Background: C. elegans acquires environmental heme through specific trafficking machinery.
Results: Heme-responsive gene-2 (HRG-2) is a heme-binding, type I membrane protein specifically expressed in the hypodermis; cytochrome distribution is abnormal in HRG-2-deficient worms.
Conclusion: HRG-2 facilitates heme utilization in the hypodermis.
Significance: Regulation of heme homeostasis by an HRG-2 prototype could be a general mechanism employed by metazoans.
Keywords: C. elegans, Gene Regulation, Heme, Iron Metabolism, Metals, Helminth, Tetrapyrrole
Abstract
The roundworm Caenorhabditis elegans is a heme auxotroph that requires the coordinated actions of HRG-1 heme permeases to transport environmental heme into the intestine and HRG-3, a secreted protein, to deliver intestinal heme to other tissues including the embryo. Here we show that heme homeostasis in the extraintestinal hypodermal tissue was facilitated by the transmembrane protein HRG-2. Systemic heme deficiency up-regulated hrg-2 mRNA expression over 200-fold in the main body hypodermal syncytium, hyp 7. HRG-2 is a type I membrane protein that binds heme and localizes to the endoplasmic reticulum and apical plasma membrane. Cytochrome heme profiles are aberrant in HRG-2-deficient worms, a phenotype that was partially suppressed by heme supplementation. A heme-deficient yeast strain, ectopically expressing worm HRG-2, revealed significantly improved growth at submicromolar concentrations of exogenous heme. Taken together, our results implicate HRG-2 as a facilitator of heme utilization in the Caenorhabditis elegans hypodermis and provide a mechanism for the regulation of heme homeostasis in an extraintestinal tissue.
Introduction
Heme is a redox-active cofactor that plays critical roles in various biological processes (1, 2). In most metazoans heme is synthesized in the mitochondrial matrix using glycine, succinyl-coenzyme A, and ferrous iron as substrates. However, numerous hemoproteins such as globins, catalases, cytochrome P450s, and heme-regulated transcription factors are present in extramitochondrial compartments (2, 3). As an iron-containing amphipathic porphyrin, free heme can catalyze the production of reactive oxygen species and intercalate into lipid bilayers (4, 5). Accordingly, heme is unlikely to diffuse freely within the cell, but instead, specific molecules and pathways must exist to facilitate heme delivery to distinct cellular destinations.
In animals, only two membrane-bound heme transporters have been characterized genetically. The heme permease HRG-12 (SLC48A1), initially identified from a transcriptomic analysis in Caenorhabditis elegans, imports heme and is conserved in vertebrates (6, 7). The feline leukemia virus subgroup C cellular receptor (FLVCR), a major facilitator superfamily protein, is involved in heme export in red blood cells and macrophages (8, 9). Additionally, a number of proteins such as hemopexin (10, 11), p22 HBP (12), HBP23 (13), and certain classes of glutathione S-transferases (GSTs) (14–16) have been shown to associate with heme, and correspondingly, these proteins have been implicated in heme homeostasis.
C. elegans is a heme auxotroph and thus serves as a unique animal model for identifying inter- and intracellular heme trafficking pathways (17). Worms acquire environmental heme by importing heme into the intestine by the coordinated actions of HRG-1 and HRG-4 heme transporters (6, 7). Heme from the intestine is mobilized by HRG-3, a secreted protein that exports maternal heme to extraintestinal tissues and the embryo (18). How is heme import facilitated in extraintestinal tissues? In this study, we have identified HRG-2, a type I membrane protein localized to the hyp7, the major hypodermal syncytium in C. elegans, as a critical player in heme homeostasis. Our studies in worms, yeast, and mammalian cells support this conclusion.
EXPERIMENTAL PROCEDURES
Worm Culture, Strains, and Experiments
Routine maintenance, synchronization, genetic crosses, and microscopic observation were described by Epstein and Shakes (19). N2 Bristol strain, deletion strains, and transgenic worms were maintained at 20 °C either on nematode growth medium (NGM) agar plates or in axenic modified C. elegans habitation and reproduction (mCeHR-2) medium supplemented with hemin chloride (19, 20). Continuous shaking was provided for all liquid worm cultures. In CdCl2 induction assays, synchronized stage 1 (L1) larvae were grown at 1.5 and 20 μm hemin for 7 days. The worms were then treated with 0 or 100 μm CdCl2 for 24 h before harvesting.
The deletion strain hrg-2 (tm3798) was isolated in mutagenesis screens by the National Bioresource Project in Japan (21). The allele was confirmed by sequencing and was outcrossed eight times with the N2 Bristol strain. Progeny from genetic crosses were genotyped by PCR on individual worms using sense primer 5′-TTTATGCTCTTCCTGCGAG-3′ and antisense primer 5′-TATACCATGCATCCTCTGC-3′. During the final out-cross, both homozygous mutants and their wild type brood mates were saved for further analysis. Transcriptional (hrg-2::gfp) and translational (hrg-2::HRG-2-YFP) reporter constructs were generated either by fusion PCR or by the multisite Gateway system (Invitrogen). Worms with extrachromosomal arrays or stable transgenic lines were obtained by microinjection or by microparticle bombardment (supplemental Table S1) (22).
Methods for RNA extraction, Northern blotting, qRT-PCR, and rapid amplification of cDNA ends (RACE) were described previously (6). The hrg-2 primers used in this study were sense (5′-GCCTGGCTGATAATCATCTCTTG-3′) and antisense (5′-ATGGACCTTCTTCATAAATAACTTTCG-3′) for qRT-PCR and sense (5′-GCTGAAATGTTATGTCACAAAG-3′) and antisense (5′-TTATTGCCACAGAGATACAGG-3′) for the synthesis of [α-32P]dCTP-labeled cDNA probe.
DNA Cloning
Total worm RNA was first reverse-transcribed into cDNA using oligo(dT) primers. hrg-2 ORF was amplified with primers flanked by BamHI and XhoI restriction sites. Following restriction digestion and DNA purification, the PCR products were cloned into the pcDNA3.1(+)zeo vector (Invitrogen) and the pEGFP-N1 vector as well as its equivalent GFP variant living color vectors (Clontech). Truncated constructs, including HRG-2N, HRG-2ΔGST-N, HRG-2ΔGST-C, and HRG-2ΔN, were introduced into mammalian expression plasmids in a similar way. For yeast studies, untagged or tagged versions of the hrg-2 ORF were cloned into the 2 μ plasmid pYES-DEST52 (Invitrogen) by Gateway cloning or into a modified pYES-DEST52 plasmid (provided by Dr. Caroline Philpott, National Institutes of Health) using primers engineered with BamHI and XbaI restriction sites. cdr-1 and truncated hrg-2 constructs were cloned into the modified pYES-DEST52 plasmid.
Immunofluorescence and GFP Fluorescence
Transfected HEK293 cells grown on coverslips were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.2% (v/v) Triton X-100, and blocked in 3% (w/v) bovine serum albumin and 50% (v/v) SuperBlock solution (Pierce). Samples were incubated in a primary polyclonal anti-HA antibody (Sigma) at a 1:2000 dilution followed by goat anti-rabbit IgG secondary antibodies conjugated to either Alexa 488 or Alexa 568 at a 1:6000 dilution. Coverslips were mounted onto slides using ProLong Antifade (Invitrogen). For GFP fluorescence studies, transfected cells were mounted directly onto slides after fixation. Transformed yeast cells were grown under inducing conditions in liquid synthetic complete (SC) medium to mid-log phase. After fixing with 4% (w/v) formaldehyde, cells were treated with zymolyase-100T (United States Biological) to create spheroplasts. Aliquots of the spheroplasts were added to 8-well slides followed by incubation with rabbit polyclonal anti-HA antibody at a 1:2000 dilution (Sigma) and then with Alexa 488-conjugated polyclonal goat anti-rabbit IgG antibody at a 1:5000 dilution. Fluorescent worms were paralyzed in 10 mm levamisole and mounted on 1.2% (w/v) agarose pads on glass slides. GFP, mCherry, and Alexa fluorophores were examined in an LSM 510 laser-scanning confocal microscope with argon (458 and 488 nm) and helium/neon (543 and 633 nm) lasers (Zeiss). Samples of mammalian cells, worms, and yeast were examined using ×63 and ×100 oil immersion objective lenses. Images with a z resolution of 1 μm were acquired and processed in the LSM image browser (Zeiss).
