Background: β-Arrestin-1 is present in the nucleus but lacks an identifiable nuclear localization signal.
Results: Using sequence comparison and mutagenesis, we found Lys157 is critical to β-arrestin-1 nuclear localization and its regulation of NF-κB activation.
Conclusion: β-Arrestin-1 uses a novel basic sequence for its entry into the nucleus.
Significance: This work provides a structural basis for the nuclear function of β-arrestin-1.
Keywords: G Protein-coupled Receptors (GPCR), NF-κB Transcription Factor, Nuclear Translocation, Post-translational Modification, Scaffold Proteins
Abstract
A mounting body of evidence suggests that β-arrestin-1 plays important roles in the nucleus, but how β-arrestin-1 enters the nucleus remains unclear because no nuclear import signal has been identified in the β-arrestins. We sought to characterize the cellular localization of wild type β-arrestin-1 and a series of N domain mutants to determine the structural basis and functional implications of β-arrestin-1 nuclear localization. A seven-residue candidate nuclear localization sequence (NLS) was identified based on sequence analysis. Mutation of the NLS led to a loss of β-arrestin-1 nuclear localization in transfected cells. Exogenous expression of wild type β-arrestin-1 enhanced the transcriptional activity of nuclear factor κB (NF-κB) induced by bradykinin, whereas mutation of the NLS reduced this effect by two-thirds relative to wild type controls. Loss of β-arrestin-1 nuclear localization was accompanied by reduced recruitment of the CREB-binding protein and altered post-translational modification profile of p65/RelA. Further mutational analysis identified Lys157 within the putative NLS as being critical to nuclear localization of β-arrestin-1. Substitution of Lys157 to Ala led to reduced nuclear localization, decreased promoter binding by p65/RelA and decreased IL-1β gene transcription. These results demonstrate a critical role for β-arrestin-1 nuclear localization in scaffolding and transcriptional regulation.
Introduction
The β-arrestin family is comprised of β-arrestin-1 (also known as arrestin-2) and β-arrestin-2 (arrestin-3), which act to regulate a diverse array of cell functions. Initially identified for their roles in desensitization of G protein-coupled receptors (GPCRs)3 (1, 2), the β-arrestins are now appreciated as critical scaffolds in a variety of cellular signaling cascades. One or both of the β-arrestins have been shown to interact with the Src family tyrosine kinases (3, 4), PI3K (5), Akt (6, 7), and other proteins including heat shock proteins (8, 9). β-Arrestins direct specific cellular functions by scaffolding the MAP kinases including JNK3 and ERK (10, 11). Of specific interest to this work, β-arrestins interact with several members of the nuclear factor κB (NF-κB) signaling pathway, including TRAF6 (12), p105 (13), and the inhibitory protein IκBα (14). β-Arrestins also interact with the upstream activating kinases, IκB kinase α, β, and NF-κB inducing kinase (15). It is reported that both β-arrestins work in the cytosol to stabilize IκBα at resting state, thus minimizing NF-κB activation. However, other published data have shown an enhancement of NF-κB activity in cells stimulated by GPCR ligands (7, 16, 17), suggesting that the scaffolding functions of the β-arrestins are able to overcome the IκBα stabilizing effect and promote NF-κB activation.
In addition to its role in scaffolding signaling pathways within the cytosol, emerging evidence suggests that β-arrestin-1 mediates signaling within the nucleus. In studies conducted by different groups, β-arrestin-1 was found to serve as an activator of CREB-mediated transcription following δ opioid receptor activation (18), but act as an inhibitor of STAT1-mediated transcription following IFN-γ stimulation (19). These data indicate that β-arrestin-1 can facilitate both activation and repression depending on cell type and the nature of stimulation. In all reported cases, β-arrestin-1 acts essentially by a scaffolding mechanism in the nucleus and the cytosol, recruiting specific proteins for complex formation. In these examples it is by the recruitment of the histone acetyltransferase p300 (18) or the tyrosine phosphatase TC45 (19) that β-arrestin-1 serves as an activator or inhibitor, respectively. Work on the nuclear function of the β-arrestins has been primarily limited to β-arrestin-1, because β-arrestin-2 has a strong nuclear export signal (NES) in its C terminus, which excludes it from the sustained presence in the nucleus (20).
Although mounting evidence demonstrates the nuclear functions of β-arrestin-1, the regulatory mechanism for its nuclear localization is still unknown. At ∼48 kDa the β-arrestin proteins are too large to passively diffuse into the nucleus and must be actively transported in an energy-dependent process (21). Although there is a large family of proteins involved in nuclear import, the most studied and well characterized are those involved in importing proteins containing a classical or canonical nuclear localization signal (NLS). Classical NLSs consist of either monopartite or bipartite clusters of basic amino acids and, in the latter case, these residues are separated by a linker of ∼10 amino acids (22). These classical NLS-containing proteins are recognized by a carrier protein importin α, which brings them to the transport protein importin β1. Importin β1 is responsible for shuttling NLS-containing proteins through the nuclear pore complex in an energy-dependent process involving GTP hydrolysis (23).
To further explore the nuclear functions of β-arrestin-1, we analyzed the structural determinants required for β-arrestin nuclear localization. Using DNA mutagenesis, we identified a region of seven amino acids (Lys157–Arg161/Arg169 and Lys170) as being critical to nuclear localization of β-arrestin-1. We identified these residues as necessary to mediate nuclear import of β-arrestin-1 via interaction with importin β1. Further characterization of this region led to the identification of Lys157 as being required for β-arrestin-1 nuclear localization. Functionally, nuclear localization of β-arrestin-1 is important for enhanced NF-κB activation following bradykinin (BK) stimulation, and β-arrestin-1 mutants are not capable of potentiating NF-κB activation. The loss of function by the β-arrestin-1 mutants is due to a lack of post-translational modification to the DNA-binding subunit p65/RelA. These modifications seem to influence DNA binding, as there is a decrease in overall protein-DNA interaction as well as a specific loss of binding of p65/RelA. This loss of promoter binding ultimately lead to a decrease in transcription of NF-κB targets, specifically IL-1β.
EXPERIMENTAL PROCEDURES
DNA Constructs
All constructs were generated using previously described human β-arrestin-1 cDNA (NM_0020251) as a template (16). β-Arrestin-1 Δ107–191 was generated by PCR amplification of fragments encoding amino acids 1–107 and 191–409, with a C-terminal FLAG tag incorporated in the sequence. The DNA fragments, after digestion with the restriction enzyme HindIII that cut at an endogenous site, were then ligated into the pCI expression vector. The β-arrestin-1 mNLS and NES+ constructs were generated using the QuikChangeTM mutagenesis kit (Stratagene, La Jolla, CA). The mNLS mutant was generated using primers for region 1 (157–161) and region 2 (170–171) of the putative bipartite NLS in two consecutive rounds of PCR. The QuikChange mutations introduced substitutions of Ala for the corresponding residues and for Lys157 substitution in β-arrestin-1. The NES+ mutant was generated with a primer converting Gln386 to Leu. Both constructs used the previously generated β-arrestin-1-FLAG as a template. β-Arrestin-1-GFP-tagged constructs were generated by PCR amplification of WT, Δ107–191, mNLS, and NES+ plasmids. The PCR products were then subcloned into pEGFP-N1 (Clontech, Mountain View, CA). HA-CBP was a kind gift from Dr. J. R. Lundblad (Vollum Institute, Oregon Health Sciences University, Portland, OR). The constructs for the B2 bradykinin receptor (B2BKR) and the NF-κB luciferase reporter were described previously (16).
Antibodies
Antibodies against p65/RelA, p50, GFP, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-FLAG was from Sigma. Phospho-p65 Ser276 was from Cell Signaling Technology (Beverly, MA). Anti-acetyl-lysine, HDAC1, and histone H1 were from Millipore (Temecula, CA). Anti-importin β1 was from Calbiochem (La Jolla, CA).
Cell Culture and Transfection
HeLa cells were purchased from ATCC. Cells were maintained in DMEM containing 10% heat-inactivated FBS, 2 mm glutamine, 100 units/ml of penicillin, and 50 μg/ml of streptomycin. Cells were transfected with DNA constructs listed above using Lipofectamine 2000 (Invitrogen) at a 3:1 reagent:DNA ratio. The DNA input was adjusted to prevent overexpression of the protein of interest relative to endogenous β-arrestins.
