Background: Neurexins and CIRL/latrophilin-1 (CL1) are independent synaptic receptors for α-latrotoxin.
Results: Neurexins and CL1 form a high affinity complex that mediates intercellular adhesion and is regulated by neurexin alternative splicing.
Conclusion: Thus, two independent α-latrotoxin receptors interact trans-cellularly to form a connection between neurons.
Significance: The neurexin-CL1 complex may be involved in trans-synaptic cell adhesion and mediate α-latrotoxin toxicity.
Keywords: Autism, Cell Adhesion, Cell Surface Receptor, G-protein-coupled Receptors (GPCR), Neurological Diseases, Neuroscience, Neurotoxin, Protein/Protein Interactions, Receptor Structure-Function, Synapses
Abstract
The G-protein-coupled receptor CIRL1/latrophilin-1 (CL1) and the type-1 membrane proteins neurexins represent distinct neuronal cell adhesion molecules that exhibit no similarities except for one common function: both proteins are receptors for α-latrotoxin, a component of black widow spider venom that induces massive neurotransmitter release at synapses. Unexpectedly, we have now identified a direct binding interaction between the extracellular domains of CL1 and neurexins that is regulated by alternative splicing of neurexins at splice site 4 (SS4). Using saturation binding assays, we showed that neurexins lacking an insert at SS4 bind to CL1 with nanomolar affinity, whereas neurexins containing an insert at SS4 are unable to bind. CL1 competed for neurexin binding with neuroligin-1, a well characterized neurexin ligand. The extracellular sequences of CL1 contain five domains (lectin, olfactomedin-like, serine/threonine-rich, hormone-binding, and G-protein-coupled receptor autoproteolysis-inducing (GAIN) domains). Of these domains, the olfactomedin-like domain mediates neurexin binding as shown by deletion mapping. Cell adhesion assays using cells expressing neurexins and CL1 revealed that their interaction produces a stable intercellular adhesion complex, indicating that their interaction can be trans-cellular. Thus, our data suggest that CL1 constitutes a novel ligand for neurexins that may be localized postsynaptically based on its well characterized interaction with intracellular SH3 and multiple ankyrin repeats adaptor proteins (SHANK) and could form a trans-synaptic complex with presynaptic neurexins.
Introduction
α-Latrotoxin, a component of black widow spider venom that induces massive exocytosis of synaptic vesicles (for a review, see Ref. 1), targets neurons via two major receptors: neurexins and CLs (calcium-independent receptors for latrotoxin or latrophilins)4 (2–6). Both receptors were affinity-purified from rat brain using immobilized α-latrotoxin and can independently mediate the actions of α-latrotoxin when expressed in transfected neuroendocrine cells (2–6).
Neurexins are presynaptic type-1 membrane proteins. Vertebrates contain three neurexin genes (neurexin-1, -2, and -3), each of which is expressed from independent promoters as two principal isoforms, longer α-neurexins and shorter β-neurexins (1, 7–10). The extracellular sequences of α-neurexins are composed of six laminin-neurexin-steroid-binding hormone (LNS) domains, three EGF-like domains, and a stalk region, whereas the extracellular sequences of β-neurexins contain only a short β-neurexin-specific sequence followed by a single LNS domain (corresponding to the sixth LNS domain of α-neurexins) and the stalk region (8). Both α- and β-neurexins have a short cytoplasmic tail terminated by a PDZ domain binding motif.
The primary transcripts of the neurexin genes are subject to extensive alternative splicing at five canonical sites (SS1–SS5), thereby potentially generating thousands of isoforms (9–11). Among the alternative splice sites, SS4 has been studied most intensely because it regulates the binding of neurexins to α-latrotoxin (12) and to three major endogenous ligands, neuroligins (13–16), leucine-rich repeat transmembrane proteins (LRRTMs) (17–19), and cerebellins (20, 21). The binding of α-latrotoxin, some neuroligins, and all LRRTMs to neurexins is optimal when the insert in SS4 is deleted, whereas the binding of cerebellins requires an insert in SS4 (12–17, 19–21).
Neurexins likely perform multiple functions at synapses, although their precise roles and mechanisms of action remain unclear. Overexpression of neuroligins in non-neuronal cells induces presynaptic specializations in co-cultured neurons, creating artificial synapses (22, 23); this activity requires binding of endogenous neurexins in the co-cultured neurons to the overexpressed neuroligin (24). Overexpression of neurexins, conversely, in non-neuronal cells induces postsynaptic specializations in the dendrites of co-cultured neurons (25, 26), although it is unclear which neurexin ligand is involved. Collectively, these observations indicated a role for neurexins in synapse formation. Deletion of α-neurexins in mice, however, does not produce a major loss of synapses but instead severely impairs synaptic function, killing the animals at birth (27, 28). The importance of neurexins is further buttressed by recent human genetics studies, which revealed that heterozygous deletions of neurexin-1α cause a strong predisposition to autism and schizophrenia, whereas the presence of two mutant neurexin-1α alleles (i.e. a quasi-homozygous mutation) produces severe neurological disorders (for example, see Refs. 29–34; for a review, see Ref. 35).
Different from neurexins, CLs are GPCRs of the cell adhesion family that are generated from three genes (CL1–3) and are also subject to alternative splicing, although not as extensively as neurexins (4–6). CL1 and CL3 are expressed almost exclusively in neurons, whereas CL2 is expressed ubiquitously (6). Cell adhesion-type GPCRs are characterized by large N-terminal extracellular sequences containing “cell adhesion”-type domains in addition to the typical GPCR sequences (36, 37). Although the extracellular domains of cell adhesion GPCRs vary, all include a recently identified large juxtamembranous region called the GPCR autoproteolysis-inducing (GAIN) domain that contains an integral “GPCR proteolysis site” at its C terminus and mediates the autoproteolysis of cell adhesion GPCRs at a single site (53). Among multiple families of adhesion-type GPCRs found in mammals, only CLs and a second class (the cadherin, EGF-like, Laminin G-like seven-pass receptor (CELSR) class) are evolutionarily conserved (36, 37).
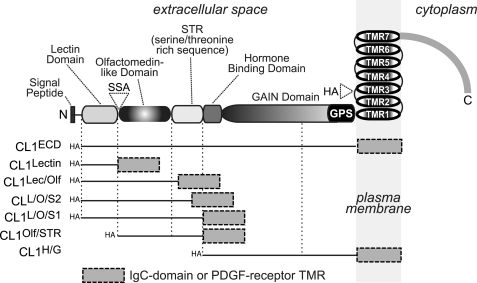
The extracellular sequences of CLs are composed of a lectin domain, an olfactomedin-like domain, a serine/threonine-rich region that may be glycosylated, a hormone-binding domain that is highly homologous to that of the otherwise unrelated corticotropin-releasing factor receptor, and a GAIN domain that includes the GPCR proteolysis site (Fig. 1). In addition to these extracellular domains, CLs contain the typical seven transmembrane regions (TMRs) of GPCRs followed by a rather long cytoplasmic tail. Most of the alternative splicing of CLs affects its cytoplasmic tail except for splice site A (SSA) in its N-terminal sequence (Fig. 1).
FIGURE 1.
CL1 domain structure and CL1 fragments used for current study. The domain structure of CL1 is shown on top (SSA (KVEQK) after Tyr131; the GAIN domain contains the GPCR proteolysis sequence (GPS) as an integral component; in addition, the positions of N and C termini as well the insertion site of the HA epitope tag in full-length recombinant CL1 are indicated). The straight lines below the domain structure indicate the regions that are included in the respective recombinant proteins used for analyses; these proteins are fused at the C terminus either to the Fc region of human IgG or to the TMR of the PDGF receptor preceded by a myc epitope.
The discovery of neurexins and CLs as putative neuronal cell adhesion molecules and α-latrotoxin receptors triggered a search for endogenous ligands of these receptors. As mentioned above, numerous ligands for neurexins have been described, in particular GABA receptors, neuroligins, LRRTMs, cerebellins, neurexophilins, and dystroglycan (13–21, 39–42). In contrast, a putative ligand for CLs was described (teneurin2/ten-m2/Odz2) only recently (43), although the modular domain structure of CLs suggests that additional ligands are likely to exist. Despite the fact that the high α-latrotoxin binding affinity of both neurexins and CL1 suggested that they do not act as co-receptors for α-latrotoxin, knock-out studies indicated complementary roles as α-latrotoxin receptors (44). Here, we reasoned that if neurexins and CL1 are part of a common pathway, they might also act as interacting partners, and we show that they indeed interact with a high affinity in a manner consistent with an intercellular adhesion complex.
EXPERIMENTAL PROCEDURES
Antibodies
Mouse monoclonal anti-hemagglutinin (HA) antibody was from Covance (Clone 16B12). Rabbit polyclonal anti-FLAG antibody was from Sigma. Monoclonal GFP antibody (Clone 3E6) was from Invitrogen. Rabbit polyclonal synapsin (E028) antibody was described previously (23).
Construction of Expression Vectors
pDisplay Vectors
All pDisplay vectors contain an Igκ signal peptide, an N-terminal HA epitope, and a C-terminal c-myc epitope followed by the PDGF receptor TMR (Invitrogen). The various pDisplay-CL1 expression plasmids were constructed after subcloning the respective PCR-amplified fragments into the SacII-SalI sites of the pDisplay vector and encode the following rat CL1 residues fused to the PDGF receptor TMR: pDisplay-CL1ECD, Ser26–Glu856; pDisplay-CL1Lectin, Ser26–Tyr131; pDisplay CL1L/O/S1, Ser26–Ser475; pDisplay-CL1L/O/S2, Ser26–Ser427 lacking an insert in splice site A; pDisplay-CL1L/O/S2+SSA, Ser26–Ser427 with inclusion of an insert in splice site A; pDisplay-CL1Lec/Olf, Ser26–Ser402; pDisplay-CL1Olf/STR, Val137–Ser475; and pDisplay-CL1H/G, Leu472–Glu856.
pCMVIg-CL1 Plasmids
pCMVIg-CL1 plasmids were generated by subcloning the EcoRI-SalI fragments from the various CL1 pDisplay vectors into the same sites of pCMVIg9 vector (8), resulting in constructs encoding the Igκ signal peptide and HA epitope from the pDisplay vector as well as the different CL1 sequences fused to the human IgG Fc domain. Only pCMVIg-CL1Lectin that encodes CL1 residues Met1–Tyr131 fused to human IgG Fc fragment was cloned by direct insertion of a PCR fragment into the EcoRI-SalI site of pCMVIg vector.