Fluorescence Protease Protection Assay
The procedure for fluorescence protease protection assay was modified from the protocol by Lorenz et al. (23). HRG-2-GFP and control plasmid pCFP-CD3δ-YFP (a gift from Dr. Jennifer Lippincott-Schwartz, National Institutes of Health) were transfected into HEK293 cells grown on Lab-Tek chambered coverglasses (Nunc). After 24 h, the cells were washed with KHM buffer (110 mm potassium acetate, 2 mm MgCl2, and 20 mm HEPES, pH 7.3), and the cell chambers were moved to a DMIRE2 epifluorescence microscope (Leica) connected with a Retiga 1300 Mono 12-bit cooled camera. The plasma membrane was permeabilized with 30 μm digitonin for 2 min, and then the cells were immediately incubated in 50 μg/ml proteinase K for 2 min. Images were taken before digitonin treatment, after digitonin treatment, and after proteinase K digestion.
Immunoblotting
Transfected HEK293 cells were lysed in cell lysis buffer (150 mm NaCl, 0.5% (v/v) Triton X-100, and 20 mm HEPES, pH 7.4) for 5 min on ice. Yeast cells were harvested and resuspended in breaking buffer (1 mm dithiothreitol, 20% (v/v) glycerol, and 100 mm Tris-HCl, pH 8.0) followed by disruption using a FastPrep-24 (MP Biomedicals) Bead Beater (three 30-s pulses at 6.5 m/s) in the presence of acid-washed glass beads. The protein concentration was quantified with Bradford reagent (Bio-Rad). Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membrane (Bio-Rad). After blocking in 5% (w/v) nonfat dry milk, the membranes were incubated in rabbit anti-HA (Sigma) at a 1:2000 dilution or mouse anti-GFP at a 1:5000 dilution. HRP-conjugated secondary antibodies diluted to 1:20,000 were applied to the membranes, and the signals were detected by SuperSignal chemiluminescence reagents (Thermo Scientific) using the gel documentation system (Bio-Rad).
Hemin-Agarose Chromatography
Hemin-agarose pulldown assays were performed according to the procedure outlined by Rajagopal et al. (6). HEK293 cells transfected with hrg-2 or control constructs were treated with heme-depleted medium with or without 10 μm heme. Each binding reaction contained 300 nmol of hemin-agarose and 500 μg of HRG-2 cell lysate or the equivalent amount of target proteins from other cell lysates. After removing the unbound cell lysates, the hemin-agarose pellets were washed three times with 1 ml of wash buffer (150 mm NaCl, 1% (v/v) Nonidet P-40, and 50 mm Tris-HCl, pH 8.0) and three times with 1 ml of radioimmune precipitation assay buffer (150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, and 50 mm Tris-HCl, pH 7.9). The bound proteins were eluted by incubating them in 8 m urea and Laemmli sample-loading buffer containing 100 mm dithiothreitol for 5 min at room temperature and then boiling for 3 min. Equivalent amounts of input protein (input), the flow-through after radioimmune precipitation assay buffer washes (wash), and the eluted protein (bound) were subjected to electrophoresis in 4–20% polyacrylamide gels and immunoblotting with HA antibodies. Each heme binding assay was performed at least twice.
Mammalian Cell Culture
HEK293 cells were maintained in DMEM (Invitrogen) supplemented with 10% (v/v) fetal bovine serum and penicillin/streptomycin/glutamine. DNA constructs were transiently transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen) for Western blotting studies and FuGENE 6 (Roche Applied Science) for immunofluorescence assays. To deplete intracellular heme, HEK293 cells were grown in heme-depleted growth medium supplemented with 0.5 mm succinylacetone, an inhibitor of the heme synthesis pathway, for 24 h. The fetal bovine serum (10%) for this medium was depleted of endogenous heme by incubating it with 10 mm ascorbic acid for 7 h at 37 °C followed by dialysis three times in phosphate-buffered saline (24).
In Vitro Transcription and Translation
HA-tagged hrg-2 and the pcDNA3.1(+) Zeo vector were transcribed and translated in vitro using the TnT-coupled wheat germ extract system (Promega). One microgram of each plasmid DNA was added to wheat germ lysates in the presence of amino acids and TnT RNA polymerase. The reactions were incubated at 30 °C for 2 h. The samples were subjected to SDS-PAGE and immunoblotting.
Yeast Experiments
The heme-deficient Saccharomyces cerevisiae strain DY1457 hem1Δ(6D) was kindly provided by Dr. Caroline Philpott. This hem1Δ strain lacks the gene encoding δ-aminolevulinic acid synthase, which is the rate-limiting enzyme in the heme biosynthesis pathway (25). hem1Δ yeast were maintained on enriched yeast extract-peptone-dextrose (YPD) or SC media (−Ura) supplemented with 250 μm δ-aminolevulinic acid. Expression plasmids containing control or hrg-2 constructs were transformed into hem1Δ yeast using polyethylene glycol and lithium acetate. Positive clones were isolated by plating the transformants onto the selective SC medium, lacking uracil. Residual δ-aminolevulinic acid was removed by incubating the transformants in SC liquid medium without δ-aminolevulinic acid for 16 h. Equal numbers of transformed yeast were inoculated onto growth assay plates containing SC medium with 2% (w/v) raffinose and 0.4% (w/v) galactose for gene induction as well as different concentrations of hemin chloride. Yeast growth was analyzed after incubation at 30 °C for 3–5 days as described (7).
Oxygen Consumption Assay
Oxygen consumption was measured in wild type W303 and hem1Δ (6D) S. cerevisiae transformed with pYES-DEST52 containing hrg-2, hrg-4, cdr-1, or vector. Transformants were induced in SC medium containing 2% (w/v) raffinose with 0.8% (w/v) galactose and 5 μm hemin chloride for 16 h in triplicate. Cultures were then diluted to an A600 of 1.0 in the same growth medium and assayed in a 3-ml volume in the electrode chamber with constant stirring at room temperature (23 °C). Oxygen consumption was monitored with a Clark-type electrode, YSI model 5300. Oxygen consumption was linear, and the slope was calculated for each sample. The total protein content of the whole yeast lysate was determined by BCA assay, and the rate was calibrated to μM O2/min/mg of protein. Statistical analysis was carried out using one-way ANOVA and the Student-Newman-Keuls multiple comparison test in GraphPad Instat.
Microarray Experiments
Triplicate biological replicates of hrg-2 (tm3978) and wild type brood mate animals obtained after eight back-crosses were grown in axenic mCeHR-2 liquid medium supplemented with 20 μm hemin chloride for one generation (P0). Synchronized F1 worms were subsequently grown at either 4 or 20 μm heme, and the ensuing F2 progeny were synchronized, grown at their respective heme concentrations, and harvested at the late L4 larval stage. Total RNA was extracted from these populations as reported previously and subjected to Affymetrix whole genome expression microarray probing with the resulting data analyzed by both MAS5.0 (Affymetrix) and Robust Multichip Average (RMA; Partek Software Suite).
C. elegans Membrane Fractionation
Approximately 1 × 106 hrg-2 (tm3798) and wild type brood mate L1 larvae were grown for one generation in axenic liquid mCeHR-2 medium with 4 or 20 μm heme. F2 progeny obtained from these F1 mothers were grown subsequently to the L4 larval stage in the corresponding concentration of heme in mCeHR-2 medium. Worm pellets were lysed in 0.5× buffer (50 mm HEPES, pH 7.4, 100 mm KCl, and 250 mm sorbitol) with protease inhibitors (1 mm PMSF, 0.02 mg/ml leupeptin, and 0.01 mg/ml pepstatin) at 16,000 psi using a French pressure cell. The crude extract was cleared by centrifugation at 1000 × g for 15 min at 4 °C. The supernatant was then fractionated by centrifugation at 100,000 × g for 1 h at 4 °C to obtain a membrane-enriched pellet and a cytosol-enriched supernatant. The membrane fraction was resuspended in buffer with fresh protease inhibitors. Both the membrane and cytosolic fractions were assayed for protein concentration using Bradford protein assay prior to cytochrome quantitation.
Quantitation of Cytochromes and Heme
Triton X-100 (1.0% (w/v)) was added to membrane fractions, and samples were sonicated for 30 s in ice, the material centrifuged at 10,000 × g for 10 min, and the supernatant fraction collected for analysis. Soluble cell fractions were analyzed directly. Protoheme was quantitated by the pyridine hemochromogen assay (26). Cytochrome content was determined by oxidized minus reduced spectroscopy with a Cary 1G spectrophotometer (27). Samples were reduced by dithionite immediately prior to determinations. Although ferricyanide was employed as an oxidant, it was found that prepared samples were maximally oxidized.
Bioinformatics and Statistics
BLAST searches were applied to identify homologous genes of hrg-2. A putative ortholog was assigned when it had a significant E value (<10−4) and met the criterion of reciprocal best BLAST hit. Molecular weights and isoelectric points were calculated in the Compute pI/Mw program (28). Transmembrane domains and protein motifs were predicted using the transmembrane prediction with the Hidden Markov Model program (TMHMM) and the conserved domain database (CDD), respectively. Multiple sequence alignment was performed using ClustalW (29) and was visualized with the BoxShade program. Following multiple sequence alignment of HRG-2 and CDR-1 proteins, a phylogenetic tree was constructed using the neighbor-joining method in MEGA 4 (Molecular Evolutionary Genetics Analysis, version 4.0) with 1000 pseudoreplicates (30, 31).