Cell Fractionation and Western Blot Analysis
Cells were fractionated using an IGEPAL ca630 and high salt lysis method (24). Briefly, 5 × 106 cells were harvested by scraping in Hanks' balanced salt solution and then pelleted by centrifugation at 1,000 × g. Pellets were then lysed in nuclear extract buffer A (NEBA; 10 mm HEPES pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride (PMSF) containing protease inhibitor mixture set III (Calbiochem) and to which 10% IGEPAL ca630 was added). Cell lysate was incubated on ice for 10 min, vortexed, and then spun to pellet the nuclei. The supernatant was removed and saved as the cytosolic fraction. The nuclei pellets were then resuspended in nuclear extract buffer B (NEBB; 20 mm HEPES pH 7.9, 0.4 m NaCl, 1 mm EGTA, 1 mm EDTA) and incubated on ice for 30 min with occasional vortexing. Nuclear debris was pelleted by centrifugation at 12,000 × g for 30 s. The supernatant was recovered as the nuclear fraction. Fractions were then directly analyzed by Western blotting. Protein concentration of samples was determined using the DC protein assay system (Bio-Rad). Equivalent protein concentrations were separated on 4–12% BisTris precast SDS-acrylamide gels (Bio-Rad) at 60 mA. Proteins were then transferred to nitrocellulose membranes at 100 V for 1 h. Membranes were then incubated with 5% nonfat milk in TBS-T (20 mm Tris-HCl, pH 7.5, 120 mm NaCl, and 0.01% Tween 20) for 1 h at room temperature. Afterward, membranes were incubated with primary antibodies against proteins of interest overnight at 4 °C. Following removal of primary antibody and a washing step, membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Bound antibodies were detected using the SuperSignal West PicoTM chemiluminescence substrate (Pierce).
Immunoprecipitation
For whole cell immunoprecipitation (IP), cells were harvested in IP lysis buffer (20 mm Tris-HCl pH 7.4, 1 mm DTT, 100 mm NaCl, 1 mm EDTA, 5 mm MgCl2, 1% Triton X-100, 1 mm PMSF) containing protease/phosphatase inhibitor mixture III (Calbiochem). For nuclear IP, cells were fractionated as described above. Following lysis, samples were pre-cleared by incubation with protein A/G-agarose beads (Santa Cruz Biotechnology) for 30 min at 4 °C. Samples were centrifuged to pellet the beads, and the supernatant was transferred to a new tube and incubated overnight with primary antibodies. Protein complexes were then precipitated with the addition of 25 μl of protein A/G beads and incubated for 1 h. Following five washes, the samples were analyzed by Western blotting as described above.
Microscopy
Cells were plated on a glass coverslip (18 mm diameter, 1 mm thickness) coated with 0.1% gelatin. Cells were transfected as above with β-arrestin-GFP constructs. For basal localization studies, samples were starved for 4 h and then prepared for imaging. For BK-stimulated samples, cells were serum starved followed by stimulation with 500 nm BK for the indicated time. To prepare samples for imaging, cells were first washed with Hanks' balanced salt solution and then fixed by incubation in 4% paraformaldehyde for 20 min at room temperature. Free paraformaldehyde was quenched by the addition of 100 mm glycine. Samples were washed with Hanks' balanced salt solution and then mounted onto glass coverslips using ProLong Gold Anti-fade Reagent with DAPI (Molecular Probes, Eugene, OR). Samples were allowed to cure overnight at room temperature and then imaged on a Zeiss LSM 310 Meta Confocal Microscope. Images were analyzed using the LSM Imaging and ImageJ software (National Institutes of Health, Bethesda, MD).
Luciferase Assay
HeLa cells in 12-well plates were transfected with vectors for the β-arrestin-1 constructs, B2BKR, a 3× κB luciferase reporter plasmid, and β-galactosidase as described above. Prior to being assayed, cells were serum starved for 4 h and then stimulated with 200 nm BK for 4 h. Following stimulation, cells were washed and harvested in reporter lysis buffer (Promega). Luciferase expression was analyzed by incubating samples with luciferase substrate (Stratagene) and reading luminescence in a Femtomaster FB12 luminometer (Zylux, Huntsville, AL). The β-galactosidase activity resulting from a co-transfected construct in the same cells was used for normalization of the NF-κB reporter data. All assays were performed three times with triplicate samples. Data were analyzed using GraphPad Prism software (version 4, GraphPad, La Jolla, CA).
Electromobility Shift and Supershift Assays
Nuclear extracts from transfected HeLa cells were generated as described under the “Cell Fractionation” section except that supershift samples were lysed without dithiothreitol (DTT) in the buffer. Nuclear samples were then incubated for 10 min with a [γ-32P]ATP-labeled NF-κB consensus oligonucleotide probe (Promega). Supershift samples were preincubated with specific antibodies to p50 or p65/RelA for 30 min prior to incubation with the probe. Samples were then separated on a 4% acrylamide gel for 3–4 h. The gels were dried and an autoradiograph image was taken using a phosphorimager (GE Healthcare).
Quantitative Real Time PCR
HeLa cells (5 × 106) transfected with WT or mNLS β-arrestin-1 and the B2 receptor were stimulated with 500 μm BK for different periods of time. Total RNA was isolated with a RNeasy isolation kit obtained from Qiagen. RNA was then reverse transcribed to create complimentary DNA (cDNA) using the SuperScript III RT Kit (Invitrogen). Real time quantitative PCR was performed using ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). 1 μg of cDNA from stimulated HeLa cells was analyzed using the GoTag qPCR Master Mix (Promega) containing 20 pmol of forward and reverse primers. The thermocycling program was 40 cycles of 95 °C for 30 s, 60 °C for 45 s, and 72 °C for 45 s, with an initial cycle of 95 °C for 2 min. Accumulation of PCR products was detected by monitoring the increase in fluorescence of SYBR Green after each cycle. A dissociation curve was constructed in the range of 60 to 95 °C, to test for primer specificity. All data were analyzed with the ABI PRISM 7000 SDS software (version 1.1). Primers for the housekeeping gene GAPDH were used to normalize sample loading. Relative levels of mRNA for IL-1β were determined by using the Ct value and the formula: fold-increase = ((1 + ηtarget)ΔCttarget (unstimulated − stimulated))/((1 + ηGAPDH)ΔCt GAPDH (unstimulated − stimulated)).
The sequences of the oligonucleotides used in this study are as follows: 5′-CTTCGGTCCAGTTGCCTTCTC-3′ and 5′-TTCTGCCAGTGCCTCTTTGC-3′ for IL-1; and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH.
Statistical Analysis
Quantification of Western blot data were performed using ImageJ software (National Institutes of Health). Statistical analysis was conducted using the GraphPad Prism software.
RESULTS
β-Arrestin-1 Translocates to Nucleus and Complexes with p65/RelA following Bradykinin Stimulation
We have previously shown that β-arrestin-1 enhances the activation of the transcription factor NF-κB following bradykinin stimulation via a Gβγ- and Akt-dependent mechanism (7). Recent reports have shown that in addition to scaffolding signaling events in the cytosolic compartment, β-arrestin-1 is able to act as a scaffold within the nucleus and has a direct effect on transcriptional responses (18, 25). Based on these reports we sought to determine whether β-arrestin-1 might influence NF-κB-mediated transcription directly in the nucleus in addition to regulating components in the cytosolic compartment, such as IκBα (14, 15), IKK, and p65/RelA (15). To test this we first determined if β-arrestin-1 moved into the nucleus following GPCR activation by agonists such as bradykinin, which activates NF-κB (26). The HeLa cell line was chosen because it has been widely used in studies of NF-κB and, unlike HEK293 and COS-7, it expresses modest levels of exogenous proteins upon DNA transfection. In β-arrestin-1- and B2BKR-transfected HeLa cells there was an appearance of β-arrestin-1 in the nuclear fraction (Fig. 1A, bottom panel). Although some β-arrestin-1 was present in the nucleus before BK stimulation, its concentration increased in the nucleus within 15 min after stimulation, and the increase continued for at least 60 min. Because β-arrestins have been shown to interact with members of the NF-κB family in the cytosolic compartment (7, 14), we sought to determine whether β-arrestin-1 also complexes with p65/RelA in the nucleus. Nuclear fractions were made from transfected and BK-stimulated HeLa cells, incubated with an anti-p65 antibody for IP, and analyzed by Western blot for the co-IP of the FLAG-tagged β-arrestin-1. Following stimulation there was a time-dependent increase in the association between β-arrestin-1 and p65/RelA (Fig. 1A), along with increased β-arrestin-1 nuclear accumulation. These results suggest that β-arrestin-1 forms a complex with p65/RelA in the nucleus following BK stimulation.
