Background: Two SH3 domains investigated are cross-reactive in peptide binding, yet also display highly divergent specificities.
Results: These domains interact with peptides in a structurally similar manner, but differ in interaction strength formed with different regions of peptides.
Conclusion: Distinct specificities of these domains arise from differences in local binding energetics.
Significance: A new mechanism for generating binding specificity is demonstrated.
Keywords: Adaptor Proteins, Energetics, Ligand-binding Protein, Protein Structure, SH3 Domains, Binding Specificity, Inhibitory Interactions, Protein Interaction Domains
Abstract
The yeast Nbp2p SH3 and Bem1p SH3b domains bind certain target peptides with similar high affinities, yet display vastly different affinities for other targets. To investigate this unusual behavior, we have solved the structure of the Nbp2p SH3-Ste20 peptide complex and compared it with the previously determined structure of the Bem1p SH3b bound to the same peptide. Although the Ste20 peptide interacts with both domains in a structurally similar manner, extensive in vitro studies with domain and peptide mutants revealed large variations in interaction strength across the binding interface of the two complexes. Whereas the Nbp2p SH3 made stronger contacts with the peptide core RXXPXXP motif, the Bem1p SH3b domain made stronger contacts with residues flanking the core motif. Remarkably, this modulation of local binding energetics can explain the distinct and highly nuanced binding specificities of these two domains.
Introduction
Protein-protein interactions are often mediated by conserved modular domains that bind to short linear peptide motifs. Although the members of a single domain family generally recognize the same consensus motif in their targets, individual domains possess unique binding specificities and recognize different variations of this motif. In many cases, this intrinsic specificity of individual domains plays a crucial role in directing them to their relevant biological targets and preventing aberrant cross-reactivity with nonphysiological targets (1, 2). Thus, to comprehend fully the contribution of protein interaction domains to the functioning of cellular pathways, insight into the principles that govern their intrinsic binding specificity is required. This knowledge will ultimately allow the accurate prediction of binding specificities, which will greatly increase our ability to map and understand the signal transduction pathways within cells. In this study, we explore the factors controlling intrinsic binding specificity by investigating the specificity determinants of two yeast SH32 domains that can bind to the same target peptides.
SH3 domains are among the most common protein interaction domains and are present in proteins with diverse cellular roles including signal transduction, cytoskeleton regulation, and membrane trafficking (3). These domains are ∼60 residues long and are composed of five β-strands, which are sequentially joined by the RT, N-Src, and distal loops, and a short 310-helix. SH3 domains generally recognize peptide sequences containing either +XXPXXP (class I) or PXXPXX+ (class II) “core” motifs (where X can be variety of residues and + represents either a Lys or Arg residue). The prolines of the core motif are accommodated in conserved hydrophobic grooves on one surface of the SH3 domain, which has been referred to as Surface I (1, 4). The positively charged residue is packed against a highly conserved tryptophan and also interacts with negatively charged residues in the RT loop. Although interactions with the core motif serve as a common anchor for binding by most SH3 domains, the specificity of individual SH3 domains is strongly influenced by interactions between target peptide residues flanking the core motif and a variable surface in the SH3 domain located between RT and N-Src loops (1, 5–8), which has been referred to as Surface II (1, 4). The interaction of this “extended” region of target peptides with Surface II is complicated because this surface is relatively flat and displays little conservation across the SH3 domain family. In addition, the conformation of peptides interacting with this surface is variable (4), and different peptides binding the same domain may adopt distinct conformations when binding this surface (9, 10). Due to the complex nature of Surface II and the relative dearth of studies on the interaction of SH3 domains with extended peptide sequences, our understanding of these interactions is limited. Thus, accurate prediction of the specificities of SH3 domains is also hampered.
We have investigated the specificity of the yeast Nbp2p SH3 (NbpSH3) and Bem1p SH3b (BemSH3b) domains. This pair of SH3 domains is unusual because they recognize the same consensus binding motif (see Fig. 3A), even though they are present in nonhomologous proteins and have a relatively low sequence identity of 36% (the sequence identity of any two randomly chosen SH3 domains is ∼30%) (11). Nbp2p is an adaptor protein that recruits Ptc1p phosphatase through interactions mediated by its SH3 domain. Binding of the NbpSH3 domain to PXXP-containing sites in the Pbs2p and Bck1p kinases is required for down-regulation of the high osmolarity glycerol and cell wall integrity MAP kinase pathways, respectively (12–14). Bem1p acts as an adaptor for multiple proteins involved in establishing cell polarity including Cdc42p GTPase and the Cdc42p-activated kinases, Ste20p and Cla4p (15–19). The BemSH3b domain binds to the PXXP motifs within Ste20p and Cla4p kinases and also binds Cdc42p itself in a PXXP-independent manner (16, 17, 19, 20). The interaction between the BemSH3b domain and Cdc42p involves a unique C-terminal extension (Cdc42-interacting (CI) subdomain) in the BemSH3b domain, which is also required for the folding of the whole domain (19, 21). The structure of the BemSH3b domain solved in complex with a peptide from Ste20p showed that the CI subdomain is intimately packed against the SH3 domain and is involved in the interaction with the peptide (21).
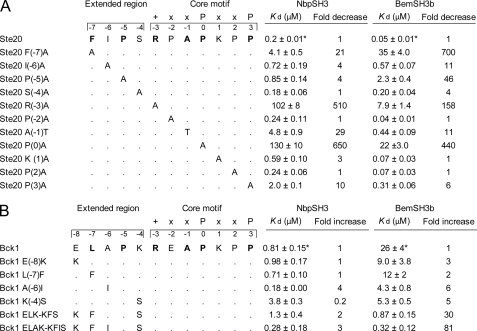
FIGURE 3.
Mutational analysis of Ste20 and Bck1 peptides. A, binding of the NbpSH3 and BemSH3b domains to wild-type and mutant Ste20 peptides. Ste20 residues that are part of the ΨXPXRXAPXXP consensus recognized by NbpSH3 and BemSH3b domains are shown in bold. The Kd values of interactions and the -fold decrease in binding relative to the wild-type Ste20 peptide are indicated. The Kd values are represented as means ± S.E. All measurements were repeated at least twice. The values marked by asterisks were taken from our previous study (23). B, binding of the NbpSH3 and BemSH3b domains to wild-type and mutant Bck1 peptides represented in the same manner as in A.
