FIH.
Throughout the course of these experiments, it was observed that the more concentrated the lysate, the more effective it was at promoting RLL-dependent binding of FIH. This is consistent with a specific, limiting factor in the lysate or, alternatively, a role for molecular crowding in RLL motif function. To examine the latter possibility, we performed similar pulldown experiments in the presence of high concentrations of molecular crowding agents, such as PEG (14%) and BSA (26%). Remarkably, these crowding agents were able to recapitulate the effects of the lysate (Fig. 7C), as were numerous other crowding agents that we have tested (data not shown). These observations suggest that molecular crowding influences the RLL motif, possibly by promoting the adoption of a conformation that binds FIH with a higher affinity.
To further explore the RLL motif, sequence analysis of ARD substrates was performed to locate any similar motifs and revealed that a number of confirmed ARD substrates contain a conserved dileucine motif (Fig. 1). However, the ARD LL motif is not in an analogous position to the RLL motif in HIF; thus, it is likely to mediate binding to FIH in a distinct manner. It is noteworthy that one of the leucinyl residues in the ankyrin motif, located at position −8, is highly conserved in both HIF and ARD substrates and consequently constitutes part of the FIH consensus sequence (9). Crystallographic analyses indicate that this −8 Leu makes a distinct interaction with a hydrophobic pocket on the surface of FIH that is likely to be important for binding and catalysis (10, 19). Furthermore, the LL motif is highly conserved at this position in ankyrin repeats, as it forms part of a hydrophobic core that facilitates the correct stacking of inter-repeat ankyrin helices (7). Thus, it is probable that the LL motifs in HIF and ARD substrates perform different tasks and mediate distinct interactions with FIH. Consistent with this interpretation, the provision of cell lysate did not improve the efficiency of the interaction between FIH and Gankyrin, as an equivalent amount of FIH was detected in pulldown assays with Gankyrin performed in the presence and absence of lysate (Fig. 7D). Similar results were also obtained with Notch (data not shown). It is, therefore, likely that the LL motif within the HIF ID-CAD is a HIF-specific means by which FIH-mediated hydroxylation can be regulated in vivo.
DISCUSSION
There is a growing body of research showing that ARD proteins are common targets for hydroxylation by FIH. Despite this recent expansion in our knowledge of substrate repertoire, the molecular requirements for hydroxylation by FIH have remained somewhat elusive. The results presented in this study demonstrate that substrate recognition differs significantly between HIF and ARD substrates. For ARD substrates, specificity is determined by more than the primary amino acid sequence and relies on the ability of the substrate to adopt a conformation that is recognized by FIH at the secondary/tertiary structural level. These data clearly demonstrate that not only is FIH able to bind a fully folded ARD, but the robust affinity of this interaction likely reflects a preference for this folded conformation. However, an intact ankyrin fold is not permissive to hydroxylation, and transient unfolding must occur to enable catalytically productive binding to FIH. As such, the conformational preference for FIH binding may be distinct from that of hydroxylation.
Previous work highlighting the importance of helix 3A in Notch, positioned C-terminal to the hydroxylation site, suggests that this is a likely site of recognition by FIH (18). Our data support a mechanism in which FIH binds initially to a folded ARD, potentially through a high affinity interaction with a helix A exposed on the convex surface of the folded ARD. N-terminal to this helix lies a potential hydroxylation site within the β-hairpin turn of the previous repeat. However, the Asn would be inaccessible to FIH in the context of a fully folded ARD. However, the structural nature of the ARD is such that it can interconvert between a number of differentially folded states, a subset of which may be favorable for catalysis. Already bound to the ARD, FIH is poised to take advantage of transient localized separation or unfolding of nearby ankyrin repeats, which would briefly elongate the β-hairpin turn and expose the target Asn for hydroxylation. FIH would then make further contacts in the vicinity of the target Asn residue and facilitate unfolding of the helix N-terminal to the hydroxylation site leading to the extended conformation observed in the crystal structure of the Notch-FIH interaction (10).