The CdCl2 induction assays used a 2 × 2 factorial design with two levels of heme concentrations and two levels of CdCl2 concentrations. The main effects of heme and CdCl2 on the expression of hrg-2 or cdr-1 were tested by two-way ANOVA in GraphPad Prism, version 5. Statistical significance in all other experiments was tested using one-way ANOVA followed by the Tukey-Kramer multiple comparisons test in GraphPad INSTAT, version 3.01 (GraphPad, San Diego). All data are presented as mean ± S.E., and a p value of <0.05 was considered statistically significant.
RESULTS
Heme Deficiency Induces the Expression of hrg-2 in C. elegans
C. elegans requires environmental heme to sustain growth and development. In response to changes in heme concentrations, worms have been shown to regulate the expression of 288 heme-responsive genes (hrg) of which 80 genes are up-regulated at low heme (6, 32). To identify candidate genes that may function in heme homeostasis, we categorized genes which encoded proteins with homology to other heme-binding proteins and were highly up-regulated at low heme.
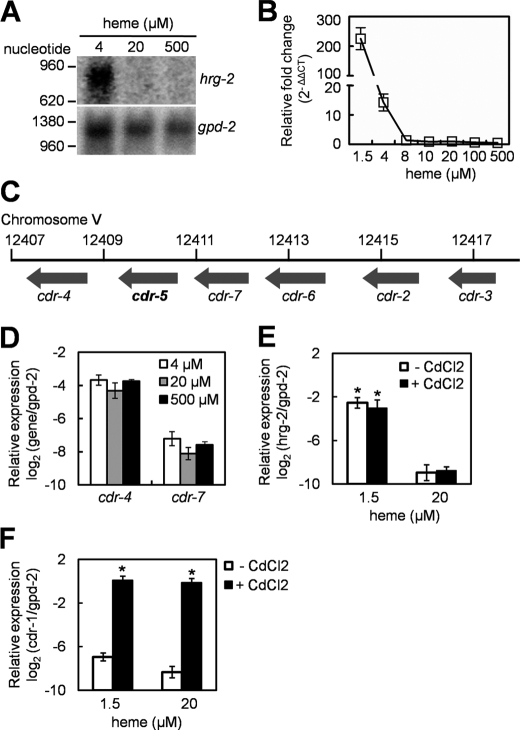
The gene K01D12.14 was up-regulated more than 70-fold when worms were grown at 4 μm heme (Table 1). K01D12.14 was previously annotated as cadmium-responsive gene-5 (cdr-5) because of its amino acid sequence homology to cdr-1, although K01D12.14 expression, unlike cdr-1, was neither induced nor repressed by cadmium (33, 34). To validate our microarray results, we analyzed K01D12.14 mRNA levels in response to heme concentrations in culture. A single ∼900-bp mRNA was detected by Northern blotting only at low heme supplementation (Fig. 1A). Quantitative RT-PCR confirmed that K01D12.14 mRNA expression was up-regulated by more than 200-fold at 1.5 μm heme compared with the heme concentration (20 μm) that is normally supplemented in mCeHR-2 medium for optimal growth (17) (Fig. 1B). Significantly, K01D12.14 mRNA was undetectable at concentrations ≥8 μm heme.
TABLE 1.
Expression profiles of hrg-2 and cdr genes by microarray analysis
The Affymetrix C. elegans genome array experiments were performed in triplicates using RNA extracted from worms grown at 4, 20, or 500 μm heme in axenic mCeHR-2 medium. Gene expression was compared with the optimal heme concentration of 20 μm, and the values are -fold change ± S.E.
| Gene name | Accession number | Heme |
||
|---|---|---|---|---|
| 4 | 20 | 500 | ||
| μm | ||||
| cdr-1 | F35E8.11 | 1.3 ± 0.1 | 1 | 1.2 ± 0.1 |
| cdr-2 | C54D10.1 | 1.7 ± 0.4 | 1 | 1.5 ± 0.2 |
| cdr-3 | C54D10.2 | 1.6 ± 0.4 | 1 | −5.7 ± 5.3 |
| cdr-4 | K01D12.11 | 1.3 ± 0.1 | 1 | 0.7 ± 0.9 |
| hrg-2//cdr-5 | K01D12.14 | 72.1 ± 12.4 | 1 | 3.8 ± 1.0 |
| cdr-6 | K01D12.12 | 1.2 ± 0.1 | 1 | 1.1 ± 0.1 |
| cdr-7 | K01D12.13 | 1.2 ± 0.1 | 1 | 0.4 ± 0.8 |
FIGURE 1.
hrg-2 is induced by heme deficiency. A, Northern blot analysis of hrg-2 expression using total RNA isolated from worms grown at different heme concentrations. The blot was reprobed with the internal control gpd-2 gene. B, quantification of hrg-2 mRNA by qRT-PCR. Relative -fold changes were derived by normalizing the cycle threshold values to gpd-2 and then to the control heme level of 20 μm using ΔΔCT methods. C, genomic structure of six cdr genes in C. elegans. cdr-5 (K01D12.14) and five other cdr genes are clustered within a 10-kb region on chromosome V (adapted from WormBase). D, quantification of cdr-4 and cdr-7 mRNA by qRT-PCR. cdr-4 and cdr-7 are adjacent to hrg-2 in C. elegans genome. The gene expression was normalized to the internal control, gpd-2. No significant difference was observed across heme levels (p > 0.05). E and F, transcriptional responses of hrg-2 (E) and cdr-1 (F) to heme and CdCl2. N2 worms grown at 1.5 and 20 μm heme were treated with 0 or 100 μm CdCl2 for 24 h. qRT-PCR experiments were performed on at least two independent worm preparations. Asterisks indicate the significant effects of heme on the expression of hrg-2 (p < 0.01 (E)) and of CdCl2 on the expression of cdr-1 (p < 0.01 (F)).
With the exception of cdr-1, the remaining six cdr genes are clustered within 10 kb on chromosome V in the C. elegans genome (Fig. 1C). In C. elegans, mRNAs from genes that lie within an operon are trans-spliced with the SL2 splice leader sequence (35). However, it is reported that none of mRNAs from the clustered cdr genes was trans-spliced with SL2 (34). Indeed, our microarray and qRT-PCR results revealed that the expression of cdr-4 (K01D12.11) and cdr-7 (K01D12.13), two cdr genes that flank K01D12.14, were not responsive to heme (Table 1 and Fig. 1D). Furthermore, 5′- and 3′-RACE experiments demonstrated that K01D12.14 mRNA was not trans-spliced, confirming that it is not within an operon (not shown).
To directly determine the relationship between heme and cadmium regulation of K01D12.14, we analyzed mRNA from wild type N2 worms grown in medium supplemented with 1.5 or 20 μm heme in the presence or absence of 100 μm cadmium chloride. qRT-PCR results show that cadmium specifically induced cdr-1, but not K01D12.14, expression by at least 70-fold (Fig. 1, E and F). By contrast, heme deficiency resulted in the specific up-regulation of K01D12.14 expression but not cdr-1. Together, these data show conclusively that K01D12.14 is an hrg and not a cdr gene. Thus, we renamed K01D12.14 as hrg-2.
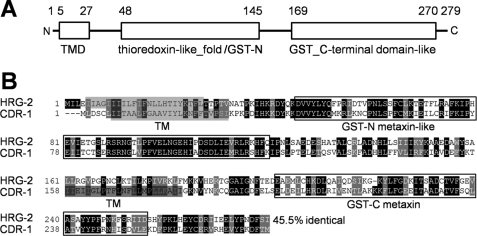
The hrg-2 gene encodes a 279-amino acid predicted protein with a molecular mass of 31.9 kDa. Putative homologs of HRG-2 are present in Caenorhabditis species with ∼75% identity at the amino acid level (supplemental Fig. S1). Protein database analysis predicts two signature domains, a thioredoxin-like (position 48–145; GST-N) and a glutathione S-transferase-C-terminal domain-like (position 169–270; GST-C) folds (Fig. 2A). In addition, a single transmembrane domain (position 5–27) was predicted at the N terminus. Within the C. elegans genome, HRG-2 shows homology to CDR-1 and five other putative CDRs with ∼45% identity (34) (Fig. 2B). BLAST searches with protein sequences identified putative homologs for hrg-2 or cdr genes in Caenorhabditis briggsae, Caenorhabditis remanei, Caenorhabditis brenneri, and Pristionchus pacificus (supplemental Fig. S2). In addition, a putative homolog exists in vertebrate species with ∼20% identity across the entire primary sequence (supplemental Fig. S3).