Full-length CL1 Vectors
pCMV-CL1-1 and pCMV-CL1-5 encode full-length CL1 lacking both splice inserts or containing an insert in SSA, respectively (CL1FL and CL1FL+SSA) and were described previously (6). pCMV-CL1HA encodes full-length CL1 lacking both splice inserts in which the HA epitope was inserted by PCR between Tyr918 and Glu919 in the first extracellular loop of CL1. pCMV-CL1mVenus encodes full-length CL1 fused to mVenus in the cytoplasmic tail between Ala1344 and Lys1345.
Full-length Neurexin Vectors
pCMV-N1α-1 FLAG and pCMV-N1α-41 FLAG encode full-length neurexin-1α fused to a FLAG epitope lacking or containing an insert in splice site 4 (Nrx1α−SS4 and Nrx1α+SS4), respectively, and were described previously (15). pCMV-N1β-1 FLAG and pCMV-N1β-3 FLAG encode full-length neurexin-1β fused to a FLAG epitope lacking or containing an insert in splice site 4 (Nrx1β−SS4 and Nrx1β+SS4), respectively, and were described previously (15).
Ig-Neurexin Vectors
pCMVIg-N1α-1 and pCMVIg-N1β-1 encode extracellular domains of neurexin-1α and neurexin-1β, respectively, both lacking an insert in splice site 4 (IgNrx1α−SS4 and IgNrx1β−SS4) and were described elsewhere (8).
Full-length Neuroligin-1
pCMV FLAG NL1ΔAB encodes full-length neuroligin-1 fused to a FLAG epitope and lacking a splice insert in both splice sites A and B (NL1ΔAB) and was described previously (15).
Soluble Neuroligin-1 Vector
pCMV NL1ΔAB* encodes the extracellular esterase-like domain of neuroligin-1 lacking both inserts in splice sites A and B (NL1ΔAB*) fused to a FLAG epitope at the N terminus and was described previously (15).
Cell Culture and Transfection
For immunoblotting and immunocytochemistry experiments, HEK293T cells were cultured in 6-well plates until they reached 70–80% confluence. The cells were then transfected using 4 μl of FuGENE 6® (Roche Diagnostics) and 2 μg of the respective CL1 DNA constructs. For immunoblotting experiments, cells were harvested 48 h post-transfection. For immunocytochemistry experiments, the cells were trypsinized 24 h post-transfection and split into 12-well plates containing coverslips that had been previously coated with 0.5 g/liter poly-l-lysine in borate buffer. The cells were grown on the coverslips for an additional 24 h before further experiments were done.
Production and Purification of Recombinant Protein
For recombinant Ig protein expression, HEK cells were cultured in 10-cm dishes until they reached 80% confluence. The medium was changed to fresh DMEM containing 25 mm chloroquine, and cells were incubated for 3 h and then transfected using calcium phosphate with 20 μg of cDNA corresponding to the various Ig proteins. Media containing the soluble Ig proteins were harvested 4 days post-transfection and cleared by centrifugation at 1,000 × g. The supernatant was then adjusted to 10 mm Hepes-NaOH, pH 7.4, 1 mm EDTA, and protease inhibitors (Roche Applied Science) and incubated overnight with protein A-Sepharose (GE Healthcare) to bind the human IgG Fc domain. The beads were then washed to remove unbound proteins.
Pulldown and Immunoblotting Experiments
HEK293T cells expressing full-length FLAG-neurexin-1β−SS4 were harvested 48 h post-transfection and incubated for 1 h at 4 °C in solubilization buffer (100 mm Hepes-NaOH, pH 7.4, 0.1 mm EDTA, 2 mm CaCl2, 2 mm MgCl2, 1% Triton X-100, and 100 mm NaCl). The solubilizate was centrifuged at 20,000 × g to remove insoluble materials, and the supernatant was incubated with 0.15 μm CL1-Ig fusion proteins supplemented with protein A-Sepharose beads for a period of 16 h at 4 °C with gentle agitation. Protein A beads were washed three times with solubilization buffer, solubilized in SDS sample buffer, and loaded onto an 8% SDS-polyacrylamide gel. Gels were then transferred onto nitrocellulose membranes and processed using standard procedures. Bound FLAG-neurexin was detected using a rabbit polyclonal anti-FLAG antibody followed by horseradish peroxidase-coupled secondary antibody, incubated with ECL reagents, and revealed on x-ray films.
Immunocytochemistry and Image Acquisition
Cells transfected with CL1 constructs were washed once with PBS and fixed with 4% paraformaldehyde for 10 min on ice. Cells were washed again three times with cold PBS and incubated at room temperature for 30 min in a blocking solution containing 3% BSA in PBS with or without 0.1% Triton X-100, respectively, depending upon whether cells were permeabilized or not. Mouse anti-HA antibody was then added (1:500 ratio), and the incubation continued for another 2 h. Cells were washed three times with blocking solution and incubated for 1 h at room temperature with anti-mouse Alexa Fluor 488 fluorescent antibody to label CL1 receptors. Cells were finally washed again three times with blocking solution and once with water before mounting on slides using medium containing DAPI for nuclear staining. Slides were then analyzed by confocal microscopy. Images were acquired using the confocal microscope Leica TCS2. The same confocal acquisition settings were applied to all samples of the experiment. Collected z-section images were analyzed blindly using Leica confocal software.
Cell Adhesion Assays
Cell adhesion assays were performed with HEK293T cells as described with slight modifications (17). HEK293T cells were individually transfected with the expression vectors as indicated in the figures. After 48 h, the cells were detached using 1 mm EDTA in PBS, mixed, and incubated under gentle agitation at room temperature in DMEM containing 10% FBS, 50 mm Hepes-NaOH, pH 7.4, 10 mm CaCl2, and 10 mm MgCl2. The extent of cell aggregation was assessed at 90 min by removing aliquots, spotting them onto culture slides (BD Falcon), and imaging by epifluorescence microscopy. The resulting images were then analyzed by counting the number and size of particles in the field using Wright Cell Imaging Facility ImageJ. An arbitrary value for particle size was then set as a threshold based on negative control values. The aggregation index was calculated by expressing the number of particles exceeding this threshold as a percentage of the total particles in the individual fields.
Saturation Ligand Binding Assays
Saturation ligand binding assays were performed essentially as described (17). Briefly, transfected HEK293 cells were incubated in DMEM containing 50 mm Hepes-NaOH, pH 7.4, 2 mm CaCl2, 2 mm MgCl2, 0.1% BSA, and the indicated amount of Ig fusion protein in serial dilutions for a period of 16 h at 4 °C with gentle agitation. Cells were then washed three times with cold DMEM to remove excess Ig proteins and fixed with 4% paraformaldehyde for 10 min on ice. Cells were washed again with cold PBS and incubated at room temperature for 15 min in a blocking solution containing PBS and 3% BSA. Rabbit anti-human IgG antibody coupled to horseradish peroxidase was then added (1:80,000 ratio in blocking solution), and the incubation continued for another hour. Cells were washed three times with blocking solution and once with PBS before performing the colorimetric assay according to the manufacturer's instructions. Briefly, 3,3′,5,5′-tetramethylbenzidine peroxidase enzyme immunoassay solution (Bio-Rad) was added to each well and incubated at room temperature for 10 min under vigorous agitation until a blue coloration appeared. The reaction was stopped by adding an equal volume of 1 n sulfuric acid, which produced a yellow coloration, and absorbance at 450 nm was measured in 96-well plates using an Apollo-8 LB912 plate reader (Berthold Technologies). The ligand concentration was plotted against the difference in absorbance measured between transfected and mock-transfected cells. The Kd was determined using Scatchard plot analysis.
Cell Surface Labeling
Cell surface labeling was performed essentially as described (15). HEK293T cells transfected with the indicated expression vector were incubated in DMEM containing 20 mm Hepes-NaOH, pH 7.4, 0.1% BSA, and 0.15 μm Ig fusion protein for a period of 16 h at 4 °C with gentle agitation. Cells were then washed three times with cold DMEM to remove excess Ig proteins and fixed with 4% paraformaldehyde for 10 min on ice. Cells were washed again three times with cold PBS and incubated at room temperature for 15 min in a blocking solution containing PBS and 3% BSA. Rabbit anti-human IgG antibody was then added (1:500 ratio), and the incubation continued for another 2 h. Cells were washed three times with blocking solution and incubated with anti-rabbit Alexa Fluor fluorescent antibody (red emission) in the blocking solution for 1 h at room temperature to label bound Ig-neurexins. Cells were finally washed again three times with blocking solution and once with water before mounting on slides using medium containing DAPI for nuclear staining. Slides were then analyzed by confocal microscopy. Images were acquired using the confocal microscope Leica TCS2. The same confocal acquisition settings were applied to all samples of the experiment. Collected z-section images were analyzed blindly using Leica confocal software.
Ligand Tracking Experiments
HEK293 cells were transfected using FuGENE 6 with plasmid encoding CL1 and cultured for 48 h. The cells were then incubated in Eagle's minimum essential medium containing 3% BSA, 2 mm CaCl2, and 2 mm MgCl2 for 5 min before adding 200 nm Ig-neurexin-1β, and incubation continued for 10, 30, or 60 min at 37 °C. Internalization was then stopped by washing with cold PBS while maintaining the cells on ice. Cells were then fixed with 4% paraformaldehyde in PBS for 10 min on ice and blocked with PBS containing 3% BSA, 2% goat serum, and 0.1% Triton X-100 for 30 min at room temperature. Immunocytochemistry was performed using antibody against human IgG Fc fragment (1:500) and secondary antibody coupled to Alexa Fluor 633 (1:500) for detection. CL1-expressing cells were monitored by detecting the presence of Ig-neurexin1b fluorescence using confocal microscopy.