The similarity in binding specificities of the NbpSH3 and BemSH3b domains was first detected when they were found to both bind the same PXXP-containing motif in Ste20p (20). To analyze this cross-reactivity further, we previously tested binding of the NbpSH3 and BemSH3b domains to peptides from six different yeast proteins that are either proven or likely biologically relevant target proteins for these domains (Ste20p, Cla4p, Pbs2p, Bck1p, Skm1p, and Boi2p). These peptides share a class I consensus sequence, ΨXPXRXAPXXP (where Ψ is a hydrophobic residue), which was derived for the NbpSH3 domain using phage display (22). We found that although the NbpSH3 and BemSH3b domains bound to all of these sites, some sites were bound with equally strong affinities, whereas other sites were bound with remarkably different affinities by the two domains (23). For example, both domains bound to the Ste20 peptide with submicromolar affinities (see Fig. 3A), yet the NbpSH3 domain bound the Bck1 peptide with a Kd of 0.8 μm, whereas the Kd of the BemSH3b domain for this peptide was only 26 μm (see Fig. 3B). These data demonstrated that even though the NbpSH3 and BemSH3b domains share a consensus binding sequence and interact with equally high affinity with some peptides, each domain possesses its own unique binding specificity. We also identified a residue within the BemSH3b domain (Lys14), which serves to prevent high affinity binding to certain sites (e.g. Bck1) and plays a general role in maintaining the BemSH3b domain specificity required for its optimal function (23).
The ability of the NbpSH3 and BemSH3b domains to bind the same extended peptides with high affinity while strongly discriminating between other peptides provides a unique model system to examine the determinants of SH3 domain specificity. In this study, we address two main questions regarding the binding behavior of these domains. First, how do these two domains with very different sequences still recognize the same consensus motif and bind some peptides with almost identical affinities? Second, how do these domains also exhibit distinctive specificities toward other peptides? To answer these questions, we have solved the structure of the NbpSH3 domain in complex with the Ste20 peptide and compared it with the previously determined structure of the BemSH3b-Ste20 complex (21). Using a combination of structural analysis and in vitro assays involving mutant domains and peptides, we have dissected the binding mechanisms of these two SH3 domains. Based on our results, we describe a new general mechanism that explains the unique binding specificities of the NbpSH3 and BemSH3b domains. Our analysis also allowed us to identify previously unrecognized sites in yeast proteins that are highly selective for the BemSH3b domain.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Yeast NbpSH3 (Nbp2p residues 110–172) and BemSH3b (Bem1p residues 155–252) domains were expressed from pET21d (Novagen) vector with a C-terminal His6 tag. Ste20(468–483), Bck1(800–815), Cla4(15–25), Cla4(451–461), and Boi1(391–401) peptidesSample B contained 0.5 used in the binding assays were expressed as C-terminal fusions to bacteriophage λ cI repressor carrying a C-terminal His6 tag, as described previously (24). All proteins were purified using Ni-affinity chromatography and dialyzed against 50 mm sodium phosphate, pH 6.8, 100 mm NaCl buffer.
NMR Spectroscopy and Structure Calculation
A peptide corresponding to Ste20p residues 468–484 (GKFIPSRAPKPPSSA) was chemically synthesized (CanPeptide). For NMR experiments, two samples were prepared. Sample A contained 0.7 mm 15N13C-labeled NbpSH3 domain and 1.5 mm unlabeled Ste20p peptide in 50 mm phosphate, pH 6.8, 100 mm NaCl, 0.05% NaN3. Sample B contained 0.5 mm 15N13C-labeled NbpSH3 and 0.5 mm unlabeled Ste20p peptide in 50 mm phosphate buffer, pH 6.8, 100 mm NaCl, and 0.05% NaN3. Backbone and side chain chemical shifts for the NbpSH3 domain were assigned using standard triple-resonance experiments (25) (HNCO, HN(CO)CA, CBCA(CO)NH, HNCACB, C(CO)NH, HCCH-TOCSY, and HCCH-COSY) with sample A. Interproton distances for the NbpSH3 domain were obtained with 15N-edited and 13C-edited NOESY spectra (sample A). Chemical shift assignments and structural restraints for the Ste20p peptide were derived from double-half-filtered two-dimensional 1H-1H TOCSY, COSY, and NOESY experiments (26, 27) (sample B). Intermolecular NOEs between the NbpSH3 domain and Ste20p peptide were obtained from three-dimensional NOESY experiments (sample A) and 13C-edited 13C-filtered three-dimensional NOESY (28) (sample B). All NMR experiments were carried out at 25 °C on a Varian INOVA 500 or 800 MHz spectrometers equipped with pulsed field gradients at the Quebec/Eastern Canada High Field NMR Facility. All data were processed using NMRPipe (29) and subsequent analysis performed using SPARKY (30). Structure calculations were performed using CYANA 2.1 (31). The input constraints included dihedral angle restraints derived using TALOS (32) and hydrogen bond upper and lower distance limits derived from examination of short range NOE data. Manually assigned NOE peaks were fixed in the first cycle of structure calculation and peak assignments. In the remaining cycles, all NOE peak assignments were made by CYANA. The 20 lowest energy structures, of 100 calculated in the final iteration, were used to derive the structure. Structural coordinates and NMR restraints have been deposited in the Protein Data Bank (PDB) with code 2LCS.
Fluorescence-based Binding Assays
SH3 domains at a concentration of 1 μm were titrated with peptide fusion proteins. Binding was monitored by measuring intrinsic tryptophan fluorescence of the SH3 domain (λexcitation = 295 nm, λemission = 326 nm), which increased significantly upon binding of the peptide. The experiments were carried out in 50 mm phosphate, pH 6.8, 100 mm NaCl at 25 °C. All titrations and fluorescence measurements were carried out on an Aviv ATF105 spectrofluorometer equipped with a Microlab 500 series automated titrator. Dissociation constants were calculated as described previously (24).