Based on this model, we propose that the ankyrin fold is a key determinant of the kinetic parameters of hydroxylation of ARD substrates by FIH. The ability of FIH to bind an intact ARD enables efficient recruitment of ARD substrates and a corresponding low Km for hydroxylation. The poor turnover rate, reflected in the low Vmax observed for Notch ARD substrates, may be due to a slow and limiting off rate as a result of this high affinity interaction. Alternatively, it may reflect the equilibrium of ARD folding that favors a more folded conformation, such that at any given time FIH is predominantly in complex with a folded and catalytically unproductive conformation of the ARD. Furthermore, FIH could potentially bind to the C-terminal helix in other repeats within an ARD, not all of which contain candidate residues for hydroxylation in the adjacent β-turn region.
Ultimately, hydroxylation of ARD substrates by FIH requires localized unfolding of ankyrin repeats, and so the stability of a particular repeat is likely to influence how readily it will be hydroxylated by FIH. A number of factors dictate the stability of an ankyrin repeat, including its overall position with the ARD as well as the primary amino acid sequence. Consistent with this, single point mutations within the β-hairpin region containing Asn-1656 in Notch4 were found to destabilize the Notch4 ARD and also promote hydroxylation by FIH. The higher thermodynamic stability of the wild type Notch4 ARD relative to Notch1 may account for its lack of hydroxylation in vitro. However, we cannot rule out the possibility that the point mutations that we introduced, which flank Asn-1656, have an effect independent of altering stability. In both Notch and HIF substrates, the amino acids directly adjacent to the target Asn form a tight turn that is important for projecting the Asn into the active site of FIH (10, 19). Based on the substrate consensus sequence, FIH shows a preference for small, hydrophobic amino acids at the +1 and −1 positions (9); however, the consensus is likely to be biased due to strong conservation of hydrophobic residues at these same positions in most ankyrin repeats (7). Although FIH is clearly able to tolerate chemically disparate residues, such as proline, in these positions, molecular modeling suggests that it is not the specific identity of a particular residue per se that is important but rather the combined ability of the residues proximal to the Asn to maintain a tight turn within the active site of FIH. These data emphasize the importance of primary amino acid sequence in substrate selection but warn that although the primary consensus may prove useful as a general guide for substrate prediction, it is by no means absolute.
Another notable result from the site-directed mutagenesis is that Notch variants that were not hydroxylated by FIH were nonetheless able to bind with a relatively high affinity. This result combined with the observation that FIH can bind stable ARDs in a catalytically unproductive manner point to a possible role for FIH that is dependent on binding as opposed to hydroxylation. Previous studies have suggested that ARD proteins may function as a pool to indirectly regulate hypoxic signaling by sequestering FIH and limiting its availability for HIF (10, 12). Although our data support this hypothesis, they also suggest that non-substrate ARD proteins may also be involved in HIF regulation. However, due to the ubiquitous nature of the ARD proteins and their high affinity interaction with FIH relative to HIF, it is unclear how efficient hydroxylation of HIF in vivo is actually achieved.
A potential explanation is provided by the RLL motif in the HIF inhibitory domain, characterized in this study as a crucial in vivo binding determinant for FIH. The RLL motif influences FIH binding by a mechanism that requires the provision of mammalian cell lysate or molecular crowding agents, strongly suggesting that HIF is likely to have a much higher affinity for FIH in cells than is apparent from in vitro kinetic analyses (18). The ability of molecular crowding to facilitate a high affinity interaction between the HIF-α RLL motif and FIH is likely to rely on the autonomously unstructured nature of the CAD, which gives it the capacity to form multiple diverse conformational states. Although our assays do not rule out the existence of a specific factor in lysate, such as a chaperone protein, which facilitates RLL-dependent FIH binding in cells, the ability of nonspecific crowding agents to mimic the lysate together with the affinity difference detected by binding assays in dilute buffer suggests that the RLL motif is autonomously capable of mediating FIH binding. We propose that a crowded cellular environment can promote HIF to adopt a secondary structure that involves the RLL motif and forms a recognition interface for FIH. There is precedence for crowding to induce structure in intrinsically disordered proteins (44), which in turn can facilitate protein-protein interactions (45). Akin to our model of ARD hydroxylation, this structural motif may be required during initial FIH binding, which would then be followed by the well defined catalytic interaction occurring within the CAD region. As such, the kinetic parameters of HIF hydroxylation are dictated largely by sequence determinants within the hydroxylation site, which appear to promote an efficient catalytic rate regardless of structural context.