FIGURE 1.
β-Arrestin-1 complexes with p65/RelA in the nucleus following BK stimulation and identification of a novel nuclear localization sequence in β-arrestin-1. HeLa cells were transfected with expression vectors for B2BKR and FLAG-tagged β-arrestin-1. In A, following stimulation with 500 nm BK for the indicated times, cells were fractionated. The nuclear fractions were immunoprecipitated with an anti-p65 antibody, and the co-immunoprecipitated β-arrestin-1-FLAG was determined by Western blotting. The level of total β-arrestin-1-FLAG in the nuclear lysate was also determined (lower gel image). Data are representative of at least 3 independent experiments. Shown in B is a schematic representation of β-arrestin-1 and its deletion and substitution mutants, Δ107–179, mNLS, PY, and NES+, tagged with GFP as detailed in the text and under “Experimental Procedures.” In C, confocal microscopy images showing the localization of β-arrestin-1-GFP in HeLa cells transfected with the WT and mutant β-arrestin-1 constructs shown in B. Forty-eight hours after transfection, cells were serum starved for 4 h and then fixed and images were taken. DAPI was used to visualize nuclei. Images representative of 5 independent experiments are shown.
Identification of Novel Nuclear Localization Signal in β-Arrestin-1
Based on our observation that β-arrestin-1 is able to complex with p65/RelA in the nucleus, we determined the structural basis for nuclear localization of β-arrestin-1. A previously published report indicated that the region between residues 1 and 185 of β-arrestin-1, known as the N domain, is required for its nuclear localization (27). To further examine this region, we created a series of N-terminal truncation mutants and their impact on nuclear localization was determined using confocal microscopy and Western blotting. Although truncation of the first 90 residues of β-arrestin-1 had no impact on nuclear localization, truncating to residue 180 led to a predominantly cytosolic distribution of β-arrestin-1 (data not shown). Based on this observation, we generated a β-arrestin-1 construct in which amino acids 107 and 191 were removed (Δ107–191, Fig. 1B). The available restriction sites in the β-arrestin-1 cDNA favors the choice of this region. Visualization of this construct revealed absence of the Δ107–191 β-arrestin-1 from nuclei (Fig. 1C, second row). Attempts to utilize available predictive algorithms to identify consensus protein localization domains within this region were unsuccessful. Manual analysis of the sequence, however, revealed two candidate regions of interest, one which bore similarity to the recently identified PY-NLS, and a second with similarity to the classical bipartite NLS (reviewed in Ref. 28). These sequences are summarized in Table 1. A PY-NLS is characterized by a hydrophobic core of residues followed by a linker leading to the critical PY residues. The candidate PY-NLS in β-arrestin-1 is found between amino acids 86 and 114, with the critical PY (YP) residues located at 113–114 (Table 1). Although a portion of the PY-NLS is outside of our original Δ107–191 mutant, the critical PY (YP) residues are within the N-terminal portion of this region. The second candidate NLS has similarity to a classical bipartite NLS, which is characterized by the presence of contiguous arginine and lysine residues separated by a ∼10-residue linker followed by a cluster of 3–5 basic residues. In β-arrestin-1, this NLS is located between residues 157–161 and 169–170, separated by 7 amino acids. To investigate the roles of these regions in β-arrestin-1 nuclear localization, alanine substitutions were made to generate two mutants, YP (YP to AA at amino acids 113–114) and mNLS (KIHKR/RK to AAAAA/AA at amino acids 157–161/169–170). Only those residues that are critical to nuclear localization in their homologous NLSs were mutated, with all other residues left intact (Fig. 1B). As can be seen in Fig. 1C, substitution of PY to AA had no impact on subcellular distribution of β-arrestin-1 (Fig. 1C, fourth row). Mutation of the candidate bipartite NLS (mNLS), however, caused a profound shift of β-arrestin-1 distribution from the nucleus to the cytosol (Fig. 1C, third row). A β-arrestin-1 mutant, NES+, which contains the NES found in β-arrestin-2, was also prepared. As expected, NES+ was localized in the cytosolic compartment and was used as a control for visualization of non-nuclear β-arrestin protein (Fig. 1C, fifth row).
TABLE 1.
Common motifs found in the nuclear localization signals
The table lists examples of various families of previously identified nuclear localization signals. Monopartite NLSs such as that from SV40 and v-Jun are characterized by a cluster of basic residues. Bipartite NLSs such as those in nucleoplasmin, NIN2, Plk1, and parafibromin are characterized by 2 clusters of basic residues separated by a 7–10-residue linker. Finally, PY NLSs like those in M9 and TAP have a string of hydrophobic or basic residues followed by a R/H/K and then a PY motif, separated by a 3–5-residue linker. Candidate NLS in β-arrestin-1 are also listed.
| NLS protein | Sequence | Source |
|---|---|---|
| Simple NLSs | ||
| SV40 | PKKKRKV | 45 |
| v-Jun | KSRKRKL | 46 |
| Bipartite NLSs | ||
| Nucleoplasmin | KRPAATKKAGQAKKKKLDK | 36 |
| NIN2 | RKKRKTEEESPLKDKAKKSK | 40 |
| Plk1 | RRRSLLELHKRRK | 47 |
| Parafibromin | KRAADEVLAEAKKPR | 48 |
| PY NLSs | ||
| M9 | FGPMKGGNFGGRSSGPY | 49 |
| TAP | VAMSDAQDGPRVRYNPY | 50 |
| Candidate NLSs | ||
| β-Arrestin-1 | 157KIHKRNSVRLVIRK170 | |
| β-Arrestin-1 | 100LQERLIKKLGEHAYP114 | |
Loss of Inducible Nuclear Translocation of β-Arrestin-1 by Mutation of NLS
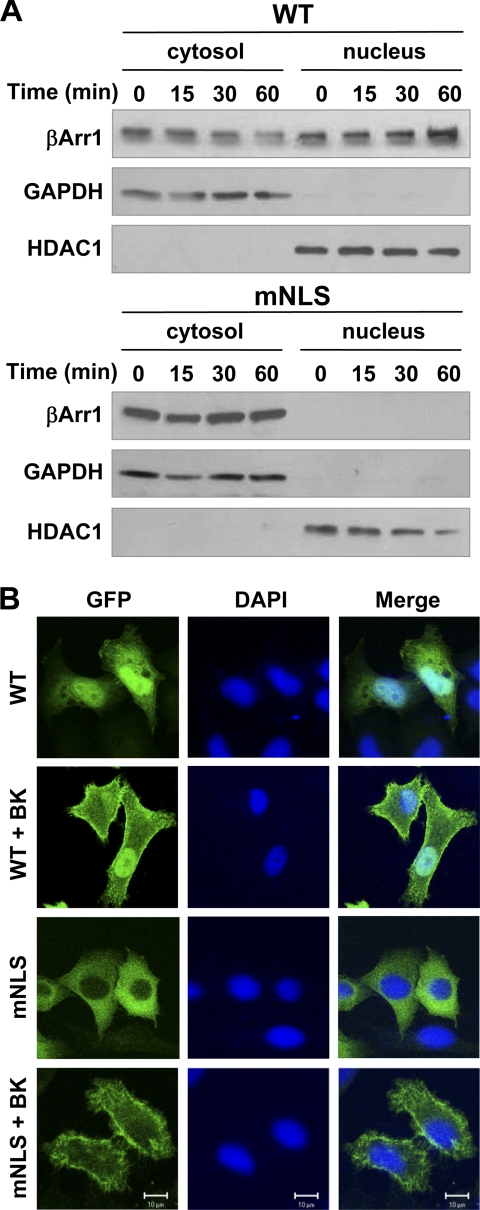
Mutation of the candidate NLS in β-arrestin-1 leads to a loss of its nuclear expression at resting state. However, this does not eliminate the possibility that activation-dependent changes to β-arrestin-1 could impact its nuclear translocation. To test this possibility, HeLa cells transfected to express the BK receptor B2BKR and either wild type or mutant β-arrestin-1 constructs were stimulated with BK and assayed for the appearance of β-arrestin-1 in the nucleus. Increased nuclear localization of the wild type β-arrestin-1 was observed following BK stimulation (Fig. 2A). Unlike wild type β-arrestin-1, there was no inducible nuclear translocation of the mNLS mutant following BK stimulation (Fig. 2A, lower panel). To ensure that the loss of function was specific and not caused by misfolding of the mutated protein, we examined whether the mNLS mutant was able to translocate to the membrane upon agonist stimulation. The results showed that both the wild type and mNLS β-arrestin-1 moved to the plasma membrane in BK-stimulated cells (Fig. 2B), suggesting that this function of mNLS was retained. This result also suggests that membrane and nuclear translocation of β-arrestin-1 might be independently regulated.