Yeast Strains and Growth Assays
The bem1Δ and bem1 sh3b F54A strains constructed in the previous study (23) were used in the growth assays. The bem1 sh3b E17Q stain was constructed as described previously (23) from the same genetic background as the bem1Δ and bem1 sh3b F54A strains. Yeast peptone dextrose medium was used for growth assays. The cells were grown overnight, diluted to an A600 of 1.0, and spotted in 5-fold serial dilutions on yeast peptone dextrose plates. The cell growth was detected after 2 days at 30, 40.5, and 41.5 °C.
RESULTS
Structure of NbpSH3-Ste20 Complex
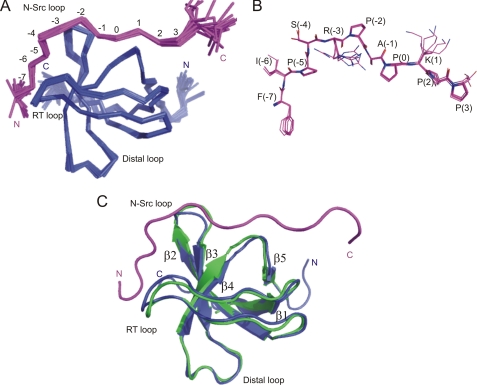
To gain insight into the specificity determinants of the NbpSH3 domain and allow comparison to the BemSH3b domain, we determined the NMR solution structure of the NbpSH3 domain in complex with the Ste20 peptide. We chose the Ste20 peptide because both domains bind it with high affinity, and the structure of a complex of the BemSH3b domain with this peptide was determined previously (21). Standard NMR experiments were used to assign chemical shifts and NOEs to the SH3 domain and the peptide (25). The structure was restrained using intrapeptide, intra-SH3, and intermolecular NOEs, hydrogen bond restraints, and dihedral angle restraints derived from TALOS (32) (Table 1). The 20 lowest energy structures overlay with a backbone r.m.s.d. of 0.22 and 0.15 Å for well defined regions of the SH3 domain and Ste20 peptide, respectively (Table 1 and Fig. 1, A and B). The NbpSH3 domain displays a typical SH3 domain fold and overlays closely (backbone r.m.s.d. of 0.93Å over residues 1–59) with a previously determined crystal structure of the free NbpSH3 domain (PDB code 1YN8, chain B, Fig. 1C). The major differences map to the RT loop and N-Src-loop regions, which are directly involved in peptide binding.
TABLE 1.
Structural statistics for the NbpSH3-Ste20 peptide complex
| Constraints | Structure | ||
|---|---|---|---|
| Input dihedral anglesa (ϕ, ψ) | 44,43 | r.m.s.d. from mean structureb (Å) | |
| NOE upper distance | Backbone atoms (SH3, peptide) | 0.22, 0.15 | |
| Intraresidual | 345 | Heavy atoms (SH3, peptide) | 0.67,0.84 |
| Medium range (1≤|i−j|<4) | 643 | r.m.s.d. from ideal geometry | |
| Long range (|i−j|>4) | 1,030 | Distance (Å) | 0.004 |
| Intermolecular | 184 | Angles (°) | 0.8 |
| Hydrogen bondc | Ramachandran plot | ||
| Lower | 32 | Favored regions | 82.3% |
| Upper | 32 | Additionally allowed regions | 17.7% |
| Generously allowed regions | 0.1% | ||
| Disallowed regions | 0.0% | ||
a Dihedral angles restraints were derived using TALOS (32).
b The r.m.s.d. is indicated for well defined regions. NbpSH3 domain, residues −1 to 61, numbering according to Larson and Davidson (11); Ste20p peptide, residues −7 to 3, numbering according to Lim et al. (37).
c Based on the predicted hydrogen bonds that were derived from the examination of secondary structure NOEs.
FIGURE 1.
Structure of NbpSH3-Ste20 complex (PDB code 2LCS). A, overlay of the 20 lowest energy structures. The NbpSH3 domain is colored blue and the Ste20 peptide magenta. Well ordered peptide positions are numbered according to Lim et al. (37). B, overlay of five low energy Ste20 peptide conformers in the NbpSH3-Ste20 complex corresponding to peptide positions −7 to 3. C, overlay of the NbpSH3 domain crystal structure (green, PDB code 1YN8 chain B) with the NMR structure of the NbpSH3 domain (blue) in complex with the Ste20 peptide (magenta).
Comparison of Nbp2SH3-Ste20 and BemSH3b-Ste20 Complexes
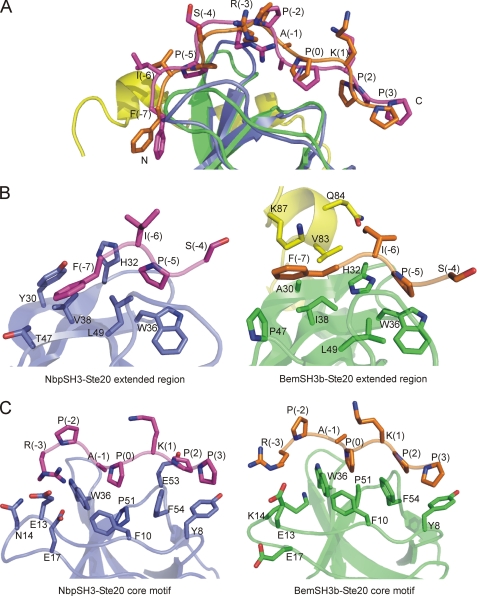
Comparison of the NbpSH3-Ste20 and BemSH3b-Ste20 complexes shows that the Ste20 peptide adopts a similar conformation in the two complexes and contacts the same surfaces in both domains (Fig. 2A). The two complexes overlay with a backbone r.m.s.d. of 1.6 Å over the well defined regions in both structures. These correspond to SH3 domain residues 2–40 and 48–58, and the peptide residues Phe−7 to Pro3. The rest of the peptide backbone (total length of the peptide used in this study was 16 amino acids) is poorly defined in both structures (Fig. 1A) (21). In each complex, the core RXXPXXP motif (positions −3 to +3) of the Ste20 peptide interacts with the SH3 domains in a canonical manner (Fig. 2C). The two XP dipeptides of the PPII helix fit into hydrophobic grooves composed of the Tyr8, Phe10, Pro51, Trp36, and Phe54 residues, which are identical in the NbpSH3 and BemSH3b domains and highly conserved across the SH3 domain family. The peptide Arg−3 residue is packed against Trp36 and positioned close enough to make electrostatic interactions with the Glu residues at positions 13 and 17, which are found in both domains. Consistent with this structural picture, Ala substitutions of Tyr8, Glu13, Glu17, and Phe54 in both domains significantly decreased their binding affinity for the Ste20 peptide (Table 2).