The importance of HIF regulation in normal physiology and disease has been clearly demonstrated together with the more recent demonstration of the role of FIH in regulating metabolism (24). Collectively, this work identifies a number of substrate-specific determinants of binding and hydroxylation by FIH and provides important insight into how FIH can recognize distinct classes of substrates in a physiological setting. The characterization of these determinants will enable a more accurate prediction of additional FIH substrates, their hydroxylation efficiency, and influence on HIF regulation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Briony Forbes and Dr. Rebecca Bilton for helpful discussions, eResearch SA for the use of high performance computing resources, and the Biophysical Characterization Facility within the Sansom Institute for Health Research at the University of South Australia for use of the CD Spectrometer.
This work was funded by the National Health and Medical Research Council of Australia and the ARC Special Research Centre for the Molecular Genetics of Development.

This article contains supplemental Methods, Figs. S1–S5, and Table S1.
- FIH
- factor inhibiting HIF
- HIF
- hypoxia-inducible factor
- ARD
- ankyrin repeat domain
- CAD
- C-terminal transactivation domain
- MBP
- maltose-binding protein
- Trx
- thioredoxin
- TEV
- tobacco etch virus
- MEF
- mouse embryonic fibroblast
- FP
- fluorescence polarization
- RAM
- RBP-Jκ-associated molecule
- ID
- inhibitory domain
- DBD
- DNA binding domain
- 6H
- His6.
REFERENCES
- 1. Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., Ratcliffe P. J., Pugh C. W., Schofield C. J. (2002) Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277, 26351–26355 [DOI] [PubMed] [Google Scholar]
- 2. Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. (2002) FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahon P. C., Hirota K., Semenza G. L. (2001) FIH-1. A novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15, 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. (2002) Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861 [DOI] [PubMed] [Google Scholar]
- 5. Cockman M. E., Webb J. D., Ratcliffe P. J. (2009) FIH-dependent asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Ann. N.Y. Acad. Sci. 1177, 9–18 [DOI] [PubMed] [Google Scholar]
- 6. Sedgwick S. G., Smerdon S. J. (1999) The ankyrin repeat. A diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 [DOI] [PubMed] [Google Scholar]
- 7. Mosavi L. K., Cammett T. J., Desrosiers D. C., Peng Z. Y. (2004) The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13, 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cockman M. E., Lancaster D. E., Stolze I. P., Hewitson K. S., McDonough M. A., Coleman M. L., Coles C. H., Yu X., Hay R. T., Ley S. C., Pugh C. W., Oldham N. J., Masson N., Schofield C. J., Ratcliffe P. J. (2006) Posttranslational hydroxylation of ankyrin repeats in IκB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH). Proc. Natl. Acad. Sci. U.S.A. 103, 14767–14772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cockman M. E., Webb J. D., Kramer H. B., Kessler B. M., Ratcliffe P. J. (2009) Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol. Cell. Proteomics 8, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coleman M. L., McDonough M. A., Hewitson K. S., Coles C., Mecinovic J., Edelmann M., Cook K. M., Cockman M. E., Lancaster D. E., Kessler B. M., Oldham N. J., Ratcliffe P. J., Schofield C. J. (2007) Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J. Biol. Chem. 282, 24027–24038 [DOI] [PubMed] [Google Scholar]