FIGURE 2.
Sequence analysis of HRG-2 protein. A, putative domains in HRG-2. The transmembrane domain (TMD) was predicted by the TMHMM program. The thioredoxin-like_fold/GST-N and the GST_C-terminal-like domains were predicted using the conserved domain database (CDD). Numbers above the schematic indicate the positions of amino acids in HRG-2. B, comparison of HRG-2 and CDR-1. Only CDR-1, but not HRG-2, has a second putative transmembrane domain. The numbers at the end of the alignment indicates the percentage identity, derived from the pairwise alignment between C. elegans HRG-2 and CDR-1.
hrg-2 Is Expressed in C. elegans Hypodermal Cells
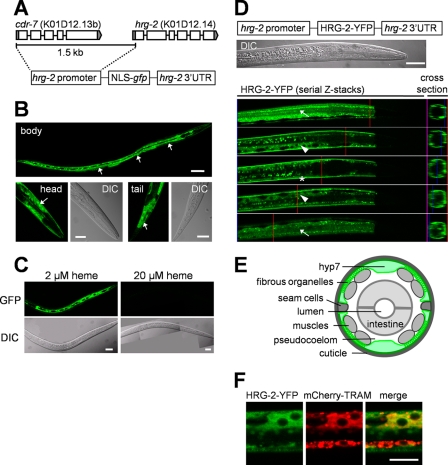
To determine the tissue-specific expression of hrg-2, we synthesized a transcriptional reporter with 1.5 kb of upstream sequence from the ATG translational start site of hrg-2 fused to gfp (Fig. 3A). hrg-2::gfp1.5 is expressed predominantly in the worm hypodermis. The major body hypodermal cell, hyp7, a 139-nuclei syncytium, and the hypodermal cells in the head and tail regions all had GFP expression when the worms were maintained at low heme, whereas GFP was undetectable at ≥20 μm heme (Fig. 3, B and C). Because the putative 1.5-kb promoter also encompassed the upstream cdr-7 (Figs. 1C and 3A), we synthesized an hrg-2::gfp0.5 transcriptional reporter in which we excluded cdr-7 and instead fused the 0.5-kb intergenic sequence with gfp. This modified hrg-2::gfp0.5 construct also showed the same expression pattern as the hrg-2::gfp1.5 and was heme-responsive (supplemental Fig. S4), indicating that the hrg-2 transgene expression was not affected by regulatory elements within cdr-7.
FIGURE 3.
hrg-2 is expressed in the hypodermal cells in C. elegans. A, schematic representation of the hrg-2::gfp1.5 reporter constructs. Genomic structures of hrg-2 and cdr-7 are shown at the top. NLS, nuclear localization signal. B, hrg-2::gfp1.5 is expressed predominantly in hypodermal cells in C. elegans. This IQ8021 worm strain contains the construct that has the 1.5-kb promoter region of hrg-2 fused with NLS-gfp. Arrows indicate GFP expression in hypodermal cells. Scale bars, top, 50 μm; bottom, 20 μm. C, responses of hrg-2::gfp1.5 reporter to heme levels. GFP can be detected only when the transgenic worms are maintained at low concentrations of heme, whereas 20 μm heme turns off the gene expression within 48 h. Scale bars, 20 μm. D, subcellular localization of HRG-2 in hypodermal cells. Schematic representation of the hrg-2::HRG-2-YFP translational reporter construct is shown at the top. Five representative fluorescence images from a confocal z-stack are displayed. HRG-2-YFP presents in fibrous organelles (arrow), the ER (arrowhead), and the apical plasma membrane (asterisk). Red vertical lines indicate the positions for cross-section images, which are shown on the right. Scale bar, 20 μm. E, schematic representation of the body hypodermal cell, hyp7 (green), in a cross-section of worm body. The schematic was modified from Labouesse (59). F, co-localization of HRG-2 with the ER marker mCherry-TRAM. The translational reporter hrg-2::HRG-2-YFP and the hypodermal ER marker dpy-7::mCherry-TRAM were introduced into the same worm by bombardment. Localization patterns of YFP and mCherry were analyzed after incubating the worms at 2 μm heme for 4 days. mCherry-TRAM is not present in fibrous organelles or plasma membrane. Scale bar, 10 μm.
HRG-2 Localizes to the Endoplasmic Reticulum and Apical Plasma Membrane of Hypodermal Cells
To determine the subcellular distribution of HRG-2 in C. elegans, we generated animals that express the translational reporter hrg-2::HRG-2-YFP. When maintained at low heme, these worms displayed a robust HRG-2-YFP signal that was localized to the fibrous organelles (structures within the hemi-adherens junction), the endoplasmic reticulum (ER), and the apical plasma membrane (Fig. 3, D and E). Fibrous organelles are composed of intermediate filament arrays that associate with the cuticle, muscle, and neurons (36). To confirm the ER localization we generated a double transgenic strain that expressed the ER marker TRAM (translocating chain-associated membrane) protein fused to mCherry from the hypodermal promoter dyp-7 (37) and hrg-2::HRG-2-YFP. As observed on Fig. 3F, HRG-2-YFP and mCherry-TRAM showed >70% co-localization, indicating that a significant portion of intracellular HRG-2 is associated with ER.
The N Terminus of HRG-2 Is Required for Membrane Targeting
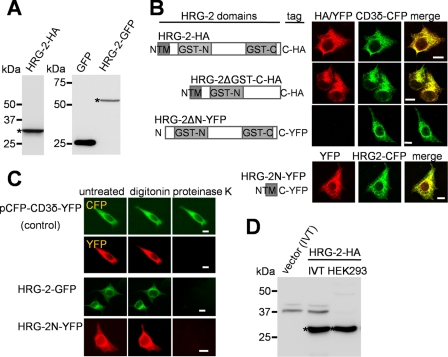
To determine the membrane targeting and topology of HRG-2, we generated HRG-2 with either an HA epitope or GFP fusion at the C terminus for expression in mammalian cell lines. Epitope or fluorescent proteins tagged at the N terminus of HRG-2 did not show any protein expression, suggesting that the tag interfered with either protein stability or targeting. Immunoblotting analyses of lysates from HEK293 cells transfected with either HRG-2-HA or HRG-2-GFP revealed that tagged HRG-2 migrated at the expected molecular weight on SDS-PAGE (Fig. 4A). Fluorescence microscopy studies showed that, as observed in C. elegans hypodermal cells, HRG-2 co-localized specifically with the ER marker CD3δ-CFP in HEK293 cells (Fig. 4B, top row) but not to the mitochondria (supplemental Fig. S5). HRG-2, however, was undetectable on the plasma membrane of mammalian cell lines.
FIGURE 4.
HRG-2 is a type I membrane protein. A, Western blot of HRG-2 transiently expressed in HEK293 cells. The lysates of transfected cells were subjected to SDS-PAGE and Western blotting using anti-HA or anti-GFP antibodies. Asterisks indicate HRG-2 proteins. B, immunofluorescence analysis of HRG-2 expressed in HEK293 cells. HA-tagged HRG-2 was detected using anti-HA and fluorophore-conjugated anti-rabbit IgG antibodies in the fixed cells. Images were acquired using a confocal microscope. Scale bars, 10 μm. C, fluorescence protease protection assays on HRG-2-GFP constructs. In this assay, the transfected cells were treated with 30 μm digitonin for 2 min followed by 50 μg/ml proteinase K for 2 min. Images were acquired throughout the process by epifluorescence microscopy. Disappearance of fluorescence after digitonin-protease K treatment indicates that the C-terminal GFP or YFP is facing the cytoplasm. The control plasmid, pCFP-CD3δ-YFP, encodes a chimera protein that contains an endoplasmic reticulum luminal CFP and a cytoplasmic YFP. Scale bars, 10 μm. D, Western blot of HRG-2 proteins produced by in vitro transcription and translation (IVT) system or by expression in HEK293 cells. There is no difference in the sizes of HRG-2 proteins (asterisks).
To resolve whether HRG-2 was inadvertently mislocalized or deliberately targeted to the ER, we synthesized truncations in HRG-2 and expressed them in HEK293 cells. Deletion of the GST-C domain resulted in HRG-2ΔGST-C, which localized to the ER (Fig. 4B, second row). However, deletion of the first 27 amino acids, which comprised the N-terminal transmembrane domain, resulted in an unstable protein that was undetectable by immunofluorescence (Fig. 4B, third row) or Western blotting (not shown). By contrast, fusion of the N-terminal 27 amino acids (HRG-2N) to YFP resulted in ER localization (Fig. 4B, bottom row). These results show that the N terminus is required for targeting HRG-2 to the ER membranes.