FIGURE 2.
Effects of NLS mutation on nuclear translocation of β-arrestin-1. In A, HeLa cells were transfected with expression constructs of FLAG-tagged wild type (WT, upper panels) or NLS mutant (mNLS, lower panels) β-arrestin-1 together with the B2BKR expression construct. Forty-eight hours after transfection, cells were serum starved for 4 h and then stimulated with 500 nm BK for the indicated time points. Western blot analysis of cytosolic and nuclear fractions was performed to determine the translocation of β-arrestin-1 from the cytosolic compartment to nucleus. The protein content of the nuclear fraction was ∼4-fold higher than that of cytosolic fraction in the gel. GAPDH and HDAC1 were used as cytosolic and nuclear loading controls, respectively. In B, HeLa cells were transfected and stimulated for 5 min as described above and then fixed and stained with DAPI to visualize nuclei. Confocal images were taken for β-arrestin-1 translocation to the plasma membrane. Note the morphological changes in BK-stimulated cells due to agonist-induced activation of small GTPases.
β-Arrestin-1 Interacts with Nuclear Import Machinery
In live cells proteins below ∼40 kDa can passively diffuse into the nucleus, whereas larger proteins must be transported into the nucleus in an energy-dependent process (29, 30). Although several import pathways have been identified, many proteins use the importin β carrier proteins for nuclear transport (28). To examine if β-arrestin-1 NLS mediates nuclear translocation through this mechanism, we performed co-immunoprecipitation experiments between WT and mNLS β-arrestin-1 and members of the import machinery. As can be seen in Fig. 3A, following BK stimulation there was a stimulus-dependent increase in the association between β-arrestin-1 and importin β1, indicating active transport into the nucleus. A reciprocal immunoprecipitation showed similar results (Fig. 3C). The β-arrestin-1 NLS mutant mNLS was unable to interact with the nuclear import machinery following BK stimulation (Fig. 3, B and D). This loss of function suggests the mutated residues in mNLS β-arrestin-1 are important for mediating interaction with the nuclear import machinery.
FIGURE 3.
Interaction of β-arrestin-1 with the nuclear import machinery. HeLa cells were transfected with the B2BKR and FLAG-tagged WT (A) or mNLS (B) β-arrestin-1 constructs. After 48 h, cells were serum starved and then stimulated with 500 nm BK for the indicated time points. FLAG-tagged β-arrestin-1 was immunoprecipitated from total cell lysate, and samples were analyzed by Western blotting for detection of the co-immunoprecipitated importin β1. Samples were also analyzed for detection of FLAG as an IP control. Lysate samples were analyzed to determine equivalent expression of importin β1 in WT and mNLS samples. Reciprocal immunoprecipitation with importin β1 and either WT (C) or mNLS β-arrestin-1 (D) were also performed.
Nuclear β-Arrestin-1 Enhances NF-κB-dependent Transcriptional Activity
Previously, we have shown that β-arrestin-1 is able to influence the activity of NF-κB following various GPCR activation (7, 16). We sought to determine whether the nuclear localization of β-arrestin-1 affects NF-κB activation. To examine this we tested our nuclear transport-deficient mutants in an NF-κB luciferase assay. As shown in Fig. 4A, the wild type β-arrestin-1 is able to increase NF-κB-driven luciferase expression following BK stimulation. Both the Δ107–191 and mNLS β-arrestin-1 were significantly less capable of potentiating NF-κB compared with the WT β-arrestin-1 (p < 0.01), suggesting that nuclear localization of β-arrestin-1 is required for optimal enhancement of NF-κB activity (Fig. 4A) and cytosolic accumulation of β-arrestin-1 alone is insufficient for augmenting NF-κB activation. To exclude the possibility that mutation in this region of β-arrestin-1 alters its interaction with key components of the NF-κB activation pathway, we examined another β-arrestin-1 construct containing the NES+ from β-arrestin-2. The NES+ construct was minimally altered in its sequence (Fig. 1B), yet cells expressing this construct also showed a significant loss of enhancement in NF-κB activation (p < 0.01) (Fig. 4A). In control experiments, we found that the loss of enhancement in NF-κB activation was not due to inefficient expression of the β-arrestin-1 mutants (Fig. 4A, Western blot), nor was it the result of lack of association with p65/RelA (Fig. 4B), as this function was unaltered with the mutations. These results suggest that nuclear localization of β-arrestin-1 is critical for its ability to potentiate NF-κB transcriptional activity.
FIGURE 4.
Effect of nucleus-localized β-arrestin-1 on NF-κB activity. NF-κB luciferase assays (A) were conducted using HeLa cells transfected with a 3× κB reporter, a β-galactosidase reporter, the B2BKR, and various β-arrestin-1 constructs. Forty-eight hours after transfection, the cells were serum starved and then stimulated with BK or vehicle for 4 h. Samples were then harvested and assayed for luciferase expression. The results were normalized against the activity of a co-transfected β-galactosidase construct. **, p < 0.01. Proper protein expression in the luciferase assay was verified by Western blotting and shown in bottom panel. In B, p65/RelA binding to the mutant forms of β-arrestin-1 was analyzed by immunoprecipitation of FLAG-tagged β-arrestin-1 from HeLa cells, followed by analysis of co-immunoprecipitation of p65/RelA by Western blotting. Blots shown are representative of at least 3 independent experiments.