FIGURE 2.
Comparison of the NbpSH3-Ste20 and BemSH3b-Ste20 complexes. A, overlay of the NbpSH3-Ste20 (PDB code 2LCS) and BemSH3b-Ste20 (PDB code 2RQW) complexes focusing on the conformation of the Ste20 peptide, which is numbered as in Fig. 1. The Nbp2SH3 domain is in blue with the bound Ste20 peptide in magenta. The BemSH3b SH3 subdomain is colored green, the CI subdomain is yellow, and the bound Ste20 peptide is orange. B and C, detailed comparison of the NbpSH3 domain (B) and the BemSH3b domain (C) interactions with the core and extended regions of Ste20 peptide. Coloring is the same as in A. SH3 domain residue numbering is according to a standardized system (11).
TABLE 2.
Binding affinities of the NbpSH3 and BemSH3b domain mutants to the Ste20 peptide
| NbpSH3+Ste20 peptide |
BemSH3b+Ste20 peptide |
||||||
|---|---|---|---|---|---|---|---|
| SH3a | Kdb | -Fold | ΔTmc | SH3 | Kd | -Fold | ΔTm |
| μm | °C | μm | °C | ||||
| WT | 0.2 ± 0.01* | 1 | 0 | WT | 0.05 ± 0.01* | 1 | 0 |
| Y8ACore | 9.1 ± 2.0 | 46 | 3.1 | Y8ACore | 0.3 ± 0.1 | 6 | 3.1 |
| E13ACore | 2.5 ± 0.2 | 13 | 0.6 | E13ACore | 0.56 ± 0.1 | 11 | 1.5 |
| N14ACore | 0.13 ± 0.0 | 0.65 | −7.9 | K14ACore | <0.01* | <0.2 | −2.2 |
| E17QCore | >100 | >500 | 4.8 | E17QCore | >100 | >2,000 | 0.3 |
| F54ACore | 56 ± 8.3 | 280 | 10.3 | F54ACore | 44 ± 5 | 880 | 6.5 |
| Y30AExt | 2.5 ± 0.01 | 13 | 2.4 | ||||
| H32AExt | 0.03 ± 0.01* | 0.15 | 4.0 | H32AExt | 0.37 ± 0.05* | 7 | −2.2 |
| V38AExt | 3.7 ± 0.4 | 19 | −9.9 | ||||
| L49AExt | 7.0 ± 0.6 | 35 | −17.9 | L49AExt | 3.9 ± 0.7 | 77 | −2.7 |
a The tested amino acid substitutions are indicated. The Core superscript indicates that the substituted residue interacts with the core region of Ste20 peptide, whereas the Ext superscript indicates that the substituted residue interacts with the extended region of the peptide.
b The affinities are represented as mean value ±S.E. Each measurement was repeated at least twice. The binding affinities which were taken from the previous study (23) are marked by asterisks.
c The stabilities of the mutant domains are reported as change in the temperature-induced denaturation midpoint (Tm) with respect to the wild-type SH3 domains. The Tm values for the wild-type NbpSH3 and BemSH3b domains measured in this study were 53.7 °C and 54.6 °C, respectively.
Many similarities were also seen between the BemSH3b and NbpSH3 domains in their interactions with the extended region of the peptide (positions −7 to −4) (Fig. 2B). In both complexes, conserved peptide residues Phe−7 and Pro−5 (Fig. 3) make extensive contacts with the domain surface. Phe−7 packs against a hydrophobic surface on the NbpSH3 domain composed of Tyr30, Val38, and Thr47. The importance of this surface is supported by the >10-fold reduction in binding affinity to Ste20 peptide caused by Ala substitutions of Tyr30 and Val38 (Table 2). In the BemSH3b complex, Phe−7 packs against Ala30, Ile38, and Pro47 in a manner similar to that seen in the NbpSH3 complex, and it also packs intimately with the CI subdomain. Interestingly, the Tyr30 residue of the NbpSH3 domain spatially overlaps with Val83 in the CI subdomain and thus appears to partially recapitulate the additional hydrophobic surface provided by the CI subdomain. Finally, peptide residue Pro−5 is accommodated in both domains by a pocket composed of His32, Trp36, and Leu49. An L49A substitution significantly lowered the binding affinity of both domains to the Ste20 peptide (Table 2).
In summary, comparison of the NbpSH3-Ste20 and BemSH3b-Ste20 complexes demonstrates that the NbpSH3 and BemSH3b domains interact with the Ste20 peptide in a similar manner. The same surfaces on the SH3 domains interact with conserved residues of the core and the extended regions of the peptide, and many of the residues on these surfaces are identical in the two domains (Fig. 2, B and C). This explains how both the NbpSH3 and BemSH3b domains can recognize the same ΨXPXRXAPXXP motif and bind some peptides, including Ste20, with a similarly high affinity.
BemSH3b Domain Interacts More Strongly with Extended Region of Ste20 Peptide
In light of the data presented above, it is remarkable that the NbpSH3 and BemSH3b domains display distinct specificities when tested against certain peptides (Fig. 3B and see Fig. 5B) (23). To investigate the origins of this specificity difference, we tested the effects of amino acid substitutions in the Ste20 peptide on its affinity for the BemSH3b and NbpSH3 domains. Strikingly, all of the Ala substitutions of residues in the extended peptide region caused reductions in affinity for the BemSH3b domain that were considerably greater than those observed for the NbpSH3 domain. In particular, the F−7A substitution reduced BemSH3b domain binding affinity 700-fold, yet decreased NbpSH3 domain affinity by only 20-fold (Fig. 3A). The stronger interactions of the BemSH3b domain with the extended region of Ste20 peptide are likely due to the influence of the CI subdomain, which forms an extra hydrophobic surface contacting both Ile−6 and Phe−7 (Fig. 2B). Pro−5 is also more tightly packed in the BemSH3b complex primarily as a result of the position of His32, which is pointed toward this peptide residue in the BemSH3b complex, but away from it in the NbpSH3 complex. Supporting a role for this residue, an H32A substitution in the BemSH3b domain reduced its binding affinity for Ste20 peptide by 7-fold, whereas the same substitution in the NbpSH3 increased its binding affinity for the Ste20 peptide (Table 2) (23). The mutagenesis of the Ste20 peptide combined with structural comparison clearly demonstrates that the BemSH3b domain forms stronger interactions with the extended region of the peptide relative to the NbpSH3 domain.