Cell Surface Neurexin Recruitment Assay
Cell adhesion assays were performed with HEK293T cells as described under “Cell Adhesion Assays.” HEK293T cells were individually transfected with the expression vectors as indicated in the figures. After 48 h, the cells were detached using 1 mm EDTA in PBS, mixed, and incubated under gentle agitation at room temperature in DMEM containing 10% FBS, 50 mm Hepes-NaOH, pH 7.4, 10 mm CaCl2, and 10 mm MgCl2. Cells were then plated on coverslips and fixed with 4% paraformaldehyde for 10 min on ice. Cells were washed again three times with cold PBS and incubated at room temperature for 15 min in a blocking solution containing PBS and 3% BSA. Rabbit anti-FLAG antibody was then added (1:500 ratio), and the incubation continued for another 2 h. Cells were washed three times with blocking solution and incubated with anti-rabbit Alexa Fluor fluorescent antibody (red emission) in the blocking solution for 1 h at room temperature to label surface neurexin-1β-FLAG. Cells were finally washed again three times with blocking solution and once with water before mounting on slides using medium containing DAPI for nuclear staining. Slides were then analyzed by confocal microscopy. Images were acquired using the confocal microscope Leica TCS2. The same confocal acquisition settings were applied to all samples of the experiment. Mean fluorescence intensity was analyzed using the line scan application from Leica Lite software on neurexin-1β-FLAG-expressing cells. Regions of interest were defined as “opposed” for the region that immediately opposed the CL1mVenus-expressing cells and “adjacent” for a region that was adjacent to but not opposing CL1mVenus-expressing cells. Statistical significance was determined by Student's t test, and all the data were expressed as means ± S.E.
Artificial Synapse Formation Assays
Artificial synapse formation assays were performed with COS-7 cells as described previously (24). COS-7 cells were transfected with FuGENE 6 (Roche Applied Science) with plasmids expressing pCMV5-NL1ΔAB-mVenus, pCMV-CL1-mVenus, or mVenus alone. After 24 h, transfected COS-7 cells were trypsinized, seeded onto hippocampal neurons cultured 9 days earlier (i.e. were at 9 days in vitro), further co-cultured for 48 h, and immunostained with GFP and synapsin antibodies at 11 days in vitro. All images were acquired by confocal microscopy. For quantitations, the contours of the transfected COS-7 cells were chosen as the region of interest. Fluorescence intensity of synapsin puncta normalized to each COS-7 cell area was quantified for both red and green channels with MetaMorph software. Statistical significance was determined by Student's t test, and all the data were expressed as means ± S.E.
RESULTS
Neurexins Are Potential Endogenous Ligands for CL1
To investigate whether neurexins interact with CLs, we used a cell surface labeling assay (15). We incubated HEK293 cells expressing HA-tagged full-length CL1 with various neurexin-1-Ig fusion proteins using IgC as a negative control. The Ig fusion proteins contain the Fc domain of human IgG with IgC including only the signal peptide and a few N-terminal residues of neurexin-1α (8). The neurexin-1-Ig fusion proteins contained the full-length extracellular sequences of the respective neurexins. Bound Ig proteins were detected by immunofluorescence using an antibody against the human Ig Fc domain moiety.
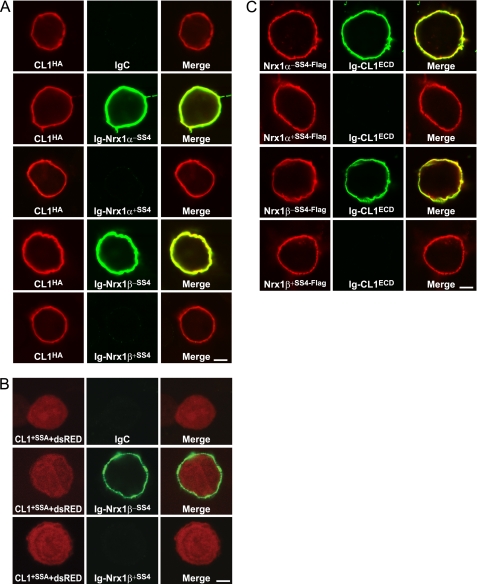
We first tested neurexin-1α and neurexin-1β both without and with an insert in SS4 (Fig. 2A). We found that both neurexins bound to surface-expressed CL1 but only when the insert in SS4 was spliced out, whereas IgC did not.
FIGURE 2.
Extracellular sequences of neurexin-1 and CL1 bind to each other: regulation by alternative splicing of neurexin-1α and -1β at SS4. A, HEK293T cells expressing full-length HA-tagged CL1 (CL1HA) without an insert in SSA were incubated with soluble Ig-neurexin-1α and -1β (0.2 μm) fusion proteins containing or lacking an insert in SS4 as indicated (Ig-Nrx1α−SS4, Ig-Nrx1α+SS4, Ig-Nrx1β−SS4, and Ig-Nrx1β+SS4, respectively) or IgC as a negative control, washed, and fixed in 4% paraformaldehyde on ice for 10 min. Cells were analyzed by immunofluorescence using primary antibodies to the Ig fusion proteins and to HA followed by fluorescent secondary antibodies (green, Alexa Fluor 488; red, Alexa Fluor 633). The fluorescence signal was visualized in a confocal microscope. B, same as A except that HEK293 cells expressed full-length CL1 with an insert in SSA but without an HA tag; to visualize the cells, HEK293 cells co-expressed Discosoma red fluorescent protein (DsRed), which fills the cytoplasm (CL1+SSA+dsRed). C, same as A except that the HEK293 cells expressed the indicated full-length neurexins containing an N-terminal FLAG epitope tag and were incubated with soluble CL1-Ig fusion protein (0.2 μm) containing the entire extracellular sequences of CL1 (Ig-CL1ECD; see Fig. 1). Data shown are representative images of experiments that were repeated at least three times. Scale bars (4 μm) apply to all images in a set.
To confirm the interaction of neurexin-1α and -1β with CL1, we repeated the same assay in the reverse orientation. We expressed full-length FLAG-tagged neurexin-1α or -1β without and with an insert in SS4 in HEK293 cells and incubated the cells with an Ig fusion protein containing the entire extracellular sequences of CL1 (CL1ECD; Fig. 1). Again, we observed binding of CL1 only to neurexin-1 splice variants lacking an insert in SS4 (Fig. 2B). These results suggest that neurexin-1α and -1β both bind to CL1 in an extracellular interaction regulated by alternative splicing of neurexins at SS4, similar to the binding of neurexins to neuroligins (15) and LRRTMs (17). Similar assays revealed that neurexin-2 and -3 also bind to CL1 (see below), suggesting that this interaction is generally applicable for neurexins. Because the extracellular sequences of β-neurexins contain only a single identified domain, the LNS domain (8), that constitutes the sixth LNS domain of α-neurexins, this domain likely mediates the binding of CL1.
CL1-containing Bound Neurexin-1β Is Internalized
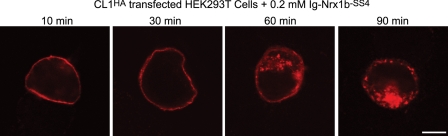
Internalization of GPCRs constitutes one of several GPCR desensitization processes that occur after activation by an agonist but is prevented by antagonists (45, 46). To investigate whether CL1 is internalized after neurexin binding, we conducted a ligand tracking experiment in which we incubated HEK293 cells expressing surface-exposed CL1 at 37 °C with either Ig-neurexin-1β or IgC. The cells were then fixed, and Ig-neurexin-1β and IgC were assayed by immunofluorescence with an antibody to the Ig Fc domain moiety. No signal was obtained with IgC (not shown), whereas the neurexin-1β signal was detectable on the surface of the HEK293 cells after only 10 min of incubation. After extended incubations of 60 and 90 min, we observed the neurexin-1β signal in vesicular and vacuolar structures inside the cells (Fig. 3). IgC used as a negative control was not detected at any step of the incubation period, suggesting that the endocytosis observed with Ig-neurexin-1β is dependent on direct binding of the neurexin to surface-expressed CL1 and is not due to nonspecific endocytosis.
FIGURE 3.
Ig-neurexin-1β is endocytosed by CL1-expressing cells. HEK293 cells expressing full-length CL1HA were incubated in internalization medium (minimum Eagle's medium supplemented with 0.5% glucose and 2 mm CaCl2) at 37 °C for 10, 30, 60, and 90 min in the presence of soluble Ig-neurexin-1β (0.2 μm to achieve receptor saturation), placed on ice, and washed with cold PBS. Cells were fixed for 10 min on ice using 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and immunostained using antibodies to human IgG followed by fluorescent secondary antibody (red, Alexa Fluor 633). Images are representative of at least three separate independent experiments. Scale bar, 4 μm.
Neurexins and CLs Interact with Nanomolar Affinities
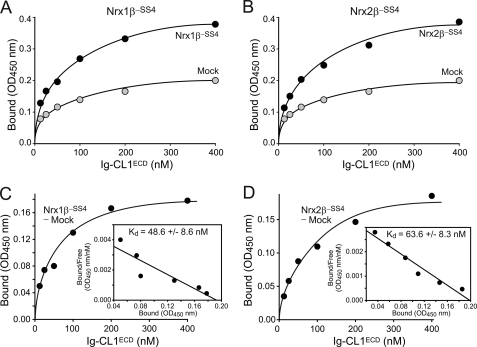
To further characterize the binding of CL1 to neurexins, we conducted quantitative cell surface binding assays that allow calculations of approximate binding affinities. We expressed full-length neurexin-1β and -2β lacking an insert in SS4 in transfected HEK293 cells and incubated the cells with soluble CL1ECD-Ig fusion protein at concentrations of 20–400 nm (Fig. 4). Mock-transfected HEK293 cells were used as negative controls, and bound Ig proteins were quantified using an HRP-tagged secondary antibody that was measured using a colorimetric substrate. To calculate binding affinities, the net binding of CL1ECD protein at any given concentration was computed and plotted as a function of the CL1ECD concentration (Fig. 4, A and B). The binding curve was then fitted to a Scatchard function, assuming a single independent binding site for CL1 in a neurexin molecule (Fig. 4, C and D).