Nuclear β-Arrestin-1 Regulates Post-translational Modification of p65/RelA
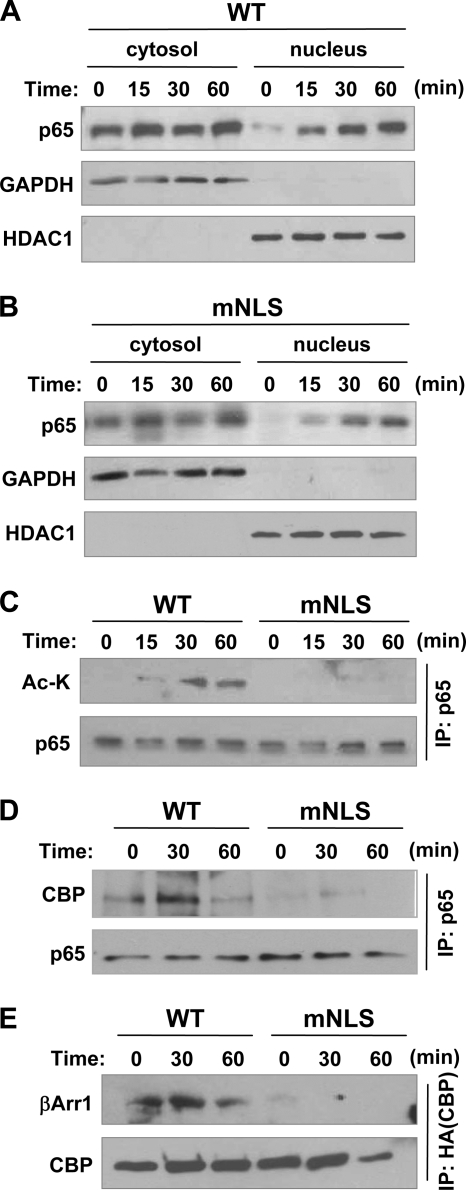
Next we determined how β-arrestin-1 regulates NF-κB activation in the nucleus. Because β-arrestin-1 formed a complex with p65/RelA in the nucleus, we investigated if mutation of the NLS in β-arrestin-1 affects the nuclear translocation of p65/RelA. Following BK stimulation, there was no apparent difference in nuclear localization of p65/RelA in cells expressing the wild type or mutant β-arrestin-1 (Fig. 5, A and B). Interestingly, there was a time-dependent accumulation of p65/RelA in the nucleus, whereas reduction in the cytosolic fraction was less obvious. This may be due to only a fraction of total p65/RelA translocating to the nucleus upon agonist stimulation. Because p65/RelA is modified in the nucleus by acetylation, which regulates its efficiency of transcriptional control (31, 32), we examined whether the nuclear β-arrestin-1 influenced acetylation of p65/RelA. We observed a significant loss of acetylation of p65/RelA in cells expressing the mNLS mutant compared with the wild type (Fig. 5C). Consistent with the loss of p65/RelA acetylation, there was a loss of association between p65/RelA and the acetyltransferase CBP (Fig. 5D) and between the mNLS mutant and CBP (Fig. 5E), which likely resulted from failed entry of the β-arrestin-1 mutant into the nucleus.
FIGURE 5.
Effect of β-arrestin-1 in regulating the post-translational profile of p65/RelA in the nucleus. HeLa cells transfected with either WT (A) or mNLS (B) β-arrestin-1 along with the B2BKR were stimulated with 500 nm BK and fractionated. Translocation of p65/RelA into the nucleus was analyzed by Western blotting. GAPDH and HDAC1 were used as cytosolic and nuclear controls, respectively. In C, cell lysate from WT β-arrestin-1 or mNLS-expressing cells stimulated with BK were immunoprecipitated with anti-p65/RelA. Acetylation of p65/RelA was then analyzed by probing p65/RelA immunoprecipitates with an anti-acetyl-lysine antibody by Western blotting. In D, HeLa cells were transfected with FLAG-tagged WT or mNLS β-arrestin-1 and the B2BKR construct. After transfection, cells were starved for 4 h and then stimulated with 500 nm BK for the indicated time points. Following stimulation, whole cell lysate was immunoprecipitated for p65/RelA. IP samples were then analyzed by Western blot for the co-immunoprecipitation of CBP. The blot for p65 serves as a loading control. Representative blots from at least 3 experiments are shown. In E, HeLa cells were transfected with FLAG-tagged WT or mNLS β-arrestin-1, the B2BKR, and HA-tagged CBP constructs. Cells were starved for 4 h and then stimulated with 500 nm BK for the indicated time points. Following stimulation, whole cell lysate was immunoprecipitated for HA-CBP. IP samples were then analyzed by Western blot for the co-immunoprecipitation of FLAG-β-arrestin-1. The blot for HA-CBP serves as a loading control. Representative blots from at least 3 experiments are shown.
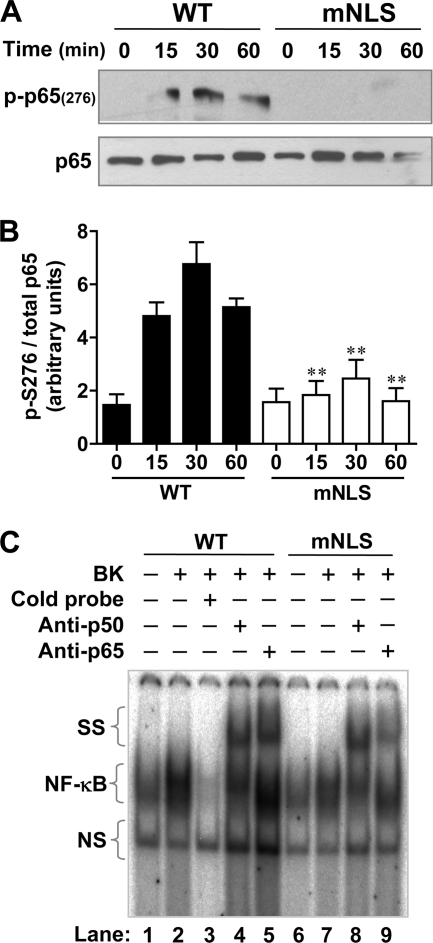
Functionally, acetylation of p65/RelA has been reported to be dependent upon its phosphorylation (33), and it is known that p65/RelA is phosphorylated at Ser276 in the nucleus by MSK1 (34). Therefore, we next examined the effect that NLS mutation of β-arrestin-1 might have on these modifications. Following stimulation with BK, there is a time-dependent increase in the phosphorylation of p65/RelA at Ser276 in cells expressing wild type β-arrestin-1, whereas in cells expressing the mNLS mutant there was a significant reduction of this phosphorylation at Ser276 (Fig. 6A). Quantification of additional blots verified these results (Fig. 6B). Acetylation and phosphorylation of p65/RelA has been shown to be important for transactivation and stable binding to the promoter (reviewed in Ref. 35). To examine this we performed electrophoretic mobility supershift assays with a canonical NF-κB binding sequence, using HeLa cells expressing the WT or mNLS mutant β-arrestin-1. As can be seen in Fig. 6C, BK stimulated an increase in NF-κB·DNA complex formation (lane 2 compared with lane 1). Consistent with the luciferase assay results (Fig. 4A), there was a decrease of protein/DNA binding in the mNLS expressing cells (lane 7) compared with cells expressing the WT β-arrestin-1 (lane 2). To identify the composition of the NF-κB dimers that bound to the DNA probe, we performed supershift assays in which the mobility of the NF-κB·DNA complex was shifted upwards with the addition of specific antibodies to p65/RelA or p50. With the addition of the individual antibody, there was a loss of the NF-κB·DNA complex corresponding to the relative size of p50 and p65/RelA. These results demonstrate the presence of a p50/RelA heterodimer, although the experiment does not rule out the presence of other NF-κB proteins. In cells expressing the mNLS mutant, there was not only decreased complex formation but also reduced complex shift. The change was more prominent in the sample containing the anti-p65/RelA antibody (lane 9), suggesting reduced p65/RelA binding to the NF-κB probe. In comparison, the shift with anti-p50 antibody (lane 8) was proportional to the NF-κB·DNA complex (lane 7).
FIGURE 6.
β-Arrestin-1 regulation of p65/RelA phosphorylation and DNA binding. In A, HeLa cells were transfected with WT or mNLS β-arrestin-1 and the B2BKR construct. Following serum starvation for 4 h, the cells were stimulated with 500 nm BK. Total cell lysate was analyzed by Western blotting using an antibody against phospho-p65/RelA at Ser276. Total p65/RelA was used as a loading control. Quantification and statistical analysis of phosphorylation of Ser276 was performed and data are shown in B. **, p < 0.01 compared WT samples at the same time points. In C, electrophoresis mobility supershift assay for NF-κB was conducted using HeLa cells expressing WT or mNLS β-arrestin-1 and the B2 BK receptor. After serum starvation for 4 h, the cells were stimulated with either vehicle or 500 nm BK for 60 min. Nuclear fractions were obtained and incubated with a radiolabeled NF-κB consensus probe. The specificity of probe binding was verified by competition with a cold (unlabeled) probe, used in 100-fold excess. Supershift with an anti-p50 or anti-p65/RelA antibody was performed to identify which NF-κB proteins were responsible for DNA binding. NS, nonspecific species. SS, supershifted bands.