HRG-2 Is a Type I Membrane Protein
To determine whether the N terminus of HRG-2 was cleaved after ER targeting or retained for tethering to the ER membranes, we performed fluorescence protease protection assays. In this assay, transfected HEK293 cells are first incubated with digitonin to permeabilize the plasma membrane followed by time-dependent exposure to proteases, which cleaves cellular proteins that are exposed to the cytoplasm (23). As a positive control, we used the ER-targeted membrane protein CFP-CD3δ-YFP, which contains CFP located in the lumen and is thus resistant to protease digestion, and a cytoplasmic YFP, which is susceptible to protease cleavage (Fig. 4C, top two rows). HEK293 cells expressing the C-terminal tagged HRG-2-GFP or HRG-2N-YFP showed a complete loss of fluorescence signal after cell permeabilization followed by protease treatment, indicating that the N terminus of HRG-2 is required for membrane targeting and insertion and that the C terminus is exposed to the cytoplasm. To ensure that HRG-2 does not undergo post-translational modifications such as signal peptide cleavage, we compared the molecular weights for HRG-2 synthesized by either an in vitro coupled transcription and translation system or expression in HEK293 cell lines (Fig. 4D). The results from SDS-PAGE revealed that the sizes for HRG-2 were identical whether expressed in a cell-free system or HEK293 cells. Taken together, our results demonstrate that HRG-2 possesses a single N terminus transmembrane domain with a cytoplasmic C terminus characteristic of a type I membrane protein, a topology similar to the ER-localized heme-containing cytochrome P450s (38).
HRG-2 Rescues Growth of Heme-deficient Yeast Strain
HRG-2 is closely related to CDRs, of which seven paralogs exist in the worm preventing a thorough examination of the role HRG-2 plays in heme homeostasis within C. elegans. Clearly, hrg-2 is highly up-regulated at ≤4 μm heme, and the GST-like domains present in HRG-2 have been shown to bind heme in other nematodes (16, 39). To dissect the function of HRG-2 in a more simply defined system, we modeled the heme auxotrophy of C. elegans in a heme-deficient strain of S. cerevisiae (40). We exploited yeast, because it does not contain hrg-2 homologs and utilizes exogenous heme poorly even in the absence of endogenous heme synthesis (25).
Strain DY1457 hem1Δ(6D) lacks HEM1, which encodes for δ-aminolevulinic acid synthase, the first enzyme of the heme synthesis pathway (25). The hem1Δ strain therefore requires exogenous heme for growth. In comparison with the yeast transformed with empty vector, the expression of HRG-2 improved the growth by almost 100-fold, whereas the C. elegans plasma membrane heme transporter HRG-4, used as a positive control, showed more than 1000-fold better growth of hem1Δ at 0.1 μm heme (Fig. 5A, left panel) (7). This result was highly reproducible and consistent between untagged and tagged HRG-2. Increasing the heme concentrations diminished the growth differences between cells transformed with hrg-2 or the vector control (Fig. 5A, center panel).
FIGURE 5.
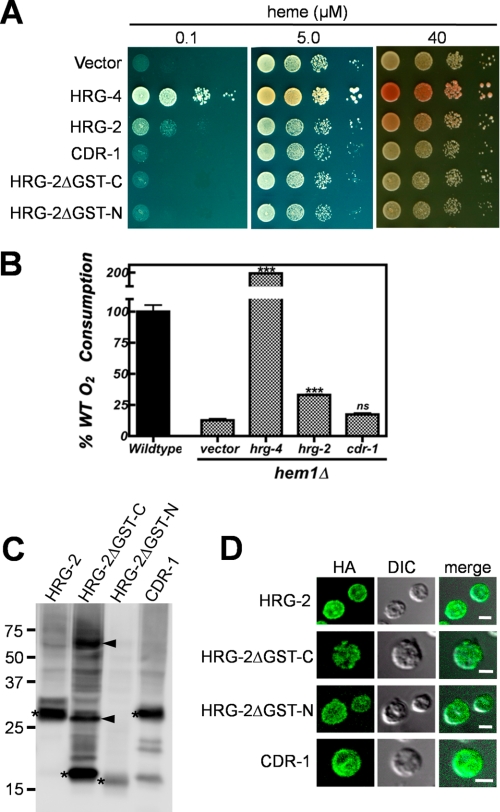
HRG-2 increases heme availability in heme-deficient S. cerevisiae. A, the DY1457 hem1Δ(6D) yeast strain transformed with indicated constructs were spotted in 10-fold serial dilutions on synthetic complete medium plates supplemented with different concentrations of heme. The vector pYES-DEST52 and the heme transporter HRG-4 were used as negative and positive controls, respectively. Yeast grown on the plate with 40 μm heme displayed red pigment accumulation due to mutation in the ADE2 locus. B, oxygen consumption rates (μm/min/mg) were measured for wild type and hem1Δ yeast grown in synthetic complete medium (2% raffinose and 0.8% galactose) with 5 μm heme using a Clark-type electrode. Rates were normalized to the wild type as the base line. HRG-2 and HRG-4 (***, p < 0.001) show significant increase in the rate of oxygen consumption in comparison with control vector or CDR-1. Error bars show ±S.E.; ns, not significant. C, Western blot of HRG-2 proteins and CDR-1 expressed in the yeast. The asterisk indicates the signal of the predicted size for each protein. Two arrowheads show putative dimers and tetramers for HRG-2ΔGST-C. The lower bands on the last lane (CDR-1) are degradation products of CDR-1 in yeast cells. D, immunofluorescence assays of HRG-2 proteins and CDR-1 expressed in yeast. The transformed yeast was fixed and spheroplasted. Anti-HA and fluorophore-conjugated anti-rabbit IgG antibodies were applied to detect the proteins. Images were acquired with different settings of laser power and detection gain on confocal microscope. Scale bars, 2 μm.
Notably, the hem1Δ strain transformed with hrg-2 accumulated red pigment at higher heme concentrations (40 μm). The red pigmentation is due to a mutation in ade2 that is in the hem1Δ genetic background (25, 41). The ade2 mutants accumulate phosphoribosylaminoimidazole, an intermediate in the adenine biosynthesis pathway, in its vacuole, and this turns red in the presence of oxygen (42, 43). Thus, greater pigment accumulation with HRG-2 is a sign that oxidative phosphorylation or mitochondrial respiration was restored in this heme-deficient mutant (Fig. 5A, right panel) (42, 43). The hem1Δ yeast transformed with hrg-2 and grown at a higher heme concentration (40 μm) consistently revealed more pigment accumulation than the vector control, indicating that the cells were able to utilize exogenous heme for oxidative metabolism and aerobic growth. To confirm this observation, we measured oxygen consumption rates using a Clark-type electrode with either S. cerevisiae hem1Δ transformed with vector, cdr-1, hrg-2, and hrg-4 or the wild type strain (Fig. 5B and supplemental Table S2). Our results show that HRG-2 and HRG-4 significantly increased the rate of oxygen consumption in the hem1Δ strain grown at 5 μm heme by 2.6 and 15-fold, respectively, compared with vector or CDR-1 controls (p < 0.001). These data are consistent with the observed differences in the growth of hem1Δ yeast transformed with the corresponding constructs (Fig. 5, A versus B).
Truncations of HRG-2 that either removed the predicted GST-N domain or the GST-C-terminal-like domain did not improve the growth of hem1Δ cells (Fig. 5A), suggesting that both domains are required for the function of HRG-2. As a positive control, the heme importer HRG-4 dramatically increased the growth of hem1Δ at all tested concentrations of heme and showed a greater accumulation of red pigment than HRG-2. Importantly, yeast transformed with CDR-1, an HRG-2 homolog in C. elegans, did not improve the growth of the hem1Δ yeast strain at any heme concentrations.
Western blot analysis revealed detectable amounts of proteins for all constructs expressed in the hem1Δ strain (Fig. 5C). Immunofluorescence results showed that the majority of HRG-2 was detected at the periphery of yeast, whereas only a small portion resided inside the cells (Fig. 5D). HRG-2ΔGST-N and CDR-1 exhibited localization patterns similar to HRG-2, whereas HRG-2ΔGST-C showed punctuate localization. Taken together, these results indicate that HRG-2 is localized to cell membranes and facilitates heme utilization in a heterologous yeast system.