FIGURE 4.
CL1 interacts with neurexin-1β and -2β with nanomolar affinities. A and B, HEK293 cells expressing neurexin-1β (A) or -2β (B; both without an insert in SS4) and control mock-transfected cells were incubated with soluble CL1-Ig fusion protein containing the entire extracellular sequences of CL1 (Ig-CL1ECD; see Fig. 1) at increasing concentrations, washed, fixed, and probed with an HRP-tagged secondary antibody. Binding incubations with the antibody were carried out for 60 min at room temperature in PBS containing 3% BSA. The amount of antibody bound was determined by colorimetry and is plotted as a function of the CL1-Ig fusion protein concentration. C and D, plot of net Ig-CL1ECD binding to neurexin-1β (C) or -2β (D) obtained by subtracting the background binding obtained with mock-transfected cells from the binding obtained with cells expressing neurexins. Insets show a Scatchard analysis of the binding results with the mean affinity ± S.E. calculated from multiple experiments (n = 3 for neurexin-1β and -2β).
The results of these experiments reveal that CL1 binds to neurexin-1β and -2β with slightly different nanomolar affinities (Fig. 4). Binding was saturable and exhibited good reproducibility in independent transfection and incubation experiments.
Neurexin/CL1 Interaction Mediates Ca2+-dependent Intercellular Adhesion
The binding of the extracellular neurexin and CL1 domains to each other raised the possibility that such binding could mediate intercellular adhesion, i.e. produce a junction between cells. Given the presumptive synaptic functions of neurexins and CL1, this attractive possibility would imply a trans-synaptic interaction.
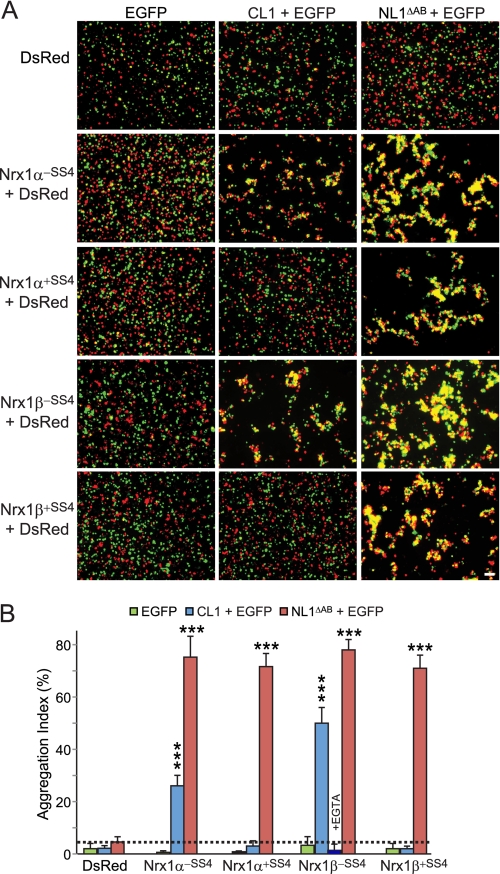
Thus, to test whether neurexins and CL1 can interact in a trans configuration and mediate cell adhesion, we co-transfected HEK293 cells separately either with neurexin-1α or -1β and DsRed or with CL1 and EGFP. As a positive control, we transfected cells with NL1ΔSSAB and EGFP; as negative controls, we transfected cells only with either DsRed or EGFP.
Red cells expressing DsRed alone or with neurexin-1 were mixed with green cells expressing either EGFP alone, EGFP with CL1, or EGFP with NL1ΔSSAB, and cell aggregation was measured after incubating the cell mixtures for up to 90 min (Fig. 5A). Aggregation was monitored by fluorescence microscopy and image analysis with “aggregation” defined as areas of clustered red and green fluorescence exceeding 3000 pixels2. It is important to note that we did not observe aggregation for individually expressed constructs nor for any combinations involving cells expressing only EGFP or DsRed (Fig. 5, A and B). Quantitations of the aggregation index revealed that CL1- and neurexin-1α- and -1β-expressing cells formed adhering clumps of cells but only when the neurexins lacked an insert in SS4 (Fig. 5B). In contrast, NL1ΔSSAB-expressing cells formed aggregates with cells expressing neurexin-1α and -1β either with or without an insert in SS4. No aggregation was ever observed in any combination of cells in which one or the other cell expressed only EGFP or DsRed (Fig. 5). At least the aggregation of neurexin-1β-containing cells with CL1-expressing cells was Ca2+-dependent as it was disrupted by addition of EGTA, which brought aggregation to the same levels as negative controls (Fig. 5), similar to the effect of EGTA on the cell adhesion induced by neuroligin binding to neurexins (47).
FIGURE 5.
CL1/neurexin-1 interaction mediates intercellular adhesion. A, HEK293T cells expressing either EGFP alone or together with CL1 or neuroligin-1 lacking an insert in splice sites A and B (NL1ΔAB) were mixed with cells expressing DsRed alone or together with neurexin-1α or -1β with or without an insert in SS4 (Nrx1α−SS4, Nrx1α+SS4, Nrx1β−SS4, and Nrx1β+SS4) and incubated at room temperature for 90 min in DMEM containing 50 mm Hepes-NaOH, pH 7.4, 10% FBS, 10 mm CaCl2, and 10 mm MgCl2. Cells were imaged by fluorescence microscopy. B, summary graphs of the aggregation index determined in independent cell adhesion assays (n = 3) such as those shown in A. The aggregation index was calculated as the percentage of the total particle surface that is present in particles exceeding a threshold of 3000 pixels2. For the samples containing cells expressing neurexin-1β−SS4 and CL1, an additional control was carried out in which the Ca2+ in the medium was replaced by EGTA. Data shown are means ±S.E. Error bars: standard error. Scale bar, 100 μm. Statistical significance was assessed by comparing CL1- or NL1ΔAB-expressing cells with the control expressing EGFP alone using one-way analysis of variance (***, p < 0.0001).
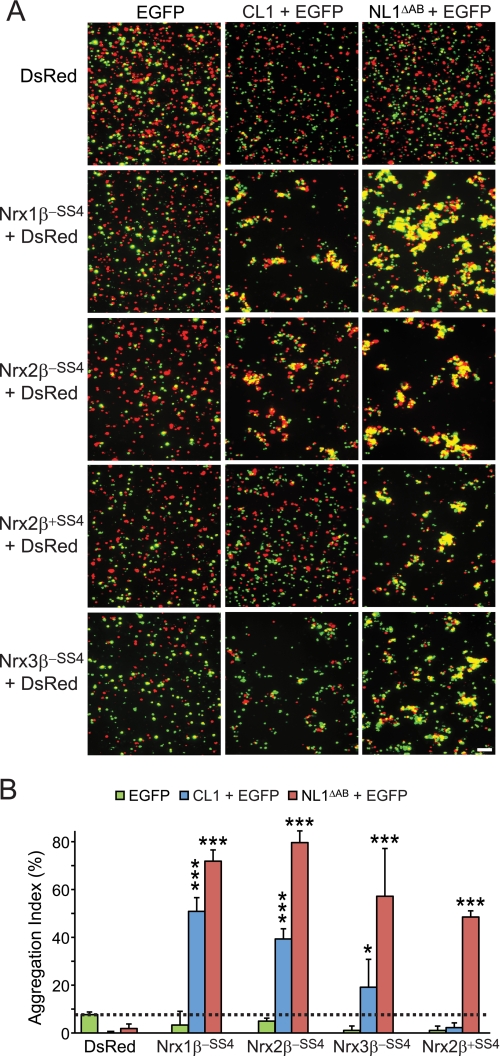
In a follow-up experiment, we used the same approach to test whether CL1 could also form cell adhesion complexes with neurexin-2β and -3β (Fig. 6). Strikingly, we found that cells expressing either of these two neurexins formed aggregates with CL1-expressing cells, again dependent on a lack of an insert in SS4 in the case of neurexin-2β (neurexin-3β containing an insert in SS4 was not tested). Again, the positive and negative control experiments showed that the observed aggregation was specific (Fig. 6). Together, these data suggest that the binding of CL1 to neurexins is capable of producing a cell adhesion complex and that this complex is regulated by alternative splicing of neurexins at SS4.
FIGURE 6.
CL1-binding to neurexin-2β and -3β also mediates intercellular adhesion. A and B, same as Fig. 5, A and B, except that cell adhesion of CL1-expressing cells to cells expressing neurexin-2β and -3β was analyzed. Data in B are means ±S.E. (n = 3 independent experiments). Error bars: standard error. Scale bar, 100 μm. Statistical significance was assessed by comparing CL1- or NL1ΔAB-expressing cells with the control expressing EGFP alone using one-way analysis of variance (*, p < 0.05; ***, p < 0.0001).