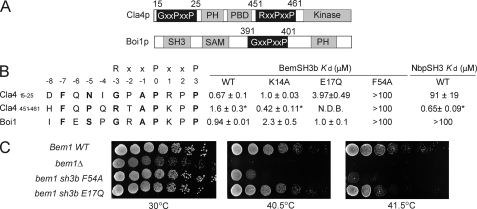
FIGURE 5.
Interaction of the BemSH3b domain with the noncanonical Cla4(15–25) and Boi1 sites. A, schematic diagram of the Cla4p and Boi1 proteins showing the location of the relevant PXXP motifs. B, interaction of the BemSH3b domain and the NbpSH3 domain with the sites derived from the Boi1p and Cla4p. The data are represented in the same way as in Fig. 3. N.D.B. (no detectable binding) indicates the absence of fluorescence shift upon addition of the peptide. C, effect of the BemSH3b E17Q substitution on cell viability relative to the BemSH3b F54A substitution. All mutations were introduced into the chromosomal copy of BEM1. Strains were grown overnight, diluted to A600 nm of 1.0, and spotted in 5-fold serial dilutions on yeast peptone dextrose plates.
Extended Region of Bck1 Peptide Interacts Suboptimally with BemSH3b Domain
The greater dependence of the BemSH3b domain binding on interaction with the extended region of the Ste20 peptide led us to predict that the weak binding of this domain to some sites that bind strongly to the NbpSH3 domain may be due to suboptimal interactions in this region. We tested this prediction for the Bck1 peptide, which binds the NbpSH3 domain with 30-fold higher affinity than the BemSH3b domain, by substituting the residues in the extended region of this site with the corresponding residues in the Ste20 site (Fig. 3B). Each of the substitutions tested increased the binding of BemSH3b severalfold, and a quadruple mutant containing all of the substitutions bound to the BemSH3b domain with an 81-fold increase in affinity. Conversely, the same substitutions caused only a 3-fold increase in affinity for the NbpSH3. The affinity of the BemSH3b domain for Bck1 peptide was increased incrementally by each substitution, suggesting that the low affinity of the BemSH3b domain for the Bck1 peptide is due to multiple suboptimal interactions with the extended region.
NpbSH3 Domain Interacts More Strongly with Core Motif of Ste20 Peptide
Whereas amino acid substitutions of the Ste20 peptide extended region reduced the binding affinity of the BemSH3b domain more than of the NbpSH3 domain, substitutions in the core motif had a greater effect on the binding of the NbpSH3 domain (Fig. 3A). For example, the R−3A substitution reduced the BemSH3b binding affinity 160-fold but reduced the NbpSH3 affinity by 510-fold. Larger effects on NbpSH3 domain affinity compared with the BemSH3b domain were also caused by the A−1T, P0A, K1A, and P3A substitutions. In total, the combined affinity reduction caused by core region substitutions was 60-fold greater for the NbpSH3 than for the BemSH3b domain. An interaction between Trp36 Hϵ of the NbpSH3 domain with the CO atom of Arg−3 of the Ste20 peptide may also play a role in mediating stronger interactions with the core motif. In all the calculated structures of the NbpSH3 complex, these atoms are within hydrogen bond distance of each other, whereas the corresponding atoms in the BemSH3b-Ste20 complex are too far apart to form a hydrogen bond (Fig. 4A). Further supporting the greater importance of core region interactions for the NbpSH3 domain, Ala substitution of Tyr8, which interacts exclusively with core peptide residues, caused an 8-fold greater reduction in binding affinity of the NbpSH3 than the BemSH3b domain (Table 2).
FIGURE 4.
Interaction of Ste20 Arg−3 with the NbpSH3 and BemSH3b domains. A, close-up of interactions with Arg−3 seen in the NbpSH3-Ste20 and BemSH3b-Ste20 complexes. In the 20 lowest energy structures of the NbpSH3-Ste20 complex the average distance between Arg−3 CO and Trp36 Hϵ is 1.8 Å, with a donor-hydrogen-acceptor angle of 170°, which suggests formation of a strong hydrogen bond (indicated by a dashed line). The average distance between the Arg−3 CO and Trp36 Hϵ in the 20 lowest energy structures of BemSH3b-Ste20 complex is 2.8, which is greater than the distance of 2.5 Å required to satisfy hydrogen bonding potential (38). Residue positions in the five lowest energy structures are shown. B, affinities of the wild-type BemSH3b domain or K14A mutant interactions with the wild-type Ste20 peptide or R−3A mutant.
Our previous work showed that Lys14 of the BemSH3b domain plays an inhibitory role in peptide binding with a K14A substitution causing a >5-fold increase in the affinity of the domain for most peptides tested (23). The BemSH3b-Ste20 peptide structure shows that the Lys14 side chain crosses the binding interface and appears to interfere with the interactions of Arg−3 through both steric and electrostatic repulsion (Fig. 4A). To investigate further the role of the Lys14-Arg−3 interaction, here we showed that the K14A substitution caused no change in affinity for the R−3A substituted Ste20 peptide (Fig. 4B). This result implies that the inhibitory effect of Lys14 on peptide binding occurs primary through unfavorable interactions with Arg−3. This interaction at least partially accounts for the weakened binding of the BemSH3b domain to the core motif.