Lys157 Is Critical to Nuclear Localization of β-Arrestin-1
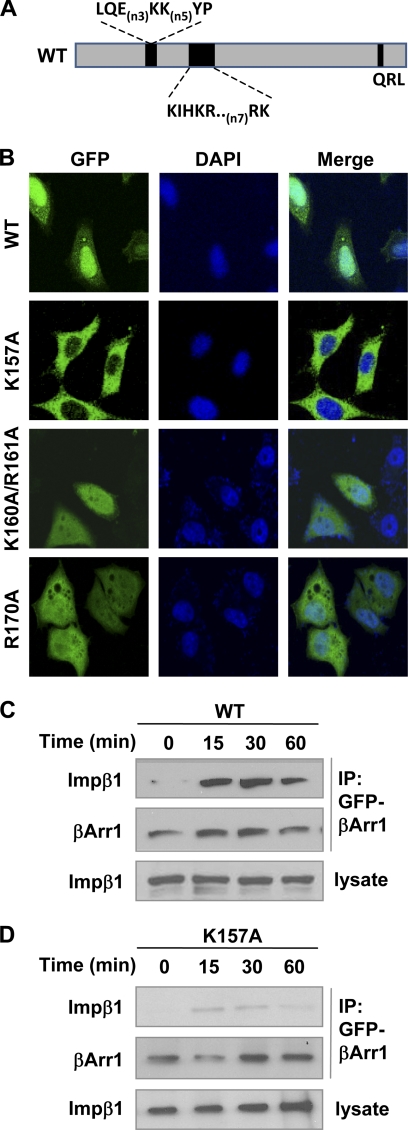
Because the construction of mNLS involves substitution of a total of 7 amino acids, potential alteration of the overall protein structure was a concern, although the mNLS mutant retained its ability to translocate to the plasma membrane upon BK stimulation (Fig. 2B) and to bind p65/RelA (Fig. 4B). Moreover, based on the crystal structures of β-arrestins, the 5 charged residues in the bipartite NLS (157–161/169,170) are located on different sides of the protein, suggesting that only one of the bipartite NLS is actually responsible for nuclear localization. To address this, we mutated charged amino acids in the bipartite NLS to Ala in combination or individually (Fig. 7A). This work identified Lys157 as being potentially important to β-arrestin-1 nuclear localization. The K157A mutant was absent from the nucleus in transfected HeLa cells, whereas Ala substitution of other charged residues in this region did not affect β-arrestin-1 nuclear localization (Fig. 7B). The inability of this mutant β-arrestin-1 to translocate to the nucleus might be attributed to a failure to interact with importin β1 (compare Fig. 7, C with D). In functional assays, the ability of the K157A mutant to enhance p65/RelA phosphorylation at Ser276 was markedly reduced compared with wild type controls (Fig. 8A, quantified in B), Another nuclear event examined was DNA binding of the NF-κB proteins. In cells transfected with the K157A mutant, BK-induced DNA binding was drastically decreased when compared with cells transfected with the wild type β-arrestin-1 (Fig. 8C). To examine a role for β-arrestin-1 in gene transcription, we measured the IL-1β mRNA level in BK-stimulated HeLa cells that were transfected with wild type β-arrestin-1, the K157A or mNLS mutant. The enhancement effect of β-arrestin-1 on IL-1β production was significantly reduced in cells expressing either of the mutants. These results show that the phenotypical changes of the K157A and mNLS mutants are very similar, suggesting that amino acids such as Lys157 are critical to nuclear localization of β-arrestin-1.
FIGURE 7.
Mutation of Lys157 abrogates β-arrestin-1 nuclear localization and binding to importin β1. A, a schematic representation of β-arrestin-1 and the identified NLS region. B, confocal microscopy images showing the localization of β-arrestin-1-GFP, K157A-GFP, K160A/R161A-GFP, and R170A-GFP in HeLa cells. Forty-eight hours after transfection, cells were serum starved for 4 h and then fixed for imaging. DAPI was used to visualize nuclei. Images are representative of at least 3 independent experiments. The interaction of wild type β-arrestin-1 (C) and the K157A mutant (D) with importin β1 was evaluated using the procedures described in the legend to Fig. 4, in HeLa cells transfected with B2BKR and FLAG-tagged wild type or K157A β-arrestin-1 constructs. Representative blots from a total of 3 independent experiments are shown.
FIGURE 8.
β-Arrestin-1 mutation at Lys157 alters p65/RelA phosphorylation and NF-κB activation. In A, phosphorylation of p65/RelA at Ser276 was detected using procedures described in the legend to Fig. 7A. Quantification and statistical analysis of phosphorylation of Ser276 was performed and results are shown in B. *, p < 0.05 and **, p < 0.01, compared with WT samples at the same time points, based on 3 experiments. C, EMSA showing reduced binding of NF-κB with the K157A mutation. A representative autoradiogram is shown. In D, quantitative RT-PCR was performed on RNA from HeLa cells expressing WT or mNLS β-arrestin-1 and the B2BKR. After serum starvation overnight, the cells were stimulated with either vehicle or 500 nm BK for the indicated time points. Total RNA was isolated as described under “Experimental Procedures” and the transcript level of IL-1β was quantified by qPCR. Results are representative of 3 independent experiments.
DISCUSSION
The presence of a nuclear localization sequence in the β-arrestins is a debated subject. However, previously published reports have clearly shown that β-arrestin-1 is actively imported into the nucleus (20), and that an intact N domain (amino acids 1–185) is required for this import (27). Due to this we limited our exploration of the molecule to that region. Initially, loss of nuclear localization of β-arrestin-1 was obtained with a mutant where 83 residues had been deleted from the N domain (Δ107–191). Presumably this level of truncation of the molecule would lead to a severe disruption of its functions due to alteration of the global structure. As such, we sought to identify a smaller region within this domain that might yield similar results. Manual analysis of the primary structure of β-arrestin-1 suggested that the residues between Lys157 and Lys170 might be important, as they bore resemblance to the canonical bipartite NLS of nucleoplasmin (36). Thus, we mutated just those residues comprising a “basic cluster” in β-arrestin-1 to alanine. The resulting mutant did indeed lose nuclear localization in both basal and stimulated conditions. Although this mutant contained a dramatic 7-residue substitution, we observed a significant loss of function with the much less dramatic single-point mutation of K157A. We feel this is an important observation as based on published reports of the structure of bovine β-arrestin-1 (37), residues Lys157, Lys160, and Arg161 are not exposed simultaneously with residues Arg169 and Lys170. In our functional assays, mutation at Lys157 produced a phenotype similar to that of the 7-residue substitution in mNLS. This is the first report of identification of a novel nuclear localization sequence in β-arrestin-1. Our data suggest that this sequence mediates active transport of β-arrestin-1 into the nucleus via interaction with importin β1, a member of the karyophorin family of nuclear import proteins. Defined importin β binding motifs include stretches of basic amino acids (38), Arg-Gly-rich sequences (39), and sequences similar to the M9 NLS containing the YP motif (40). The variety of NLS in this category shows how difficult it is to define a consensus motif for importin β binding. At this time it is unclear if β-arrestin-1 follows the more classical importin β-mediated nuclear import or bind to importin β proteins directly. Alternatively, because our experiments measure the formation of a complex rather than individual protein interaction, it may be possible that β-arrestin-1 is transported into the nucleus via binding to an unknown protein carrier that is present in the complex with importin β1. However, this is less likely because it was previously shown that β-arrestin could be actively imported into the nucleus in an in vivo import assay (20), where the unknown carrier may not exist. An additional finding is that the ability of β-arrestin-1 to translocate and function in the nucleus appears to be independent of its membrane and cytosolic functions, as our results show that mNLS is fully capable of translocating to the plasma membrane upon BK stimulation and to bind p65/RelA. We would predict that β-arrestin-2 is also able to bind to the nuclear import machinery as it contains the same sequence at its corresponding residues. In addition, importin β1 has been identified as a potential binding partner for β-arrestin-2 by proteomic analysis (8). Although both β-arrestins are imported into the nucleus, the nuclear function is most likely β-arrestin-1-specific because β-arrestin-2 is rapidly exported from the nucleus due to a strong leucine-rich nuclear export signal in its C terminus.