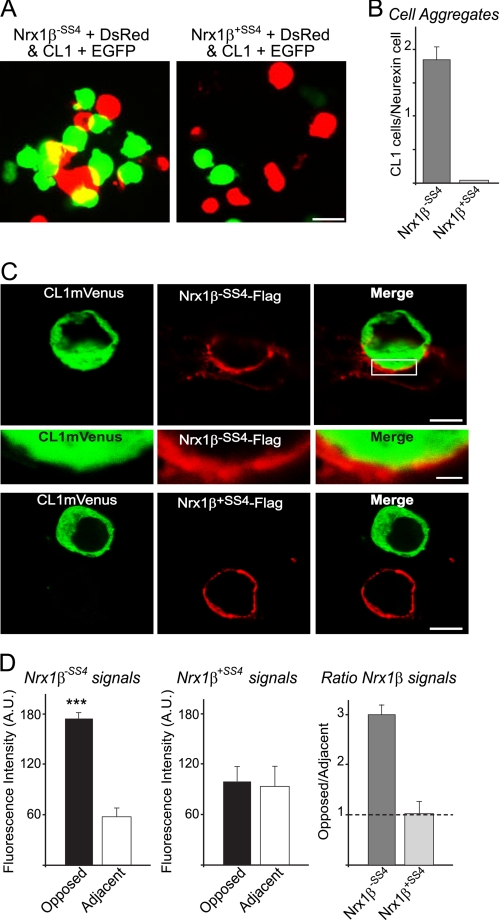
In an effort to better characterize CL1/neurexin molecular recognition events in intercellular adhesion, we sought to determine the composition of cell aggregates and the amount of neurexin-1β present at the cell-cell junction. We first analyzed high magnification pictures of mixed neurexin-1β- and CL1-expressing cells (Fig. 7A). We counted the number of CL1-expressing cells that were within 1 μm of neurexin-1β-expressing cells as a criterion for contacting cells. We determined that approximately two CL1 cells could contact one neurexin-1β cell without an insert in SS4 for a 2:1 ratio (Fig. 7B), whereas almost none was detected for neurexin-1β containing an insert in SS4. In addition, we expressed FLAG-tagged neurexin-1β and CL1-mVenus in separate cells and mixed them to perform a cell adhesion assay. The amount of cell surface immunofluorescence intensity that we detected for neurexin-1β (lacking an insert in SS4) was 3-fold higher at the junction with CL1-mVenus-expressing cells then in other regions of the same cell (Fig. 7, C and D). This suggests that the molecular interaction of neurexin-1β with CL1 causing the cell adhesion is stable and leads to the recruitment of neurexin-1β at the cell-cell junction.
FIGURE 7.
Neurexin-1β is enriched at junction of cell aggregates mediated by CL1/neurexin-1β cell adhesion. A, HEK293T cells co-expressing CL1 with EGFP were mixed with cells co-expressing DsRed with neurexin-1β with or without an insert in SS4 (Nrx1β+SS4 and Nrx1β−SS4) and incubated at room temperature for 90 min in DMEM containing 50 mm Hepes-NaOH, pH 7.4, 10% FBS, 10 mm CaCl2, and 10 mm MgCl2. Cells were imaged by fluorescence microscopy. The scale bar (15 μm) applies to all the pictures in a set. B, quantitative analysis of the number of CL1-expressing cells that directly contact one neurexin cell determined in independent cell adhesion assays (n = 3). High magnification images of the aggregation assays done in A were analyzed, and the number of CL1 +EGFP-expressing cells located within 1 μm of a neurexin-1β +DsRed-expressing cell was counted manually. The results are expressed as a ratio of CL1 cells per neurexin cell. C, HEK293T cells expressing CL1-mVenus were mixed with cells expressing neurexin-1β-FLAG with or without an insert in SS4 (Nrx1β+SS4FLAG and Nrx1β−SS4FLAG) and incubated at room temperature for 90 min in DMEM containing 50 mm Hepes-NaOH, pH 7.4, 10% FBS, 10 mm CaCl2, and 10 mm MgCl2. Cells were plated on coverslips, fixed for 10 min on ice using 4% paraformaldehyde, and analyzed by immunofluorescence using antibodies against FLAG epitope (red). Note that only neurexin-1β−SS4-FLAG leads to cell adhesion with CL1mVenus-expressing cells. Scale bar, 8 μm (top and bottom panels). The middle panel represents a high magnification of the image in the top panel. Scale bar, 1 μm. D, quantitative analysis of the fluorescence intensity distribution at the surface of cells expressing neurexin-1β-FLAG determined in cell adhesion assays such as those shown in C. The mean fluorescence intensity for cell surface neurexin-1β-FLAG was determined using the line scan application from Leica Lite software (Leica). Results from neurexin-1β-FLAG-expressing cells were acquired for the region that immediately opposed the CL1mVenus-expressing cells (Opposed) and for a region that was adjacent to but not opposing CL1mVenus-expressing cells (Adjacent). Normalized neurexin fluorescence intensity is expressed as the ratio of opposed to adjacent intensity. Note that the cell surface neurexin-1β−SS4 signal is enriched at the junction with CL1mVenus-expressing cells in comparison with the rest of the cell; no such enrichment is observed for neurexin-1β+SS4 signals. Data shown are means ±S.E. (n = 3 independent experiments) (statistical significance at p < 0.0001 (***)). Error bars: standard error. A.U., arbitrary units.
CL1 Overexpression in COS Cells is Inactive in Recruiting Presynapses in Artificial Synapse Formation Assay
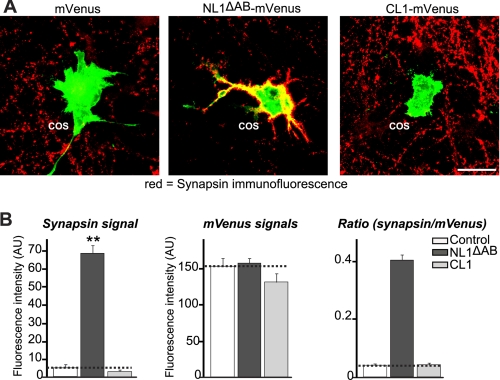
Having demonstrated that intercellular adhesion was a feature of this interaction pair, we next tested the role of CL1 adhesion properties in synapse morphology by performing an artificial synapse formation assay. COS cells were transfected either with CL1 or NL1ΔAB fused to mVenus or mVenus alone and co-cultured in the presence of neurons. The cells were stained by immunofluorescence for the presynaptic protein synapsin to monitor the formation and/or maintenance of presynaptic specialization that co-localizes with the endogenous mVenus signal from the overexpressed proteins in the COS cells. Although a very robust recruitment of synapsin puncta was observed on COS cells expressing NL1ΔAB, these puncta were completely absent in the case of CL1-expressing cells, similarly to the mVenus negative control cells (Fig. 8). These data suggest that CL1 does not stabilize presynapses in the artificial synapse formation assay.
FIGURE 8.
CL1 expressed in transfected COS cells has no presynapse induction activity. A, representative images of COS-7 cells that were transfected with mVenus alone (negative control), wild-type neuroligin-1 (NL1ΔAB; positive control), and CL1 constructs. Transfected COS-7 cells were co-cultured with hippocampal neurons and examined by double immunofluorescence using antibodies to GFP to detect the mVenus signals (green) and antibodies to synapsin to identify presynapses (red). Merged labeling of red presynapses on green transfected COS-7 cells is depicted in yellow (the scale bar (25 μm) applies to all images). B, quantitative analysis of the fluorescence intensities for synapsin over the transfected COS cells and for mVenus in the transfected COS-7 cells co-cultured with hippocampal neurons (dashed lines, mVenus signal as the base line). Normalized synapse density on transfected COS cells co-cultured with neurons is expressed as the ratio of synapsin staining to mVenus fluorescence. All data shown are means ± S.E. (n = 3 independent experiments). Only NL1-mVenus-expressing COS-7 cells induced synapsin clustering (statistical significance at p < 0.01 (**)). Error bars: standard error. AU, arbitrary units.
CL1 Olfactomedin Domain Mediates Its Interaction with Neurexins
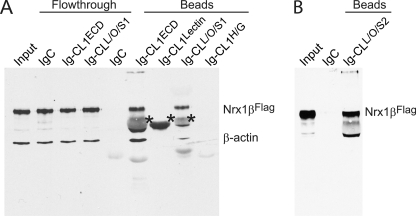
We next aimed to identify which of the extracellular domains of CL1 mediate its interaction with neurexins. For this purpose, we generated a series of expression vectors encoding different parts of the CL1 extracellular sequences (Fig. 1). Then in a first set of experiments, we performed pulldown experiments using immobilized Ig fusion proteins of CL1 domains and full-length neurexin-1β without an insert in SS4 that was solubilized from transfected HEK293 cells (Fig. 9). For the CL1 fragments, we tested various sequences for expression and chose only proteins that were well expressed. The pulldowns revealed that Ig domain fusion proteins containing both the lectin and the olfactomedin-like domains of CL1 were capable of capturing neurexin-1β, whereas Ig domain protein containing only the lectin domain (which exhibited the highest expression levels) did not, nor did Ig domain protein containing the hormone-binding and GAIN domains (Fig. 9).
FIGURE 9.
Pulldown experiments of full-length FLAG-tagged neurexin-1β (Nrx1βFLAG) with immobilized Ig-CL1 fusion proteins. A and B, full-length neurexin-1β containing an N-terminal FLAG tag and lacking an insert in splice site 4 was solubilized from expressing HEK293 cells and chromatographed over columns containing the indicated Ig fusion proteins immobilized on protein A-Sepharose (for the exact composition of the CL1-Ig fusion proteins, see Fig. 1; IgC is control Ig protein). The input, a subset of flow-through samples, and all bounds samples were analyzed by immunoblotting for the FLAG epitope and for β-actin (asterisks, cross-reactivity with IgG captured on the columns). The two separate experiments shown were performed because we initially only analyzed the CL1L/O/S1 fusion protein but found it to be poorly folded, prompting us to produce the CL1L/O/S2 fusion protein that folded better. Data show a single representative experiment that was independently performed three times.
Cell Surface Binding
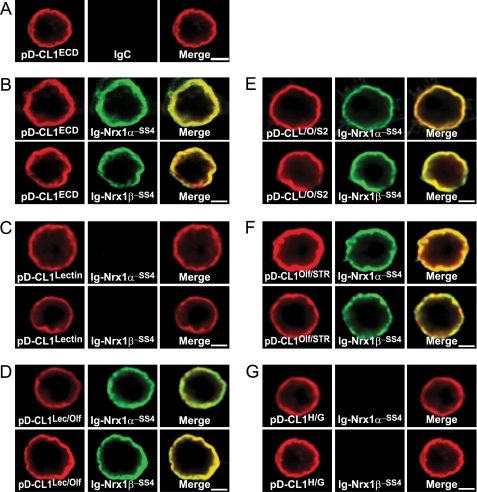
To corroborate the pulldown experiments with an independent approach and to additionally further define the CL1 interaction domain, we performed cell surface binding assays using constructs that isolate different CL1 domains (Fig. 10). We expressed different domains of CL1 on the surface of transfected HEK293 cells using the display vector in which the coding sequences of the domains are preceded by a signal peptide and followed by the TMR of the PDGF receptor (Fig. 1). We then conducted cell surface labeling assays by incubating the HEK293 cells with recombinant Ig protein fusions of neurexin-1α and -1β (always lacking an insert in SS4). The neurexins bound to the cell surface of CL1 fragment-expressing cells only when the olfactomedin-like domain was present in various combinations (Fig. 10). The smallest fragment in which this was the case was a fragment consisting of only the olfactomedin-like domain and 8 residues of the serine/threonine-rich sequence (data from pD-CL1Olf/STR and pD-CL1Lec/Olf cell surface binding assays).