HRG-2 Binds Heme in Vitro
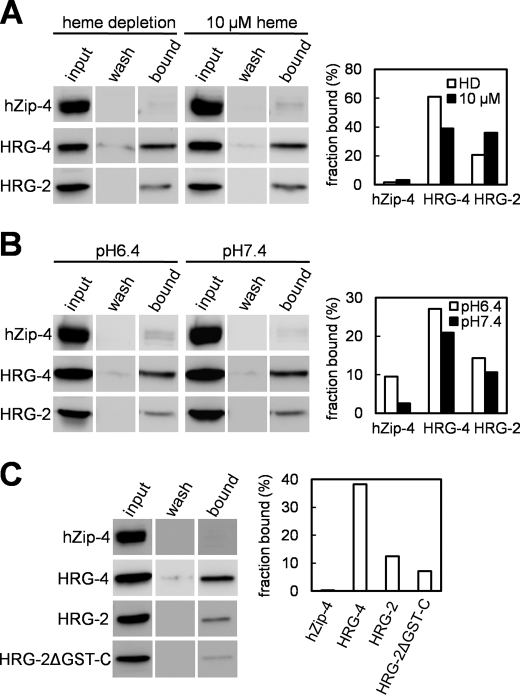
To test whether HRG-2 interacts directly with heme, we performed hemin-agarose binding assays with cell lysates from HEK293 cell lines that were transiently transfected with mammalian expression plasmids. To recapitulate the heme auxotrophy of C. elegans in mammalian cells, we either incubated HEK293 cells with heme-depleted growth medium plus succinylacetone, a heme synthesis inhibitor, or replenished the heme-depleted medium with 10 μm heme (see “Experimental Procedures”). To eliminate the possibility of nonspecific heme-protein interactions due to the hydrophobicity associated with membrane proteins, we used an eight-transmembrane domain protein, the human zinc transporter hZIP-4, as a negative control (44). We found that HRG-2 bound heme specifically, regardless of the intracellular heme status (Fig. 6A). Because a portion of HRG-2 localizes to the ER (Figs. 3 and 4), part of the secretory pathway that experiences a pH gradient from near neutral (ER, pH 7.2) to acidic (Golgi, pH 6.4; and vesicles, pH 5.5) (45), we tested whether HRG-2 binding to heme is pH-dependent. Reproducible binding was observed for HRG-2 when the assays were performed at either pH 6.4 or 7.4 (Fig. 6B), a result that was consistent with heme binding to the plasma membrane heme importer HRG-4 in C. elegans (6).
FIGURE 6.
HRG-2 binds heme. A and B, hemin-agarose pulldown assays were performed by incubating the lysates of transfected HEK293 cells with hemin-agarose. The transfected cells were either depleted of heme or supplemented with 10 μm heme (A), and the heme binding assays were performed at pH 6.4 or 7.4 (B). Equivalent amounts of input lysates (input), the final washes before elution (wash), and the eluates (bound) were subjected to SDS-PAGE and Western blotting using anti-HA antibodies. Hemin binding assays were performed 2–3 times, and one representative binding result is shown. Human zinc transporter hZip-4 and C. elegans heme transporter HRG-4 were used as the negative and positive controls, respectively. The panels to the right of the Western blots are the quantification of the signals as bound fractions relative to the input for each protein. C, heme binding assays on HRG-2ΔGST-C. The left and right panels are the Western blots and the quantification, respectively. HRG-2ΔGST-N was unstable when expressed in HEK293 and was excluded from the heme binding assays.
To determine the heme-binding region in HRG-2, we analyzed the predicted GST-N (HRG-2ΔGST-N) or the GST-C-terminal-like domain (HRG-2ΔGST-C) constructs. HRG-2 protein was still able to bind heme when the GST-C domain was removed, although this truncated protein bound lower amounts of heme (Fig. 6C). Heme binding studies could not be performed with HRG-2ΔGST-N because the truncated protein was expressed poorly in HEK293 cells.
Lack of hrg-2 in C. elegans Reveals Aberrant Cytochromes
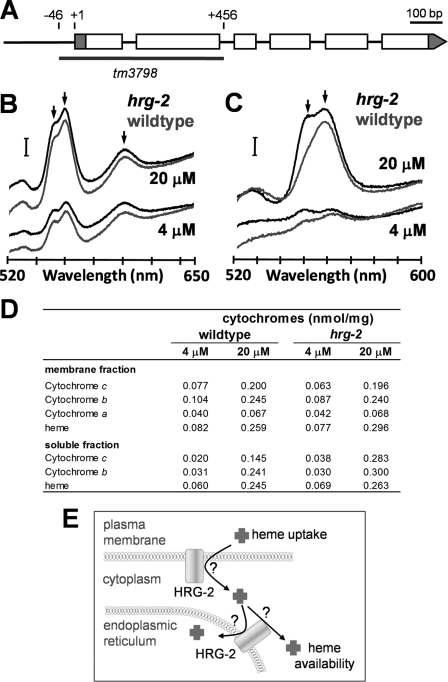
To determine the in vivo function of HRG-2, we analyzed worms containing a deletion in hrg-2. The tm3798 strain contains a 502-bp deletion that removes exons 1 and 2 plus 46 bp of the upstream sequence (Fig. 7A). Although hrg-2 mutant worms have no overt morphological defects, they consistently show >30% reduction in the total number of progeny compared with the wild type brood mate controls; heme supplementation, however, does not increase the number of progeny (supplemental Fig. S6).
FIGURE 7.
Analysis of hrg-2 deletion worm. A, location of tm3798 deletion in hrg-2 gene. In the tm3798 allele, part of the promoter region and the first two exons of the hrg-2 gene are deleted. Exons are depicted as empty boxes, and untranslated regions are shown as gray boxes. “+1” is the confirmed transcription start site. B, determination of the cytochrome content of membrane fraction. Oxidized minus reduced spectra for wild type (lower curves) and hrg-2 mutants (upper curves) at 20 and 4 μm heme. Protein concentrations for all samples were adjusted to 10 mg/ml. The arrows denote (left to right) the absorbance maxima for cytochrome c, cytochrome b, and a-type cytochromes. The bar denotes an absorbance of 0.02. C, determination of cytochrome content for cell soluble fraction. The wild type is the lower curve, and hrg-2 mutant worms are represented by the upper curve. Protein concentrations were 18 mg/ml for the wild type and 20 mg/ml for the mutant samples. Details are same as given in B. D, tabular presentation of heme and cytochrome content for the membrane and soluble fractions of the wild type and hrg-2 mutant worms. E, proposed model of HRG-2 in heme homeostasis in C. elegans. HRG-2 localizes to the apical plasma membrane and the ER in hypodermal cells. On the plasma membrane, HRG-2 may function as a hemin reductase to facilitate heme import. On the ER membrane, HRG-2 may contribute to the sequestration or redistribution of intracellular heme.
Because HRG-2 expression increased heme utilization of hem1Δ yeast, we sought to determine whether heme levels or hemoproteins were altered in hrg-2 mutant worms. Worms were grown in mCeHR-2 axenic medium supplemented with either 4 or 20 μm heme, and lysates were enriched for membrane and cytosol fractions. Total protoheme was determined as the pyridine hemochromogen and levels of cytochromes b, c, and a were quantitated via their characteristic reduced minus oxidized visible spectra (Fig. 7, B–D). The data revealed that at 20 μm heme there is essentially no difference between heme and cytochrome content between wild type and hrg-2 mutant worms in the membrane fraction, although cytochrome content, particularly cytochrome c, is increased in the soluble fraction for the mutant worms. However, at 4 μm heme, the mutant worms possessed reduced amounts of membrane-associated cytochromes b and c.
To determine whether cytochrome expression was perturbed in hrg-2-deficient animals, we conducted a transcriptomic analysis using Affymetrix microarrays on total RNA extracted from hrg-2 mutants and wild type brood mate controls grown at 4 and 20 μm heme. Of the 75 cytochrome genes annotated in the worm genome, we observed that three genes at 4 μm heme and four genes at 20 μm heme were down-regulated in the mutant compared with the wild type across all three biological replicates (supplemental Fig. S7 and supplemental “Methods”). These gene expression analyses lend further support to the biochemical studies that cytochrome levels are aberrant in the hrg-2 mutants.
DISCUSSION
As a heme auxotroph, C. elegans relies solely on environmental heme for growth and reproduction (17). The intestine acquires dietary heme via heme permeases HRG-1 and HRG-4, and heme is distributed to extraintestinal tissues by HRG-3 (6, 7, 18), but it is unclear how the hypodermis and muscles acquire intestinally derived heme and coordinate heme homeostasis at the organismal level. In the current study we show that HRG-2 may play a role in maintaining heme homeostasis in the hypodermis of C. elegans. In response to heme deficiency, hrg-2 is up-regulated by more than 200-fold. HRG-2 localizes to the ER and apical plasma membrane in hypodermal cells. Functional characterization by heterologous expression in heme-deficient yeast strains suggests that HRG-2 promotes the utilization of exogenous heme and interacts directly with heme. Furthermore, we show that hrg-2 deficiency results in aberrant cytochrome c distribution that is only partially restored in the presence of heme.
hrg-2 was previously named cdr-5 because of its sequence homology to the cadmium-responsive gene, cdr-1 (34). However, the expression of hrg-2 is specifically regulated by heme and not cadmium. Besides CDR-1, HRG-2 is also homologous to CE22631, CE02505, CE22138, and five other putative CDRs in C. elegans, all of which contain GST-N and GST-C-terminal like metaxin domains. Although CDR-1, CDR-4, and CDR-6 have been proposed to be involved in either cadmium detoxification or longevity determination (46, 47), the specific molecular functions of these proteins are unknown. We have analyzed an hrg-2 (tm3798) deletion strain in which a 502-bp region, including part of the promoter and first two exons of hrg-2, was deleted. However, this mutant did not reveal any obvious defects in morphology or growth, possibly because of the existence of several HRG-2/CDR paralogs, which may compensate for hrg-2 loss of function. BLAST searches using HRG-2 or any of the homologous proteins as a query retrieve a single uncharacterized protein in mammals, birds, and fishes. In the fruit fly, an HRG-2/CDR homolog was identified as an enhancer of the tyrosine kinase abl in a forward genetic screen (48). The gene was named “failed axon connection” or fax because flies with mutations in both fax and abl display severe disruptions in axon connections.