- 11. Ferguson J. E., 3rd, Wu Y., Smith K., Charles P., Powers K., Wang H., Patterson C. (2007) ASB4 is a hydroxylation substrate of FIH and promotes vascular differentiation via an oxygen-dependent mechanism. Mol. Cell Biol. 27, 6407–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Webb J. D., Murányi A., Pugh C. W., Ratcliffe P. J., Coleman M. L. (2009) MYPT1, the targeting subunit of smooth-muscle myosin phosphatase, is a substrate for the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH). Biochem. J. 420, 327–333 [DOI] [PubMed] [Google Scholar]
- 13. Yang M., Chowdhury R., Ge W., Hamed R. B., McDonough M. A., Claridge T. D., Kessler B. M., Cockman M. E., Ratcliffe P. J., Schofield C. J. (2011) Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 278, 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang M., Ge W., Chowdhury R., Claridge T. D., Kramer H. B., Schmierer B., McDonough M. A., Gong L., Kessler B. M., Ratcliffe P. J., Coleman M. L., Schofield C. J. (2011) Asparagine and aspartate hydroxylation of the cytoskeletal ankyrin family is catalyzed by factor-inhibiting hypoxia-inducible factor. J. Biol. Chem. 286, 7648–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hardy A. P., Prokes I., Kelly L., Campbell I. D., Schofield C. J. (2009) Asparaginyl β-hydroxylation of proteins containing ankyrin repeat domains influences their stability and function. J. Mol. Biol. 392, 994–1006 [DOI] [PubMed] [Google Scholar]
- 16. Kelly L., McDonough M. A., Coleman M. L., Ratcliffe P. J., Schofield C. J. (2009) Asparagine β-hydroxylation stabilizes the ankyrin repeat domain fold. Mol. Biosyst. 5, 52–58 [DOI] [PubMed] [Google Scholar]
- 17. Devries I. L., Hampton-Smith R. J., Mulvihill M. M., Alverdi V., Peet D. J., Komives E. A. (2010) Consequences of IκBα hydroxylation by the factor inhibiting HIF (FIH). FEBS Lett. 584, 4725–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilkins S. E., Hyvärinen J., Chicher J., Gorman J. J., Peet D. J., Bilton R. L., Koivunen P. (2009) Differences in hydroxylation and binding of Notch and HIF-1α demonstrate substrate selectivity for factor inhibiting HIF-1 (FIH-1). Int. J. Biochem. Cell Biol. 41, 1563–1571 [DOI] [PubMed] [Google Scholar]
- 19. Elkins J. M., Hewitson K. S., McNeill L. A., Seibel J. F., Schlemminger I., Pugh C. W., Ratcliffe P. J., Schofield C. J. (2003) Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 α. J. Biol. Chem. 278, 1802–1806 [DOI] [PubMed] [Google Scholar]
- 20. Dames S. A., Martinez-Yamout M., De Guzman R. N., Dyson H. J., Wright P. E. (2002) Structural basis for Hif-1 α/CBP recognition in the cellular hypoxic response. Proc. Natl. Acad. Sci. U.S.A. 99, 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Freedman S. J., Sun Z. Y., Poy F., Kung A. L., Livingston D. M., Wagner G., Eck M. J. (2002) Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1α. Proc. Natl. Acad. Sci. U.S.A. 99, 5367–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linke S., Hampton-Smith R. J., Peet D. J. (2007) Characterization of ankyrin repeat-containing proteins as substrates of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible transcription factor. Methods Enzymol. 435, 61–85 [DOI] [PubMed] [Google Scholar]
- 23. Linke S., Stojkoski C., Kewley R. J., Booker G. W., Whitelaw M. L., Peet D. J. (2004) Substrate requirements of the oxygen-sensing asparaginyl hydroxylase factor-inhibiting hypoxia-inducible factor. J. Biol. Chem. 279, 14391–14397 [DOI] [PubMed] [Google Scholar]