FIGURE 10.
Binding of neurexin-1α and -1β to different CL1 extracellular domains expressed on surface of HEK293 cells. A–G, binding of IgC, Ig-neurexin-1α, and Ig-neurexin-1β (both minus an insert in SS4) to various domains of CL1 (see Fig. 1) expressed on the surface of transfected HEK293 cells as fusion proteins with the PDGF receptor transmembrane region. CL1 fragments were expressed via the pDisplay vector (pD-CL1ECD, total extracellular CL1 sequences; pD-CL1Lectin, CL1 lectin domain; pD-CL1Lec/Olf, CL1 lectin and olfactomedin-like domains; pD-CL1L/O/S2, CL1 lectin and olfactomedin-like domains plus part of its serine/threonine-rich sequence; pD-CL1Olf/STR, CL1 olfactomedin-like domain plus its serine/threonine-rich sequence; pD-CL1H/G, CL1 hormone-binding and GAIN domains; see Fig. 1 for more information). Ig fusion proteins (0.2 μm) were incubated with the transfected cells for 16 h at 4 °C in DMEM, 50 mm Hepes-NaOH, pH 7.4, 0.1% BSA, and 2 mm CaCl2. Cells were washed with incubation buffer, fixed for 10 min on ice using 4% paraformaldehyde, and analyzed by immunofluorescence using antibodies to IgG (green) and to the HA epitope encoded by the pDisplay proteins (red). Data shown are representative images for experiments that were repeated independently at least three times. Scale bars, 4 μm.
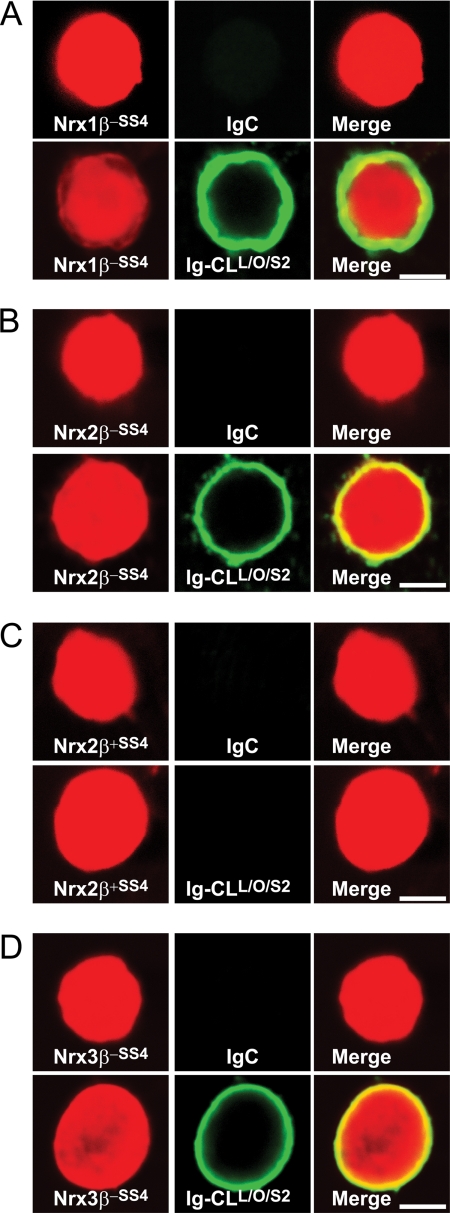
We next performed the surface labeling assay in the opposite orientation with soluble CL1-Ig Fc domain fusion proteins and HEK293 cells expressing neurexins on their surface. In these experiments, we examined the β-neurexins of all three neurexin genes (Fig. 11). In these experiments, CL1 fragments containing the olfactomedin-like domain again bound to neurexins and bound to all three β-neurexins. Just like for neurexin-1β, alternative splicing of neurexin-2β at SS4 regulated binding such that no binding was observed in the presence of an insert (Fig. 11).
FIGURE 11.
Binding of extracellular CL1 fragment containing its lectin and olfactomedin-like domains to β-neurexins expressed in HEK293 cells. A–D, HEK293 cells co-expressing DsRed with neurexin-1β without an insert in SS4 (A), neurexin-2β without (B) and with an insert in SS4 (C), and neurexin-3β without an insert in SS4 (D) were incubated with IgC (control) or Ig-CL1L/O/S2 protein (see Fig. 1), and binding of the Ig proteins to the cells was analyzed as described in the legend to Fig. 8. Data shown are representative images for experiments that were repeated independently at least three times. Scale bars, 4 μm.
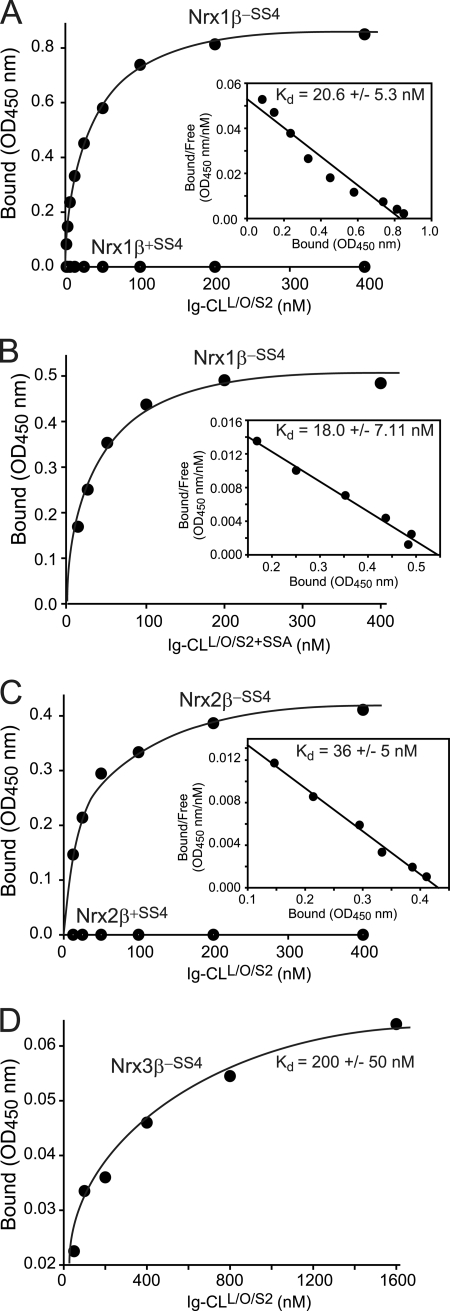
CL1 Fragments Containing Olfactomedin-like Domain Bind to Neurexins with Nanomolar Affinity
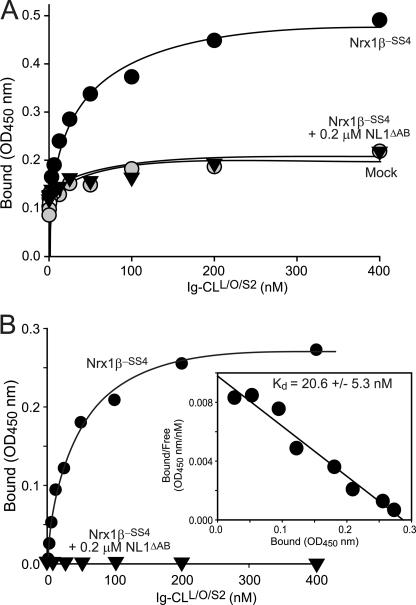
We next measured the approximate neurexin affinity of the CL1 fragment containing its lectin and olfactomedin-like domains as well as part of its serine/threonine-rich sequence. We incubated cells expressing neurexin-1β and -2β plus or minus an insert in SS4 as well as neurexin-3β lacking an insert in SS4 with increasing concentrations of the recombinant CL1-Ig fusion protein and measured binding as described above (see Fig. 4). Strikingly, the Ig fusion protein containing only a subset of extracellular domains of CL1 exhibited a similar if not higher affinity for β-neurexins than the Ig fusion protein containing all extracellular domains of CL1 (Fig. 12). No binding of neurexin-1β or -2β containing an insert in SS4 was detected. Interestingly, although neurexin-1β and -2β displayed similar affinities, the apparent binding affinity of neurexin-3β was ∼10-fold lower, suggesting a possible difference among neurexin isoforms (Fig. 12).
FIGURE 12.
Binding affinity of extracellular CL1 fragment containing its lectin and olfactomedin-like domains for β-neurexins. A–D, binding affinities were determined as described for Fig. 4 using cells expressing the indicated neurexins (neurexin-1β with or without an insert in SS4, Nrx1β+SS4 and Nrx1β−SS4 (A and B); neurexin-2β with or without an insert in SS4, Nrx2β+SS4 and Nrx2β−SS4 (C); and neurexin-3β without an insert in SS4, Nrx3β−SS4 (D)) as receptors and CL1 fragments containing its lectin and olfactomedin-like domains without (A, C, and D) or with an insert in SSA (B) as the ligand. Data show a representative experiment (insets, Scatchard analyses); the affinities listed represent the means ± S.E. of multiple independent experiments (A, n = 6; B, n = 3; C, n = 4; D, n = 3).
In the same experiments, we also probed for a possible effect of the alternative splice site A in CL1 because this site is located at the junction of the lectin and olfactomedin-like domains (Fig. 1). However, insertion of the alternatively spliced sequence in SSA had no effect on the CL1 affinity for neurexin-1β, suggesting that this site does not regulate neurexin binding (Fig. 12).