Identification of New BemSH3b-specific Sites in Cla4p and Boi1p
Because the BemSH3b domain interacts very strongly with the extended regions of target peptides, we hypothesized that it might be able to form biologically important interactions with peptide sequences lacking a canonical RXXPXXP motif. Consistent with this idea, previous studies showed that the BemSH3b domain could bind to Cla4p residues 1–448 and Boi1p residues 371–444 (12, 33) even though each region lacks canonical SH3 domain binding sites. These regions do contain sites (Cla4(15–25) and Boi1(391–401)) that closely resemble characterized BemSH3b/Nbp2 SH3 targets except they lack Arg at the −3 position (Fig. 5, A and B). The Cla4(15–25) motif should not be confused with the Cla4(451–461) that we previously characterized, which possesses an Arg at the −3 position and binds both BemSH3b and NbpSH3 domains with high affinity (23) (Fig. 5, A and B).
To investigate these putative binding sites, we measured their affinities for the BemSH3b and NbpSH3 domains. Strikingly, the BemSH3b bound to the Cla4(15–25) and Boi1 sites with high affinities of 0.67 and 0.94 μm, respectively, despite their lack of the Arg−3 residue. However, the NbpSH3 domain interacted very weakly with these sites, binding the Cla4(15–25) site with an affinity of 91 μm and the Boi1 site with such low affinity that it could not be measured accurately (Fig. 5B). Interestingly, the E17Q BemSH3b domain mutant, which is unable to bind the Arg−3 containing Ste20 and Cla4(391–401) sites (Table 2 and Fig. 5B), showed little or no reduction in binding affinity to the noncanonical Cla4(15–25) and Boi1 sites (Fig. 5B). Similarly, the K14A substitution, which increased binding affinity to sites containing Arg−3, had little effect on binding to the Boi1 and Cla4(15–25) sites (Fig. 5B). By contrast, substitution of Phe54, which interacts with the PXXP motif, caused large reductions in binding affinity to all peptides tested (Table 2 and Fig. 5B). These results are all consistent with data described above indicating the energetic importance of the Glu17-Arg−3 interaction and a repulsive interaction between Arg−3 and Lys14. Because noncanonical peptides lack Arg−3, their binding is not affected significantly by substitutions of either Lys14 or Glu17 residues in the BemSH3b domain.
The contrasting effects of the E17Q and F54A substitutions of the BemSH3b domain on binding to the noncanonical Boi1 and Cla4(15–25) sites motivated us to assess the in vivo effects of these substitutions. Strikingly, a strain expressing Bem1p bearing the E17Q-substituted BemSH3b domain, although impaired compared with wild-type, grew significantly better under restrictive conditions than one expressing the F54A-substituted protein (Fig. 5C). These results demonstrate that the E17Q BemSH3b mutant can confer growth under these conditions despite its deficiency in binding canonical sites possessing Arg−3. Thus, we conclude that a previously unrecognized subset of BemSH3b sites lacking Arg−3, which likely includes the Boi1 and/or Cla4(15–25) sites, is functionally important, and interaction with these sites is sufficient to partially fulfill the function of the BemSH3b domain under these conditions.
DISCUSSION
In this study, we investigated the peptide binding mechanisms of the NbpSH3 and BemSH3b domains, which recognize the same consensus motif despite their limited sequence similarity. Comparison of the structures of the NbpSH3-Ste20 and BemSH3b-Ste20 complexes demonstrated that each domain interacts with the same conformation of the Ste20 peptide, and utilizes similar surfaces to engage the peptide (Fig. 2). Many residues at structurally equivalent positions on these surfaces are identical or very similar in the two domains. These structural similarities explain how the NbpSH3 and BemSH3b domains can recognize the same consensus motif. However, mutagenesis of the Ste20 peptide revealed that the energetics of the NbpSH3- and BemSH3b-mediated interactions are distinct. Relative to the NbpSH3 domain, the high affinity of the BemSH3b domain is much more dependent on interaction with the extended region of the Ste20 peptide, whereas the NbpSH3 domain forms stronger interactions with the core motif (Figs. 3A and 4A). The stronger interaction of the BemSH3b domain with the extended region of the Ste20 peptide is likely due in part to the contribution of its unique CI subdomain toward peptide binding (Fig. 2B), whereas its weaker interaction with the core peptide region results from the presence of Lys14 (Fig. 4A) and possibly other factors.
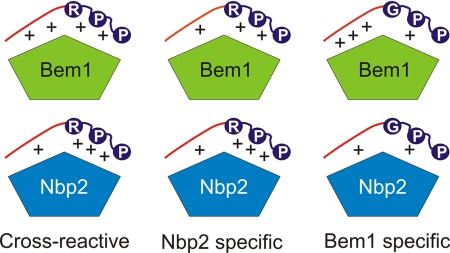
The divergent interaction strengths of the NbpSH3 and BemSH3b domains with the core and extended regions of the Ste20 peptide explain why these domains display very different affinities for certain yeast peptides and similar affinities for others (Fig. 6). For example, residues in the extended region of the Bck1 peptide are suboptimal for binding to the BemSH3b, which more strongly depends on this region for high affinity binding (Fig. 3). Because the NbpSH3 domain forms weaker interactions with the extended region, it is more tolerant to amino acid substitutions in this region (Fig. 3) and makes equally favorable interactions with the extended regions of both the Bck1 and Ste20 peptides (Fig. 6). However, the greater dependence of the NbpSH3 domain on interactions with the core region causes it to bind very poorly to peptides such as the Boi1 peptide that lack Arg−3, a key residue of the core motif (Fig. 5B). The BemSH3b domain can bind these peptides strongly despite weakened interaction with the core motif because of the very strong interactions that it can form with extended region residues (Fig. 6). In the cases of sites that both domains bind with high affinity, such as Ste20 and Cla4(451–461) (Figs. 3A and 5B), the relative contributions of core and extended motif interactions are different for the two domains, yet the sum of these interactions results in a similar strength of binding (Fig. 6).
FIGURE 6.