An interesting finding of this study is that all residues within the identified NLS, including Lys157, are located within the highly mobile “hinge” region of β-arrestin-1 (37), which has proven to be a “hot spot” of residues required for important arrestin functions such as IP6 binding (41) and receptor binding (42). In fact, in their previous report Milano et al. (41) speculated about the possibility of a 8-residue basic nuclear localization sequence in the β-arrestins that is masked by IP6 binding and oligomerization. Signal masking of the NLS is a common mechanism used to regulate the nuclear import of many proteins (43). We would predict that these residues are both necessary and sufficient for nuclear localization of β-arrestins, as it has been shown that β-arrestin-2 is actively imported into the nucleus of cells in an in vivo nuclear import assay in yeast (20). In that assay, β-arrestin-2 with its NES mutated was fused to a Gal4 activation domain that was attached to a LexA DNA binding domain lacking its NLS. The chimeric construct was introduced into cells containing a lacZ reporter gene. The β-arrestin-2 fusion protein was able to direct the expression of lacZ, indicating that β-arrestin-2 is actively imported into the nucleus (20). These findings are consistent with our results and together they indicate an important role of the NLS in β-arrestin-1 for its nuclear localization.
β-Arrestins are found to regulate NF-κB activation, but the underlying mechanisms are not fully understood. Several possibilities are suggested based on published reports and data from the present study. In the cytosolic compartment, β-arrestins bind to several NF-κB proteins including p105 (13) and the inhibitory protein IκBα (14, 15), thus stabilizing the NF-κB complex in resting cells. Stimulation of cells with various agonists for GPCRs and cytokine receptors leads to NF-κB activation, indicating that the activation signals are able to overcome the inhibitory function of endogenous β-arrestins. Exactly how β-arrestins dissociate from the NF-κB proteins and allow nuclear translocation of the NF-κB proteins has yet to be determined. In agonist-stimulated cells, both β-arrestins serve as scaffolding proteins facilitating diverse signaling events, including those leading to NF-κB activation. We have previously shown that β-arrestin-1 promotes NF-κB activation via interaction with c-Src following D2 dopamine receptor activation (16). β-Arrestin-1 also enhances NF-κB activation involving Gβγ and Akt (7). Thus, activation of NF-κB downstream of B2BKR is mediated through both G protein-dependent and β-arrestin-1-dependent signaling pathways, and there is a switch of the role for β-arrestins in regulating NF-κB activation upon agonist stimulation. A part of this switch occurs in the cytosolic compartment, whereas results shown in the present study suggest that nuclear translocation of β-arrestin-1 also contribute to the regulation of NF-κB activation. Indeed, the observed loss of β-arrestin-1 nuclear localization in cells expressing the mNLS and NES+ mutants was accompanied by reduced NF-κB activation, which could result from stabilization of the inactive NF-κB complex due to accumulation of β-arrestin-1 in the cytosolic compartment (14). However, it is notable that cytosolic accumulation of mNLS and NES+ had minimal impact on the nuclear translocation of p65/RelA. Unlike what has been described for JNK3 (44), it does not appear that β-arrestin-1 is shuttling the members of the NF-κB pathway into and out of the nucleus, and p65/RelA is able to translocate into the nucleus independent of the translocation of β-arrestin-1. Based on these observations, we believe that accumulation of β-arrestin-1 in the nucleus contributes significantly to NF-κB activation.
Our results demonstrate that one of the mechanisms by which β-arrestin-1 modulates NF-κB activation and transcriptional response involves regulating the recruitment of protein modifiers to p65/RelA in the nucleus and scaffolding them together. Specifically, β-arrestin-1 is able to recruit CBP/p300 and a protein kinase to p65/RelA, which directly contributes to the phosphorylation and acetylation of p65/RelA in the nucleus and increases transcriptional responsiveness. Our results show that mutation of the NLS in β-arrestin-1 diminishes p65/RelA phosphorylation at Ser276 and profoundly decreases acetylation of p65/RelA, thereby confirming the nuclear function of β-arrestin-1 in regulating NF-κB activation. Post-translational modifications of p65/RelA, specifically its acetylation, have been reported to regulate the duration of its occupancy on specific promoters (32). Indeed, our results from electrophoretic mobility shift assays show not only an overall decrease in NF-κB-DNA interaction but also a more pronounced loss of p65/RelA-specific binding to the DNA probe. Transcriptional regulation by β-arrestin-1 was also reported in other studies and contributes to the regulation of expression of p27 and c-fos (18) and IFN-γ (19). Taken together, these findings support an important function of β-arrestin-1 in transcriptional regulation in the nucleus.
In conclusion, our study has led to the identification of a novel, functional NLS in β-arrestin-1. We have also shown a role for nuclear-localized β-arrestin-1 in the regulation of NF-κB activation. Given the potential significance of β-arrestin-1-mediated transcriptional regulation in the nucleus, one of the future research directions is to understand the dynamic process of β-arrestin-1 nuclear translocation in activated cells as well as in primary cells. Understanding how GPCRs, including B2BKR and the δ-opioid receptor, induce β-arrestin-1 nuclear translocation is of interest in uncovering additional physiological functions of β-arrestin-1.
This work was supported, in whole or in part, by National Institutes of Health Grants AI033503, HL077806, and AI040176.
- GPCR
- G protein-coupled receptor
- BK
- bradykinin
- NF-κB
- nuclear factor-κB
- NLS
- nuclear localization sequence
- NES
- nuclear export sequence
- IP
- immunoprecipitation
- B2BKR
- B2 bradykinin receptor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- CBP
- cAMP-response element-binding protein.
REFERENCES
- 1. Benovic J. L., Kühn H., Weyand I., Codina J., Caron M. G., Lefkowitz R. J. (1987) Functional desensitization of the isolated β-adrenergic receptor by the β-adrenergic receptor kinase. Potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc. Natl. Acad. Sci. U.S.A. 84, 8879–8882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lohse M. J., Benovic J. L., Codina J., Caron M. G., Lefkowitz R. J. (1990) β-Arrestin. A protein that regulates β-adrenergic receptor function. Science 248, 1547–1550 [DOI] [PubMed] [Google Scholar]
- 3. Penela P., Elorza A., Sarnago S., Mayor F., Jr. (2001) β-Arrestin- and c-Src-dependent degradation of G-protein-coupled receptor kinase 2. EMBO J. 20, 5129–5138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miller W. E., Maudsley S., Ahn S., Khan K. D., Luttrell L. M., Lefkowitz R. J. (2000) β-Arrestin-1 interacts with the catalytic domain of the tyrosine kinase c-SRC. Role of β-arrestin-1-dependent targeting of c-SRC in receptor endocytosis. J. Biol. Chem. 275, 11312–11319 [DOI] [PubMed] [Google Scholar]
- 5. Povsic T. J., Kohout T. A., Lefkowitz R. J. (2003) β-Arrestin-1 mediates insulin-like growth factor 1 (IGF-1) activation of phosphatidylinositol 3-kinase (PI3K) and anti-apoptosis. J. Biol. Chem. 278, 51334–51339 [DOI] [PubMed] [Google Scholar]
- 6. Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., Caron M. G. (2005) An Akt·β-arrestin 2·PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 122, 261–273 [DOI] [PubMed] [Google Scholar]