CL1 Competes with Neuroligin-1 for Binding to Neurexin-1β
Because the binding of both CL1 and neuroligins to neurexins is mediated by the LNS domain of β-neurexins, which corresponds to the sixth LNS domain of α-neurexins, is Ca2+-dependent and is modulated by alternative splicing at SS4 (see Figs. 1–12 and Refs. 13–16), it is possible that these two neurexin ligands bind to similar sites on the neurexin LNS domain. Thus, we sought to determine whether these two ligands compete for binding to neurexins. We transfected HEK293 cells with neurexin-1β and performed a quantitative surface binding assay of CL1 in the absence or presence of a soluble NL1ΔSSAB protein containing a FLAG epitope. 0.2 μm concentrations of the soluble neuroligin totally blocked CL1 binding, suggesting that neuroligin-1 can effectively outcompete CL1 for binding to neurexin-1β (Fig. 13).
FIGURE 13.
Neuroligin-1 and CL1 compete for binding to neurexin-1β. A and B, HEK293 cells expressing neurexin-1β without an insert in SS4 and control mock-transfected cells were incubated with increasing concentrations of soluble CL1-Ig fusion protein containing the lectin and olfactomedin-like domains and part of the serine/threonine rich sequence of CL1 as described for Figs. 3 and 10; in addition, the incubations of neurexin-1β-expressing cells were carried out in the presence of the soluble neuroligin-1 extracellular domain lacking splice site A and B inserts as a competitor. A depicts the raw binding data; B depicts the summary plot of net Ig-CL1L/O/S2 binding under the conditions described above. Net binding was calculated by subtracting the background binding with mock-transfected cells from the binding obtained with cells expressing neurexin-1β. The inset shows a Scatchard analysis of the binding results with the mean affinity ± S.E. calculated from multiple experiments (n = 3).
DISCUSSION
Understanding the molecular events guiding synaptic cell adhesion, for example during synapse formation and elimination, is crucial for insight into how neuronal networks are generated in the brain. Cell adhesion molecules likely perform central roles in the formation and operation of synaptic circuits. Cell adhesion molecules are probably not only involved in the initial establishment of synapses but also in directing synapse specification, controlling synapse maintenance, and regulating long term synaptic plasticity. Neurexins are currently the best studied trans-synaptic cell adhesion molecules that may contribute to all of these processes. Various interaction partners of neurexins are known to influence the formation and specification of excitatory or inhibitory synapses; for example, neuroligin-1 and LRRTMs control the former, and neuroligin-2 control the latter. Moreover, the identification of neurexin genes as candidate loci for cognitive diseases such as autism, schizophrenia, and addiction further reinforced the idea that neurexin-mediated synaptic cell adhesion significantly impacts the development and plasticity of synapses in the brain (29–35). Here, we show that CL1, a GPCR of the “cell adhesion class” (36, 37), represents a novel ligand (or receptor, depending on the perspective) for neurexins. Because CL1 was identified as a second independent α-latrotoxin receptor (4–6) after neurexins were discovered based on the same activity (1–3), the finding that neurexins and CL1 interact with each other suggests the intriguing hypothesis that CL1 and neurexins form a trans-synaptic cell adhesion complex that is the actual substrate for α-latrotoxin action and that CL1 and neurexins are part of the same molecular pathway. Poignant in this regard is the finding that the CL1 homolog CL3 was recently linked to attention deficit/hyperactivity disorder, a condition that shows frequent co-occurrence with autism spectrum disorders (29, 48).
The conclusion that neurexins and CL1 interact with each other via their extracellular domains is based on the following lines of evidence.
Recombinant soluble neurexin fragments specifically bind to full-length CL1 expressed on the surface of transfected HEK293 cells; this binding is specific because it is not observed with negative controls and not even with recombinant soluble neurexins containing an insert in SS4.
Conversely, recombinant soluble CL1 fragments bind to full-length neurexins expressed on the surface of HEK293 cells, again regulated in an all-or-none manner by neurexin alternative splicing at SS4.
Only particular fragments of CL1 bind in the assays described in points 1 and 2.
Estimates of the binding affinity reveal a nanomolar apparent dissociation constant of neurexin-1 and -2 binding to CL1 and an ∼10-fold lower affinity for neurexin-3.
Pulldown experiments show that extracellular fragments of neurexins and CL1 form a complex.
Most importantly, cells expressing CL1 and neurexins on their surface aggregate with each other; this process is specific because it is only observed when appropriate combinations of neurexin- and CL1-expressing cells are mixed with each other.
The binding of CL1 to neurexins came as a surprise because this hypothesis was historically put aside based on the fact that the recombinant neurexins and CL1 exhibited similar high affinity binding to α-latrotoxin although with distinct Ca2+ requirements, suggesting that no co-receptor complex is necessary to mediate α-latrotoxin binding (12, 49). Note, however, that the affinity of CL1 for neurexins is lower than that of neuroligins as revealed among others by the ability of neuroligin-1 to outcompete CL1 for binding to neurexin-1β (Fig. 13). The higher affinity of neuroligins than of CL1 for neurexins in addition to the protease sensitivity and size of CL1 may explain why we and others could not previously identify this interaction using brain tissues in which both neuroligins and CLs compete for the same ligand but where neuroligin is the preferred entity to be purified. Additionally, the affinities reported for the neurexin/LRRTM interaction also appear to be higher than that of the neurexin/CL1 interaction (17), providing a further explanation for the lack of a previous observation of the neurexin/CL1 interaction.
We identified the olfactomedin-like domain of CL1 as its interaction site with the LNS6 domain of neurexins (the only LNS domain present in β-neurexins). Our data provide evidence that the olfactomedin-like domain may be sufficient to mediate the interaction with neurexins, although the serine/threonine-rich sequence of CL1 may contribute. Although the CL family of GPCRs along with neurexins is conserved in invertebrates, it is interesting to note that the olfactomedin-like domain is absent from the Caenorhabditis elegans and Drosophila melanogaster CL homologues (50). This observation suggests that the neurexin-CL1 complex might not be evolutionarily conserved but only developed in vertebrates. Furthermore, this observation indicates that the neurexin/CL1 interaction is not essential for basic synapse formation events per se.
The importance of neurexin alternative splicing in regulating protein/protein interaction is once more underlined by the present study. We describe that inclusion of an insert in neurexin SS4 disrupts the high affinity interaction with CL1 established by its SS4 insert-lacking counterpart. This feature seems to apply to most neurexin ligands known to date except for cerebellins, which bind better to SS4 insert-containing neurexins (20, 21), and to GABA receptors and acetylcholine receptors, which bind independently of alternative splicing (42, 51). Additionally, isoform hierarchy also seems to take place because among interacting partners we observed that there was a respective preferential binding of CL1 to neurexin-1β and -2β over neurexin-3β.
Our study has at least two significant weaknesses that need to be addressed in future experiments. First, due to the lack of avid antibodies to either neurexins or CL1, we do not know the endogenous localization of these proteins. The at least partial presynaptic localization of neurexins is well established based on functional experiments (e.g. see Refs. 24 and 27), and a postsynaptic localization of CL1 would be consistent with its avid binding to postsynaptic SH3 and multiple ankyrin repeats scaffolding proteins (SHANK) (38, 52). However, the postsynaptic localization of CL1 proposed here has not been tested directly nor can it be confirmed easily at present. For example, we found that epitope-tagged overexpressed CL1 in transfected neurons is efficiently targeted to neuronal dendrites; however, the nature of these overexpression experiments limits their interpretability, and thus we do not believe these data allow a major conclusion. Also, by opposition to a postsynaptic localization of CL1, we did not observe the recruitment of presynaptic proteins while performing a neuron-COS cell co-culture assay in which CL1 was expressed at the plasma membrane of COS cells (Fig. 8). However, this evidence does not eliminate the possibility of CL1 being localized in the postsynapse, but its role might not be related to synapse formation or maintenance.
Second, the physiological importance of the neurexin/CL1 interaction remains unclear. The fact that their interaction is sufficiently tight to produce an intercellular adhesion event indicates that they form a true cell adhesion pair, but it does not reveal the functional importance of this cell adhesion event. In particular, it is tempting to speculate that a trans-synaptic neurexin-CL1 cell adhesion complex represents the true substrate of α-latrotoxin action with the notion that the Ca2+-independent triggering of neurotransmitter release by α-latrotoxin may involve a mechanical “pull” on this complex akin to the action of hypertonic sucrose in mechanically triggered release. Proving this idea, however, has turned out to be difficult, and we cannot at present test it. However, the CL1/neurexin interaction identified here is independent of the resolution of these questions and likely represents an important molecular event in shaping synapse function.
Acknowledgments
We thank Drs. Jason Aoto, Chen Zhang, Garret Anderson, and Mikhail Khvochtchev for advice.
This work was supported in part by a fellowship grant from the Canadian Institutes of Health Research (to A. A. B.) and Human Frontier Science Program Organization Grant LT00021/2008-L (to J. K.). This study was also supported by NIH Grant R37 MH052804-18.
- CL
- calcium-independent receptor for latrotoxin and latrophilin
- LRRTM
- leucine-rich repeat transmembrane protein
- EGFP
- enhanced green fluorescent protein
- GAIN
- GPCR autoproteolysis-inducing domain
- LNS
- laminin-neurexin-steroid-binding hormone
- TMR
- transmembrane region
- SS
- splice site
- SSA
- splice site A.