Model of the BemSH3b and NbpSH3 domain peptide binding mechanisms. SH3 domains are shown as pentagons with the top two sides representing the distinct surfaces that interact with the core and extended regions of target peptides (Surface I and Surface II, respectively). The conserved Arg and Pro residues of the core motif are denoted with circles, and the extended regions of peptides are colored red. The plus signs symbolize the relative strengths of interactions between core and extended peptide regions and the domain binding surfaces. Interactions with four plus signs possess Kd values of less than 1 μm, three plus signs represent Kd values in the range of 20 μm, and two plus signs represents interactions with Kd values >90 μm. Cross-reactive sites, including Ste20 and Cla4(451–461), bind the NbpSH3 and BemSH3b domains with the same affinity despite different energetic contributions of the extended and core peptide regions toward binding. Nbp2-specific sites, including Skm1 and Bck1, bind the BemSH3b domain weakly due to suboptimal interactions with the extended region, but can still bind the NbpSH3 domain tightly because it is less dependent on these interactions. Bem1-specific sites, such as Boi1 and Cla4(15–25), interact weakly with the NbpSH3 domain due to suboptimal core motif interactions resulting from the lack of Arg−3. BemSH3b can still bind these sites with high affinity by compensating for weak interactions with the core motif through strong interactions with the extended region of these peptides.
It was previously proposed that SH3 domains may have evolved noncanonical specificities through a process of weakening interactions with the peptide core region, which is recognized by many SH3 domains, and strengthening interactions with the extended peptide region (4). The binding mechanism employed by the BemSH3b domain compared with the NbpSH3 domain illustrates such a situation. The interactions of the BemSH3b domain with the core motif are weaker than those of the NbpSH3 domain, whereas the interactions with the extended region are stronger. Relevant to evolutionary mechanisms, we observed that single amino acid substitutions in the Bck1 site with residues found in the Ste20 site resulted in an incremental and additive increase in binding affinity for the BemSH3b domain. These data imply that the lower affinity of the BemSH3b domain toward the Bck1 site relative to the Ste20 site is due to multiple suboptimal interactions with the extended region of the peptide, and no single peptide residue acts as a “specificity switch.” This example demonstrates how large changes in peptide binding specificity could evolve slowly through multiple single substitutions, each of which has a relatively small effect on binding. It is also notable that even though Lys14 plays a key role in specificity determination, introduction of this residue alone would not lead to the distinct binding specificity of the BemSH3b domain without a concomitant increase in binding strength toward the extended region of the peptide. This point is vividly illustrated by our previous data showing that introduction of Lys at position 14 in the NbpSH3 domain severely abrogates binding to all target peptides (23). This domain does not interact strongly enough to the extended region of peptides to overcome the detrimental effect of Lys14 on peptide core interaction.
Our results demonstrating the crucial role of extended peptide regions in regulating affinity and specificity of the NbpSH3 and BemSH3b interactions add to the growing number of examples (1, 5–8) showing the importance of peptide residues flanking the core motif in SH3 domain-peptide interactions. The contribution of flanking residues has also been observed for other classes of protein interaction domains including PDZ domains (34) and SH2 domains (35). A question arises as to whether the contribution of extended peptide sequences is limited to specific cases or is a general property of domain-peptide interactions. One study, which examined available peptide-domain complexes, estimated that interactions with the residues outside the core motif contribute on average 20% of total binding energy and 30% in the case of SH3 domains (36). Our binding studies on the Ala substitutions of Ste20 peptide (Fig. 3A) indicate that, consistent with the above mentioned findings, ∼25% of the binding energy of the NbpSH3 interaction is contributed by flanking residues, and the BemSH3b domain relies even more strongly on interaction with flanking residues (∼50% of the total energy). The proven general importance of flanking residues emphasizes that these positions must be carefully considered in any attempts to predict or characterize functionally important protein interactions.
In summary, our work provides the first demonstration that the specificity of SH3 domains can be modulated by varying local interaction energetics across the domain-peptide interface. This mechanism can allow two or more domains to bind some targets with the same affinities, while maintaining distinct affinities for other targets. The flexibility of this mechanism and the potential it provides for the stepwise evolution of specificity suggests that it may be relevant for many other protein-peptide interaction modules. By illustrating the importance of knowing both the structure of domain-peptide complexes and the energetic contributions across the binding interface, our work emphasizes that detailed mutagenesis and quantitative binding studies are required for understanding protein binding specificity. It is clear that further investigation into both the structural and energetic factors affecting domain-peptide interactions will be required to achieve the goals of predicting and designing protein interaction specificity.
Acknowledgments
We thank Paul Sadowski and Karen Maxwell for critical reading of the manuscript and Tara Sprules for NMR data acquisition.
This work was supported by Canadian Institutes of Health Research Operating Grant MOP-13609 and by a Natural Sciences and Engineering Research Council PGS-D scholarship (to M. G.). All NMR experiments were recorded at the Québec/Eastern Canada High Field NMR Facility, supported by the Natural Sciences and Engineering Research Council of Canada, the Canada Foundation for Innovation, the Québec ministère de la recherche en science et technologie, and McGill University.