Expression of HRG-2 in hem1Δ yeast increased cell viability as well as the oxidative metabolism by more than 100-fold (Fig. 5A). However, compared with HRG-4, a multi-span transmembrane permease, the growth rescue of HRG-2 is lower. Because HRG-2 contains only a single predicted transmembrane domain, it is unlikely that HRG-2 itself is a heme transporter. Because HRG-2 has a thioredoxin-like fold, we speculate that it may function as a membrane-associated oxidoreductase (Fig. 7E). Reductases have been shown to be essential for the uptake of metals. For example, duodenal cytochrome b (Dcytb) and six-transmembrane epithelial antigen of the prostate-3 (Steap3) were identified as ferric reductases associated with efficient iron uptake (49, 50). Additionally, studies have demonstrated that oxidized heme (hemin) needs to be reduced for its covalent attachment to apocytochrome c (51, 52). In Gram-negative bacteria, the cytochrome c synthetase CcmF was proposed to function as a quinol:heme oxidoreductase (52). In addition, the lipocalin α1-microglobulin has the ability to reduce hemin in cytochrome c and methemoglobin (53). Thus, under heme-limiting conditions, HRG-2 may function as a dedicated hemin reductase to increase the efficiency of heme import or its availability in hypodermal cells. The presence of HRG-2 on ER membranes further suggests that HRG-2 may mediate heme delivery to membrane-bound or luminal hemoproteins (Fig. 7E). Indeed, in the absence of hrg-2 and under heme-limiting conditions, intracellular sorting of heme is altered such that less membrane-associated cytochromes c and b are assembled. Interestingly, under heme-sufficient conditions, there is still evidence for disordered heme sorting, as there exists an increase in soluble hemoprotein content in the hrg-2 mutant worms. Notably, we quantitated hemoprotein content as the reduced minus oxidized difference spectra and assigned the values obtained to cytochromes c and b based upon standard procedures. However, the nature and function of the soluble hemoproteins is at present unknown, and it should be noted that they may not be cytochromes per se but cytoplasmic hemoproteins with as yet unidentified functions. Regardless of their specific nature, the significant observation is that in hrg-2 mutant worms, the overall cellular hemoprotein distribution and content is altered, as demonstrated by our biochemical and microarray analysis. Although very little is known about how heme is incorporated into hemoproteins, peroxidases in the secretory pathway likely acquire heme in the ER. This assumption is based on the evidence that even when the Golgi is disrupted by brefeldin A, the lysosomal heme-containing enzyme myeloperoxidase still receives its heme moiety (54).
Our studies revealed that HRG-2 binds heme, which may be mediated by the GST-like domains. Heme-binding activities have been demonstrated for GSTs from other organisms (16, 39, 55). HRG-2 may function to transfer the bound heme to target hemoproteins. For example, the heme-binding protein Dap1p in yeast and its human ortholog, PGRMC1, can interact with certain types of cytochrome P450s and increase their activities (56, 57). HRG-2 has identical membrane topology to the microsomal cytochrome P450s, which are a family of xenobiotic detoxification enzymes that require heme for activity. The C. elegans genome contains more than 75 cytochrome genes (58), some of which are significantly perturbed in the hrg-2 deletion strain (supplemental Fig. S7) raising the possibility that HRG-2 and its CDR paralogs may function as membrane-anchored “chaperones” and regulate heme homeostasis by associating with target hemoproteins.
Supplementary Material
Acknowledgments
We thank the National Bioresource Project and S. Mitani for the hrg-2 strain and M. Colombini for use of the YSI-5300 instrument.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant R01DK74797 (extramural funding ) (to I. H.) and an NIH Intramural Research Program grant from the NIDDK (to M. K.).

This article contains supplemental “Methods,” Figs. S1–S7, and Tables S1 and S2.
- HRG
- heme-responsive gene
- mCeHR
- modified C. elegans habitation and reproduction
- RACE
- rapid amplification of cDNA ends
- CDR
- cadmium-responsive
- ER
- endoplasmic reticulum
- TRAM
- translocating chain-associated membrane
- qRT-PCR
- quantitative real-time-PCR
- ANOVA
- analysis of variance.
REFERENCES
- 1. Olea C., Jr., Kuriyan J., Marletta M. A. (2010) Modulating heme redox potential through protein-induced porphyrin distortion. J. Am. Chem. Soc. 132, 12794–12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Severance S., Hamza I. (2009) Trafficking of heme and porphyrins in metazoa. Chem. Rev. 109, 4596–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schultz I. J., Chen C., Paw B. H., Hamza I. (2010) Iron and porphyrin trafficking in heme biogenesis. J. Biol. Chem. 285, 26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balla G., Vercellotti G. M., Muller-Eberhard U., Eaton J., Jacob H. S. (1991) Exposure of endothelial cells to free heme potentiates damage mediated by granulocytes and toxic oxygen species. Lab. Invest. 64, 648–655 [PubMed] [Google Scholar]
- 5. Jeney V., Balla J., Yachie A., Varga Z., Vercellotti G. M., Eaton J. W., Balla G. (2002) Pro-oxidant and cytotoxic effects of circulating heme. Blood 100, 879–887 [DOI] [PubMed] [Google Scholar]
- 6. Rajagopal A., Rao A. U., Amigo J., Tian M., Upadhyay S. K., Hall C., Uhm S., Mathew M. K., Fleming M. D., Paw B. H., Krause M., Hamza I. (2008) Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453, 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yuan X., Protchenko O., Philpott C. C., Hamza I. (2012) Topologically conserved residues direct heme transport in HRG-1-related proteins. J. Biol. Chem. 287, 4914–4924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keel S. B., Doty R. T., Yang Z., Quigley J. G., Chen J., Knoblaugh S., Kingsley P. D., De Domenico I., Vaughn M. B., Kaplan J., Palis J., Abkowitz J. L. (2008) A heme export protein is required for red blood cell differentiation and iron homeostasis. Science 319, 825–828 [DOI] [PubMed] [Google Scholar]
- 9. Quigley J. G., Yang Z., Worthington M. T., Phillips J. D., Sabo K. M., Sabath D. E., Berg C. L., Sassa S., Wood B. L., Abkowitz J. L. (2004) Identification of a human heme exporter that is essential for erythropoiesis. Cell 118, 757–766 [DOI] [PubMed] [Google Scholar]
- 10. Hrkal Z., Vodrázka Z., Kalousek I. (1974) Transfer of heme from ferrihemoglobin and ferrihemoglobin-isolated chains to hemopexin. Eur. J. Biochem. 43, 73–78 [DOI] [PubMed] [Google Scholar]
- 11. Yang Z., Philips J. D., Doty R. T., Giraudi P., Ostrow J. D., Tiribelli C., Smith A., Abkowitz J. L. (2010) Kinetics and specificity of feline leukemia virus subgroup C receptor (FLVCR) export function and its dependence on hemopexin. J. Biol. Chem. 285, 28874–28882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taketani S., Adachi Y., Kohno H., Ikehara S., Tokunaga R., Ishii T. (1998) Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J. Biol. Chem. 273, 31388–31394 [DOI] [PubMed] [Google Scholar]
- 13. Iwahara S., Satoh H., Song D. X., Webb J., Burlingame A. L., Nagae Y., Muller-Eberhard U. (1995) Purification, characterization, and cloning of a heme-binding protein (23 kDa) in rat liver cytosol. Biochemistry 34, 13398–13406 [DOI] [PubMed] [Google Scholar]
- 14. Harvey J. W., Beutler E. (1982) Binding of heme by glutathione S-transferase: a possible role of the erythrocyte enzyme. Blood 60, 1227–1230 [PubMed] [Google Scholar]
- 15. Tipping E., Ketterer B., Christodoulides L., Enderby G. (1976) The interactions of haem with ligandin and aminoazo-dye-binding protein A. Biochem. J. 157, 461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Rossum A. J., Jefferies J. R., Rijsewijk F. A., LaCourse E. J., Teesdale-Spittle P., Barrett J., Tait A., Brophy P. M. (2004) Binding of hematin by a new class of glutathione transferase from the blood-feeding parasitic nematode Haemonchus contortus. Infect. Immun. 72, 2780–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao A. U., Carta L. K., Lesuisse E., Hamza I. (2005) Lack of heme synthesis in a free-living eukaryote. Proc. Natl. Acad. Sci. U.S.A. 102, 4270–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen C., Samuel T. K., Sinclair J., Dailey H. A., Hamza I. (2011) An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 145, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Epstein H. F., Shakes D. C. (eds) (1995) Caenorhabditis elegans: Modern Biological Analysis of an Organism, Vol. 48, Academic Press, San Diego [Google Scholar]