- 24. Zhang N., Fu Z., Linke S., Chicher J., Gorman J. J., Visk D., Haddad G. G., Poellinger L., Peet D. J., Powell F., Johnson R. S. (2010) The asparaginyl hydroxylase factor inhibiting HIF-1α is an essential regulator of metabolism. Cell Metab. 11, 364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krivov G. G., Shapovalov M. V., Dunbrack R. L., Jr. (2009) Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77, 778–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 27. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 28. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 29. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 31. Raussens V., Ruysschaert J. M., Goormaghtigh E. (2003) Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra. A new scaling method. Anal. Biochem. 319, 114–121 [DOI] [PubMed] [Google Scholar]
- 32. Whitmore L., Wallace B. A. (2004) DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson W. C. (1999) Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35, 307–312 [PubMed] [Google Scholar]
- 34. Zheng X., Linke S., Dias J. M., Zheng X., Gradin K., Wallis T. P., Hamilton B. R., Gustafsson M., Ruas J. L., Wilkins S., Bilton R. L., Brismar K., Whitelaw M. L., Pereira T., Gorman J. J., Ericson J., Peet D. J., Lendahl U., Poellinger L. (2008) Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 105, 3368–3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greenfield N. J. (2004) Circular dichroism analysis for protein-protein interactions. Methods Mol. Biol. 261, 55–78 [DOI] [PubMed] [Google Scholar]
- 36. Dann C. E., 3rd, Bruick R. K., Deisenhofer J. (2002) Structure of factor-inhibiting hypoxia-inducible factor 1. An asparaginyl hydroxylase involved in the hypoxic response pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 15351–15356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ehebauer M. T., Chirgadze D. Y., Hayward P., Martinez Arias A., Blundell T. L. (2005) High resolution crystal structure of the human Notch 1 ankyrin domain. Biochem. J. 392, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee C., Kim S. J., Jeong D. G., Lee S. M., Ryu S. E. (2003) Structure of human FIH-1 reveals a unique active site pocket and interaction sites for HIF-1 and von Hippel-Lindau. J. Biol. Chem. 278, 7558–7563 [DOI] [PubMed] [Google Scholar]
- 39. Dawson S., Apcher S., Mee M., Higashitsuji H., Baker R., Uhle S., Dubiel W., Fujita J., Mayer R. J. (2002) Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J. Biol. Chem. 277, 10893–10902 [DOI] [PubMed] [Google Scholar]
- 40. Nakamura Y., Nakano K., Umehara T., Kimura M., Hayashizaki Y., Tanaka A., Horikoshi M., Padmanabhan B., Yokoyama S. (2007) Structure of the oncoprotein Gankyrin in complex with S6 ATPase of the 26 S proteasome. Structure 15, 179–189 [DOI] [PubMed] [Google Scholar]
- 41. Jiang B. H., Zheng J. Z., Leung S. W., Roe R., Semenza G. L. (1997) Transactivation and inhibitory domains of hypoxia-inducible factor 1α. Modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272, 19253–19260 [DOI] [PubMed] [Google Scholar]
- 42. Pugh C. W., O'Rourke J. F., Nagao M., Gleadle J. M., Ratcliffe P. J. (1997) Activation of hypoxia-inducible factor-1. Definition of regulatory domains within the α subunit. J. Biol. Chem. 272, 11205–11214 [DOI] [PubMed] [Google Scholar]
- 43. O'Rourke J. F., Tian Y. M., Ratcliffe P. J., Pugh C. W. (1999) Oxygen-regulated and transactivating domains in endothelial PAS protein 1. Comparison with hypoxia-inducible factor-1α. J. Biol. Chem. 274, 2060–2071 [DOI] [PubMed] [Google Scholar]
- 44. Dedmon M. M., Patel C. N., Young G. B., Pielak G. J. (2002) FlgM gains structure in living cells. Proc. Natl. Acad. Sci. U.S.A. 99, 12681–12684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia Z., Webster A., Du F., Piatkov K., Ghislain M., Varshavsky A. (2008) Substrate binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283, 24011–24028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.