The atomic coordinates and structure factors (code 2LCS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SH3
- Src homology 3
- CI
- Cdc42-interacting
- PDB
- Protein Data Base
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Stollar E. J., Garcia B., Chong P. A., Rath A., Lin H., Forman-Kay J. D., Davidson A. R. (2009) Structural, functional, and bioinformatic studies demonstrate the crucial role of an extended peptide binding site for the SH3 domain of yeast Abp1p. J. Biol. Chem. 284, 26918–26927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zarrinpar A., Park S. H., Lim W. A. (2003) Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 [DOI] [PubMed] [Google Scholar]
- 3. Bhattacharyya R. P., Reményi A., Yeh B. J., Lim W. A. (2006) Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 [DOI] [PubMed] [Google Scholar]
- 4. Kim J., Lee C. D., Rath A., Davidson A. R. (2008) Recognition of non-canonical peptides by the yeast Fus1p SH3 domain: elucidation of a common mechanism for diverse SH3 domain specificities. J. Mol. Biol. 377, 889–901 [DOI] [PubMed] [Google Scholar]
- 5. Bauer F., Schweimer K., Meiselbach H., Hoffmann S., Rösch P., Sticht H. (2005) Structural characterization of Lyn-SH3 domain in complex with a herpesviral protein reveals an extended recognition motif that enhances binding affinity. Protein Sci. 14, 2487–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghose R., Shekhtman A., Goger M. J., Ji H., Cowburn D. (2001) A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat. Struct. Biol. 8, 998–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kami K., Takeya R., Sumimoto H., Kohda D. (2002) Diverse recognition of non-PXXP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p. EMBO J. 21, 4268–4276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lewitzky M., Harkiolaki M., Domart M. C., Jones E. Y., Feller S. M. (2004) Mona/Gads SH3C binding to hematopoietic progenitor kinase 1 (HPK1) combines an atypical SH3 binding motif, R/KXXK, with a classical PXXP motif embedded in a polyproline type II (PPII) helix. J. Biol. Chem. 279, 28724–28732 [DOI] [PubMed] [Google Scholar]
- 9. Hoelz A., Janz J. M., Lawrie S. D., Corwin B., Lee A., Sakmar T. P. (2006) Crystal structure of the SH3 domain of βPIX in complex with a high affinity peptide from PAK2. J. Mol. Biol. 358, 509–522 [DOI] [PubMed] [Google Scholar]
- 10. Janz J. M., Sakmar T. P., Min K. C. (2007) A novel interaction between atrophin-interacting protein 4 and β-p21-activated kinase-interactive exchange factor is mediated by an SH3 domain. J. Biol. Chem. 282, 28893–28903 [DOI] [PubMed] [Google Scholar]
- 11. Larson S. M., Davidson A. R. (2000) The identification of conserved interactions within the SH3 domain by alignment of sequences and structures. Protein Sci. 9, 2170–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hruby A., Zapatka M., Heucke S., Rieger L., Wu Y., Nussbaumer U., Timmermann S., Dünkler A., Johnsson N. (2011) A constraint network of interactions: protein-protein interaction analysis of the yeast type II phosphatase Ptc1p and its adaptor protein Nbp2p. J. Cell Sci. 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 13. Mapes J., Ota I. M. (2004) Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J. 23, 302–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Du Y., Walker L., Novick P., Ferro-Novick S. (2006) Ptc1p regulates cortical ER inheritance via Slt2p. EMBO J. 25, 4413–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matsui Y., Matsui R., Akada R., Toh-e A. (1996) Yeast Src homology region 3 domain-binding proteins involved in bud formation. J. Cell Biol. 133, 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Butty A. C., Perrinjaquet N., Petit A., Jaquenoud M., Segall J. E., Hofmann K., Zwahlen C., Peter M. (2002) A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 21, 1565–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bose I., Irazoqui J. E., Moskow J. J., Bardes E. S., Zyla T. R., Lew D. J. (2001) Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J. Biol. Chem. 276, 7176–7186 [DOI] [PubMed] [Google Scholar]
- 18. France Y. E., Boyd C., Coleman J., Novick P. J. (2006) The polarity-establishment component Bem1p interacts with the exocyst complex through the Sec15p subunit. J. Cell Sci. 119, 876–888 [DOI] [PubMed] [Google Scholar]
- 19. Yamaguchi Y., Ota K., Ito T. (2007) A novel Cdc42-interacting domain of the yeast polarity establishment protein Bem1: implications for modulation of mating pheromone signaling. J. Biol. Chem. 282, 29–38 [DOI] [PubMed] [Google Scholar]
- 20. Winters M. J., Pryciak P. M. (2005) Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol. Cell. Biol. 25, 2177–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takaku T., Ogura K., Kumeta H., Yoshida N., Inagaki F. (2010) Solution structure of a novel Cdc42 binding module of Bem1 and its interaction with Ste20 and Cdc42. J. Biol. Chem. 285, 19346–19353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tong A. H., Drees B., Nardelli G., Bader G. D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., Quondam M., Zucconi A., Hogue C. W., Fields S., Boone C., Cesareni G. (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321–324 [DOI] [PubMed] [Google Scholar]
- 23. Gorelik M., Stanger K., Davidson A. R. (2011) A conserved residue in the yeast Bem1p SH3 domain maintains the high level of binding specificity required for function. J. Biol. Chem. 286, 19470–19477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maxwell K. L., Davidson A. R. (1998) Mutagenesis of a buried polar interaction in an SH3 domain: sequence conservation provides the best prediction of stability effects. Biochemistry 37, 16172–16182 [DOI] [PubMed] [Google Scholar]
- 25. Kay L. E. (1995) Pulsed field gradient multidimensional NMR methods for the study of protein structure and dynamics in solution. Prog. Biophys. Mol. Biol. 63, 277–299 [DOI] [PubMed] [Google Scholar]
- 26. Gemmecker G., O. E. T., Fesik S. W. (1992) An improved method for selectively observing protons attached to 12C in the presence of 1H 13C spin pairs. J. Magn. Reson. 96, 199–204 [Google Scholar]
- 27. Ikura M., Bax A. (1992) Isotope-filtered 2D NMR of a protein-peptide complex: study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc. 114, 2433–2440 [Google Scholar]
- 28. Zwahlen C., Legault P., Vincent S. J. F., Greenblatt J., Konrat R., Kay L. E. (1997) Methods for measurement of intermolecular NOEs by multi nuclear NMR spectroscopy: Application to bacteriophage lambda N-peptide/Box B RNA complex. J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 29. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 30. Goddard T. D., Kneller D. G. SPARKY 3, University of California, San Francisco [Google Scholar]
- 31. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 32. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 33. Hertveldt K., Robben J., Volckaert G. (2006) Whole genome phage display selects for proline-rich Boi polypeptides against Bem1p. Biotechnol. Lett. 28, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 34. Luck K., Fournane S., Kieffer B., Masson M., Nominé Y., Travé G. (2011) Putting into practice domain-linear motif interaction predictions for exploration of protein networks. PLoS One 6, e25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu B. A., Jablonowski K., Shah E. E., Engelmann B. W., Jones R. B., Nash P. D. (2010) SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell. Proteomics 9, 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stein A., Aloy P. (2008) Contextual specificity in peptide-mediated protein interactions. PLoS One 3, e2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim W. A., Richards F. M., Fox R. O. (1994) Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372, 375–379 [DOI] [PubMed] [Google Scholar]
- 38. McDonald I. K., Thornton J. M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]