- 20. Nass R., Hamza I. (2007) The nematode C. elegans as an animal model to explore toxicology in vivo: solid and axenic growth culture conditions and compound exposure parameters in Current Protocols in Toxicology (Costa L. G., Hodgson E., Lawrence D. A., Reed D. J., eds) pp. 1.9.1–1.9.18, John Wiley & Sons, Inc., Somerset, NJ: [DOI] [PubMed] [Google Scholar]
- 21. Gengyo-Ando K., Mitani S. (2000) Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64–69 [DOI] [PubMed] [Google Scholar]
- 22. Praitis V., Casey E., Collar D., Austin J. (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lorenz H., Hailey D. W., Lippincott-Schwartz J. (2006) Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods 3, 205–210 [DOI] [PubMed] [Google Scholar]
- 24. Zhu Y., Hon T., Ye W., Zhang L. (2002) Heme deficiency interferes with the Ras-mitogen-activated protein kinase signaling pathway and expression of a subset of neuronal genes. Cell Growth Differ. 13, 431–439 [PubMed] [Google Scholar]
- 25. Crisp R. J., Pollington A., Galea C., Jaron S., Yamaguchi-Iwai Y., Kaplan J. (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J. Biol. Chem. 278, 45499–45506 [DOI] [PubMed] [Google Scholar]
- 26. Smith K. M. (1975) Porphyrins and Metalloporphyrins, Elsevier Scientific Publishing Co., New York [Google Scholar]
- 27. Rieske J. S. (1967) The quantitative determination of mitochondrial hemoproteins. Methods Enzymol. 488–493 [Google Scholar]
- 28. Bjellqvist B., Basse B., Olsen E., Celis J. E. (1994) Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539 [DOI] [PubMed] [Google Scholar]
- 29. Thompson J. D., Higgins D. G., Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saitou N., Neil M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 31. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software, version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 32. Severance S., Rajagopal A., Rao A. U., Cerqueira G. C., Mitreva M., El-Sayed N. M., Krause M., Hamza I. (2010) Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLoS Genet. 6, e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cui Y., McBride S. J., Boyd W. A., Alper S., Freedman J. H. (2007) Toxicogenomic analysis of Caenorhabditis elegans reveals novel genes and pathways involved in the resistance to cadmium toxicity. Genome Biol. 8, R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dong J., Song M. O., Freedman J. H. (2005) Identification and characterization of a family of Caenorhabditis elegans genes that is homologous to the cadmium-responsive gene cdr-1. Biochim. Biophys. Acta 1727, 16–26 [DOI] [PubMed] [Google Scholar]
- 35. Blumenthal T., Evans D., Link C. D., Guffanti A., Lawson D., Thierry-Mieg J., Thierry-Mieg D., Chiu W. L., Duke K., Kiraly M., Kim S. K. (2002) A global analysis of Caenorhabditis elegans operons. Nature 417, 851–854 [DOI] [PubMed] [Google Scholar]
- 36. Cox E. A., Hardin J. (2004) Sticky worms: adhesion complexes in C. elegans. J. Cell Sci. 117, 1885–1897 [DOI] [PubMed] [Google Scholar]
- 37. Rolls M. M., Hall D. H., Victor M., Stelzer E. H., Rapoport T. A. (2002) Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell 13, 1778–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neve E. P., Ingelman-Sundberg M. (2008) Intracellular transport and localization of microsomal cytochrome P450. Anal. Bioanal. Chem. 392, 1075–1084 [DOI] [PubMed] [Google Scholar]
- 39. Zhan B., Liu S., Perally S., Xue J., Fujiwara R., Brophy P., Xiao S., Liu Y., Feng J., Williamson A., Wang Y., Bueno L. L., Mendez S., Goud G., Bethony J. M., Hawdon J. M., Loukas A., Jones K., Hotez P. J. (2005) Biochemical characterization and vaccine potential of a heme-binding glutathione transferase from the adult hookworm Ancylostoma caninum. Infect. Immun. 73, 6903–6911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Protchenko O., Shakoury-Elizeh M., Keane P., Storey J., Androphy R., Philpott C. C. (2008) Role of PUG1 in inducible porphyrin and heme transport in Saccharomyces cerevisiae. Eukaryot. Cell 7, 859–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Myers A. M., Pape L. K., Tzagoloff A. (1985) Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4, 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dorfman B. Z. (1969) The isolation of adenylosuccinate synthetase mutants in yeast by selection for constitutive behavior in pigmented strains. Genetics 61, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shadel G. S. (1999) Yeast as a model for human mtDNA replication. Am. J. Hum. Genet. 65, 1230–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim B. E., Wang F., Dufner-Beattie J., Andrews G. K., Eide D. J., Petris M. J. (2004) Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J. Biol. Chem. 279, 4523–4530 [DOI] [PubMed] [Google Scholar]
- 45. Wu M. M., Llopis J., Adams S., McCaffery J. M., Kulomaa M. S., Machen T. E., Moore H. P., Tsien R. Y. (2000) Organelle pH studies using targeted avidin and fluorescein-biotin. Chem. Biol. 7, 197–209 [DOI] [PubMed] [Google Scholar]
- 46. Dong J., Boyd W. A., Freedman J. H. (2008) Molecular characterization of two homologs of the Caenorhabditis elegans cadmium-responsive gene cdr-1: cdr-4 and cdr-6. J. Mol. Biol. 376, 621–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao V. H., Dong J., Freedman J. H. (2002) Molecular characterization of a novel, cadmium-inducible gene from the nematode Caenorhabditis elegans. A new gene that contributes to the resistance to cadmium toxicity. J. Biol. Chem. 277, 42049–42059 [DOI] [PubMed] [Google Scholar]
- 48. Hill K. K., Bedian V., Juang J. L., Hoffmann F. M. (1995) Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and failed axon connections (Fax), a novel protein in axon bundles. Genetics 141, 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McKie A. T., Marciani P., Rolfs A., Brennan K., Wehr K., Barrow D., Miret S., Bomford A., Peters T. J., Farzaneh F., Hediger M. A., Hentze M. W., Simpson R. J. (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol. Cell 5, 299–309 [DOI] [PubMed] [Google Scholar]
- 50. Ohgami R. S., Campagna D. R., Greer E. L., Antiochos B., McDonald A., Chen J., Sharp J. J., Fujiwara Y., Barker J. E., Fleming M. D. (2005) Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat. Genet. 37, 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nicholson D. W., Neupert W. (1989) Import of cytochrome c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc. Natl. Acad. Sci. U.S.A. 86, 4340–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Richard-Fogal C. L., Frawley E. R., Bonner E. R., Zhu H., San Francisco B., Kranz R. G. (2009) A conserved haem redox and trafficking pathway for cofactor attachment. EMBO J. 28, 2349–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Allhorn M., Klapyta A., Akerström B. (2005) Redox properties of the lipocalin α1-microglobulin: reduction of cytochrome c, hemoglobin, and free iron. Free Radic. Biol. Med. 38, 557–567 [DOI] [PubMed] [Google Scholar]
- 54. Nauseef W. M., McCormick S., Yi H. (1992) Roles of heme insertion and the mannose 6-phosphate receptor in processing of the human myeloid lysosomal enzyme, myeloperoxidase. Blood 80, 2622–2633 [PubMed] [Google Scholar]
- 55. Harwaldt P., Rahlfs S., Becker K. (2002) Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol. Chem. 383, 821–830 [DOI] [PubMed] [Google Scholar]
- 56. Hughes A. L., Powell D. W., Bard M., Eckstein J., Barbuch R., Link A. J., Espenshade P. J. (2007) Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 5, 143–149 [DOI] [PubMed] [Google Scholar]
- 57. Mallory J. C., Crudden G., Johnson B. L., Mo C., Pierson C. A., Bard M., Craven R. J. (2005) Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 25, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. C. elegans SequencingConsortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018 [DOI] [PubMed] [Google Scholar]
- 59. Labouesse M. (2006) Epithelial junctions and attachments. WormBook, doi/ 10.1895/wormbook.1.56.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.