- 7. Yang M., He R. L., Benovic J. L., Ye R. D. (2009) β-Arrestin-1 interacts with the G-protein subunits β1γ2 and promotes β1γ2-dependent Akt signaling for NF-κB activation. Biochem. J. 417, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao K., McClatchy D. B., Shukla A. K., Zhao Y., Chen M., Shenoy S. K., Yates J. R., 3rd, Lefkowitz R. J. (2007) Functional specialization of β-arrestin interactions revealed by proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 12011–12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rojanathammanee L., Harmon E. B., Grisanti L. A., Govitrapong P., Ebadi M., Grove B. D., Miyagi M., Porter J. E. (2009) The 27-kDa heat shock protein confers cytoprotective effects through a β2-adrenergic receptor agonist-initiated complex with β-arrestin. Mol. Pharmacol. 75, 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) β-Arrestin-2. A receptor-regulated MAPK scaffold for the activation of JNK3. Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 11. Cervantes D., Crosby C., Xiang Y. (2010) Arrestin orchestrates cross-talk between G protein-coupled receptors to modulate the spatiotemporal activation of ERK MAPK. Circ. Res. 106, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Tang Y., Teng L., Wu Y., Zhao X., Pei G. (2006) Association of β-arrestin and TRAF6 negatively regulates Toll-like receptor-interleukin 1 receptor signaling. Nat. Immunol. 7, 139–147 [DOI] [PubMed] [Google Scholar]
- 13. Parameswaran N., Pao C. S., Leonhard K. S., Kang D. S., Kratz M., Ley S. C., Benovic J. L. (2006) Arrestin-2 and G protein-coupled receptor kinase 5 interact with NFκB1 p105 and negatively regulate lipopolysaccharide-stimulated ERK1/2 activation in macrophages. J. Biol. Chem. 281, 34159–34170 [DOI] [PubMed] [Google Scholar]
- 14. Witherow D. S., Garrison T. R., Miller W. E., Lefkowitz R. J. (2004) β-Arrestin inhibits NF-κB activity by means of its interaction with the NF-κB inhibitor IκBα. Proc. Natl. Acad. Sci. U.S.A. 101, 8603–8607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao H., Sun Y., Wu Y., Luan B., Wang Y., Qu B., Pei G. (2004) Identification of β-arrestin-2 as a G protein-coupled receptor-stimulated regulator of NF-κB pathways. Mol. Cell 14, 303–317 [DOI] [PubMed] [Google Scholar]
- 16. Yang M., Zhang H., Voyno-Yasenetskaya T., Ye R. D. (2003) Requirement of Gβγ and c-Src in D2 dopamine receptor-mediated nuclear factor-κB activation. Mol. Pharmacol. 64, 447–455 [DOI] [PubMed] [Google Scholar]
- 17. Sun J., Lin X. (2008) β-Arrestin-2 is required for lysophosphatidic acid-induced NF-κB activation. Proc. Natl. Acad. Sci. U.S.A. 105, 17085–17090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kang J., Shi Y., Xiang B., Qu B., Su W., Zhu M., Zhang M., Bao G., Wang F., Zhang X., Yang R., Fan F., Chen X., Pei G., Ma L. (2005) A nuclear function of β-arrestin-1 in GPCR signaling. Regulation of histone acetylation and gene transcription. Cell 123, 833–847 [DOI] [PubMed] [Google Scholar]
- 19. Mo W., Zhang L., Yang G., Zhai J., Hu Z., Chen Y., Chen X., Hui L., Huang R., Hu G. (2008) Nuclear β-arrestin-1 functions as a scaffold for the dephosphorylation of STAT1 and moderates the antiviral activity of IFN-γ. Mol. Cell 31, 695–707 [DOI] [PubMed] [Google Scholar]
- 20. Scott M. G., Le Rouzic E., Périanin A., Pierotti V., Enslen H., Benichou S., Marullo S., Benmerah A. (2002) Differential nucleocytoplasmic shuttling of β-arrestins. Characterization of a leucine-rich nuclear export signal in β-arrestin-2. J. Biol. Chem. 277, 37693–37701 [DOI] [PubMed] [Google Scholar]
- 21. Terry L. J., Shows E. B., Wente S. R. (2007) Crossing the nuclear envelope. Hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416 [DOI] [PubMed] [Google Scholar]
- 22. Leung S. W., Harreman M. T., Hodel M. R., Hodel A. E., Corbett A. H. (2003) Dissection of the karyopherin alpha nuclear localization signal (NLS)-binding groove. Functional requirements for NLS binding. J. Biol. Chem. 278, 41947–41953 [DOI] [PubMed] [Google Scholar]
- 23. Yoneda Y., Hieda M., Nagoshi E., Miyamoto Y. (1999) Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct. Funct. 24, 425–433 [DOI] [PubMed] [Google Scholar]
- 24. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi Y., Feng Y., Kang J., Liu C., Li Z., Li D., Cao W., Qiu J., Guo Z., Bi E., Zang L., Lu C., Zhang J. Z., Pei G. (2007) Critical regulation of CD4+ T cell survival and autoimmunity by β-arrestin 1. Nat. Immunol 8, 817–824 [DOI] [PubMed] [Google Scholar]
- 26. Pan Z. K., Zuraw B. L., Lung C. C., Prossnitz E. R., Browning D. D., Ye R. D. (1996) Bradykinin stimulates NF-κB activation and interleukin 1β gene expression in cultured human fibroblasts. J. Clin. Invest. 98, 2042–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang P., Wu Y., Ge X., Ma L., Pei G. (2003) Subcellular localization of β-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J. Biol. Chem. 278, 11648–11653 [DOI] [PubMed] [Google Scholar]
- 28. Stewart M. (2007) Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 [DOI] [PubMed] [Google Scholar]
- 29. Davis L. I. (1995) The nuclear pore complex. Annu. Rev. Biochem. 64, 865–896 [DOI] [PubMed] [Google Scholar]
- 30. Panté N., Aebi U. (1996) Molecular dissection of the nuclear pore complex. Crit. Rev. Biochem. Mol. Biol. 31, 153–199 [DOI] [PubMed] [Google Scholar]
- 31. Chen Lf., Fischle W., Verdin E., Greene W. C. (2001) Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293, 1653–1657 [DOI] [PubMed] [Google Scholar]
- 32. Chen L. F., Mu Y., Greene W. C. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen L. F., Williams S. A., Mu Y., Nakano H., Duerr J. M., Buckbinder L., Greene W. C. (2005) NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25, 7966–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vermeulen L., De Wilde G., Van Damme P., Vanden Berghe W., Haegeman G. (2003) Transcriptional activation of the NF-κB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22, 1313–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Perkins N. D., Gilmore T. D. (2006) Good cop, bad cop. The different faces of NF-κB. Cell Death Differ. 13, 759–772 [DOI] [PubMed] [Google Scholar]
- 36. Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence. Identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 [DOI] [PubMed] [Google Scholar]
- 37. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Crystal structure of β-arrestin at 1.9 Å. Possible mechanism of receptor binding and membrane translocation. Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 38. Mühlhäusser P., Müller E. C., Otto A., Kutay U. (2001) Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2, 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leslie D. M., Zhang W., Timney B. L., Chait B. T., Rout M. P., Wozniak R. W., Aitchison J. D. (2004) Characterization of karyopherin cargoes reveals unique mechanisms of Kap121p-mediated nuclear import. Mol. Cell. Biol. 24, 8487–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kleinschmidt J. A., Seiter A. (1988) Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 7, 1605–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Milano S. K., Kim Y. M., Stefano F. P., Benovic J. L., Brenner C. (2006) Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J. Biol. Chem. 281, 9812–9823 [DOI] [PubMed] [Google Scholar]
- 42. Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W. (2011) Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual subtypes. J. Mol. Biol. 406, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaffman A., O'Shea E. K. (1999) Regulation of nuclear localization. A key to a door. Annu. Rev. Cell Dev. Biol. 15, 291–339 [DOI] [PubMed] [Google Scholar]
- 44. Song X., Raman D., Gurevich E. V., Vishnivetskiy S. A., Gurevich V. V. (2006) Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J. Biol. Chem. 281, 21491–21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kalderon D., Richardson W. D., Markham A. F., Smith A. E. (1984) Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311, 33–38 [DOI] [PubMed] [Google Scholar]
- 46. Chida K., Vogt P. K. (1992) Nuclear translocation of viral Jun but not of cellular Jun is cell cycle dependent. Proc. Natl. Acad. Sci. U.S.A. 89, 4290–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taniguchi E., Toyoshima-Morimoto F., Nishida E. (2002) Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J. Biol. Chem. 277, 48884–48888 [DOI] [PubMed] [Google Scholar]
- 48. Hahn M. A., Marsh D. J. (2005) Identification of a functional bipartite nuclear localization signal in the tumor suppressor parafibromin. Oncogene 24, 6241–6248 [DOI] [PubMed] [Google Scholar]
- 49. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Truant R., Kang Y., Cullen B. R. (1999) The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J. Biol. Chem. 274, 32167–32171 [DOI] [PubMed] [Google Scholar]