REFERENCES
- 1. Südhof T. C. (2001) α-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu. Rev. Neurosci. 24, 933–962 [DOI] [PubMed] [Google Scholar]
- 2. Ushkaryov Y. A., Petrenko A. G., Geppert M., Südhof T. C. (1992) Neurexins: synaptic cell surface proteins related to the α-latrotoxin receptor and laminin. Science 257, 50–56 [DOI] [PubMed] [Google Scholar]
- 3. Sugita S., Khvochtev M., Südhof T. C. (1999) Neurexins are functional α-latrotoxin receptors. Neuron 22, 489–496 [DOI] [PubMed] [Google Scholar]
- 4. Krasnoperov V. G., Bittner M. A., Beavis R., Kuang Y., Salnikow K. V., Chepurny O. G., Little A. R., Plotnikov A. N., Wu D., Holz R. W., Petrenko A. G. (1997) α-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron 18, 925–937 [DOI] [PubMed] [Google Scholar]
- 5. Davletov B. A., Shamotienko O. G., Lelianova V. G., Grishin E. V., Ushkaryov Y. A. (1996) Isolation and biochemical characterization of a Ca2+-independent α-latrotoxin-binding protein. J. Biol. Chem. 271, 23239–23245 [DOI] [PubMed] [Google Scholar]
- 6. Sugita S., Ichtchenko K., Khvotchev M., Südhof T. C. (1998) α-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J. Biol. Chem. 273, 32715–32724 [DOI] [PubMed] [Google Scholar]
- 7. Ushkaryov Y. A., Südhof T. C. (1993) Neurexin III α: extensive alternative splicing generates membrane-bound and soluble forms. Proc. Natl. Acad. Sci. U.S.A. 90, 6410–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ushkaryov Y. A., Hata Y., Ichtchenko K., Moomaw C., Afendis S., Slaughter C. A., Südhof T. C. (1994) Conserved domain structure of β-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J. Biol. Chem. 269, 11987–11992 [PubMed] [Google Scholar]
- 9. Tabuchi K., Südhof T. C. (2002) Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics 79, 849–859 [DOI] [PubMed] [Google Scholar]
- 10. Rowen L., Young J., Birditt B., Kaur A., Madan A., Philipps D. L., Qin S., Minx P., Wilson R. K., Hood L., Graveley B. R. (2002) Analysis of the human neurexin genes: alternative splicing and the generation of protein diversity. Genomics 79, 587–597 [DOI] [PubMed] [Google Scholar]
- 11. Ullrich B., Ushkaryov Y. A., Südhof T. C. (1995) Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron 14, 497–507 [DOI] [PubMed] [Google Scholar]
- 12. Davletov B. A., Krasnoperov V., Hata Y., Petrenko A. G., Südhof T. C. (1995) High affinity binding of α-latrotoxin to recombinant neurexin I α. J. Biol. Chem. 270, 23903–23905 [DOI] [PubMed] [Google Scholar]
- 13. Ichtchenko K., Hata Y., Nguyen T., Ullrich B., Missler M., Moomaw C., Südhof T. C. (1995) Neuroligin 1: a splice site-specific ligand for β-neurexins. Cell 81, 435–443 [DOI] [PubMed] [Google Scholar]
- 14. Ichtchenko K., Nguyen T., Südhof T. C. (1996) Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 271, 2676–2682 [DOI] [PubMed] [Google Scholar]
- 15. Boucard A. A., Chubykin A. A., Comoletti D., Taylor P., Südhof T. C. (2005) A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron 48, 229–236 [DOI] [PubMed] [Google Scholar]
- 16. Chih B., Gollan L., Scheiffele P. (2006) Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 51, 171–178 [DOI] [PubMed] [Google Scholar]
- 17. Ko J., Fuccillo M. V., Malenka R. C., Südhof T. C. (2009) LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 64, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Wit J., Sylwestrak E., O'Sullivan M. L., Otto S., Tiglio K., Savas J. N., Yates J. R., 3rd, Comoletti D., Taylor P., Ghosh A. (2009) LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 64, 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siddiqui T. J., Pancaroglu R., Kang Y., Rooyakkers A., Craig A. M. (2010) LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 30, 7495–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uemura T., Lee S. J., Yasumura M., Takeuchi T., Yoshida T., Ra M., Taguchi R., Sakimura K., Mishina M. (2010) Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 21. Matsuda K., Yuzaki M. (2011) Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur. J. Neurosci. 33, 1447–1461 [DOI] [PubMed] [Google Scholar]
- 22. Scheiffele P., Fan J., Choih J., Fetter R., Serafini T. (2000) Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669 [DOI] [PubMed] [Google Scholar]
- 23. Chubykin A. A., Liu X., Comoletti D., Tsigelny I., Taylor P., Südhof T. C. (2005) Dissection of synapse induction by neuroligins: effect of a neuroligin mutation associated with autism. J. Biol. Chem. 280, 22365–22374 [DOI] [PubMed] [Google Scholar]
- 24. Ko J., Zhang C., Arac D., Boucard A. A., Brunger A. T., Südhof T. C. (2009) Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 28, 3244–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Graf E. R., Zhang X., Jin S. X., Linhoff M. W., Craig A. M. (2004) Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nam C. I., Chen L. (2005) Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc. Natl. Acad. Sci. U.S.A. 102, 6137–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Missler M., Zhang W., Rohlmann A., Kattenstroth G., Hammer R. E., Gottmann K., Südhof T. C. (2003) α-Neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 423, 939–948 [DOI] [PubMed] [Google Scholar]
- 28. Zhang W., Rohlmann A., Sargsyan V., Aramuni G., Hammer R. E., Südhof T. C., Missler M. (2005) Extracellular domains of α-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J. Neurosci. 25, 4330–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rujescu D., Ingason A., Cichon S., Pietiläinen O. P., Barnes M. R., Toulopoulou T., Picchioni M., Vassos E., Ettinger U., Bramon E., Murray R., Ruggeri M., Tosato S., Bonetto C., Steinberg S., Sigurdsson E., Sigmundsson T., Petursson H., Gylfason A., Olason P. I., Hardarsson G., Jonsdottir G. A., Gustafsson O., Fossdal R., Giegling I., Möller H. J., Hartmann A. M., Hoffmann P., Crombie C., Fraser G., Walker N., Lonnqvist J., Suvisaari J., Tuulio-Henriksson A., Djurovic S., Melle I., Andreassen O. A., Hansen T., Werge T., Kiemeney L. A., Franke B., Veltman J., Buizer-Voskamp J. E., GROUP Investigators, Sabatti C., Ophoff R. A., Rietschel M., Nöthen M. M., Stefansson K., Peltonen L., St Clair D. (2009) Disruption of the neurexin 1 gene is associated with schizophrenia. Hum. Mol. Genet. 18, 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ching M. S., Shen Y., Tan W. H., Jeste S. S., Morrow E. M., Chen X., Mukaddes N. M., Yoo S. Y., Hanson E., Hundley R., Austin C., Becker R. E., Berry G. T., Driscoll K., Engle E. C., Friedman S., Gusella J. F., Hisama F. M., Irons M. B., Lafiosca T., LeClair E., Miller D. T., Neessen M., Picker J. D., Rappaport L., Rooney C. M., Sarco D. P., Stoler J. M., Walsh C. A., Wolff R. R., Zhang T., Nasir R. H., Wu B. L.; Children's Hospital Boston Genotype Phenotype Study Group (2010) Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zweier C., de Jong E. K., Zweier M., Orrico A., Ousager L. B., Collins A. L., Bijlsma E. K., Oortveld M. A., Ekici A. B., Reis A., Schenck A., Rauch A. (2009) CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am. J. Hum. Genet. 85, 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harrison V., Connell L., Hayesmoore J., McParland J., Pike M. G., Blair E. (2011) Compound heterozygous deletion of NRXN1 causing severe developmental delay with early onset epilepsy in two sisters. Am. J. Med. Genet. A 155A, 2826–2831 [DOI] [PubMed] [Google Scholar]
- 33. Yan J., Noltner K., Feng J., Li W., Schroer R., Skinner C., Zeng W., Schwartz C. E., Sommer S. S. (2008) Neurexin 1α structural variants associated with autism. Neurosci. Lett. 438, 368–370 [DOI] [PubMed] [Google Scholar]
- 34. Kim H. G., Kishikawa S., Higgins A. W., Seong I. S., Donovan D. J., Shen Y., Lally E., Weiss L. A., Najm J., Kutsche K., Descartes M., Holt L., Braddock S., Troxell R., Kaplan L., Volkmar F., Klin A., Tsatsanis K., Harris D. J., Noens I., Pauls D. L., Daly M. J., MacDonald M. E., Morton C. C., Quade B. J., Gusella J. F. (2008) Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 82, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Südhof T. C. (2008) Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yona S., Lin H. H., Siu W. O., Gordon S., Stacey M. (2008) Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem. Sci. 33, 491–500 [DOI] [PubMed] [Google Scholar]
- 37. Schiöth H. B., Nordström K. J., Fredriksson R. (2010) The adhesion GPCRs; gene repertoire, phylogeny and evolution. Adv. Exp. Med. Biol. 706, 1–13 [DOI] [PubMed] [Google Scholar]
- 38. Kreienkamp H. J., Zitzer H., Gundelfinger E. D., Richter D., Bockers T. M. (2000) The calcium-independent receptor for α-latrotoxin from human and rodent brains interacts with members of the ProSAP/SSTRIP/Shank family of multidomain proteins. J. Biol. Chem. 275, 32387–32390 [DOI] [PubMed] [Google Scholar]
- 39. Petrenko A. G., Ullrich B., Missler M., Krasnoperov V., Rosahl T. W., Südhof T. C. (1996) Structure and evolution of neurexophilin. J. Neurosci. 16, 4360–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Missler M., Südhof T. C. (1998) Neurexophilins form a conserved family of neuropeptide-like glycoproteins. J. Neurosci. 18, 3630–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugita S., Saito F., Tang J., Satz J., Campbell K., Südhof T. C. (2001) A stoichiometric complex of neurexins and dystroglycan in brain. J. Cell Biol. 154, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang C., Atasoy D., Araç D., Yang X., Fucillo M. V., Robison A. J., Ko J., Brunger A. T., Südhof T. C. (2010) Neurexins physically and functionally interact with GABAA receptors. Neuron 66, 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silva J. P., Lelianova V. G., Ermolyuk Y. S., Vysokov N., Hitchen P. G., Berninghausen O., Rahman M. A., Zangrandi A., Fidalgo S., Tonevitsky A. G., Dell A., Volynski K. E., Ushkaryov Y. A. (2011) Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl. Acad. Sci. U.S.A. 108, 12113–12118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tobaben S., Südhof T. C., Stahl B. (2002) Genetic analysis of α-latrotoxin receptors reveals functional interdependence of CIRL/latrophilin 1 and neurexin 1α. J. Biol. Chem. 277, 6359–6365 [DOI] [PubMed] [Google Scholar]
- 45. Calebiro D., Nikolaev V. O., Persani L., Lohse M. J. (2010) Signaling by internalized G-protein-coupled receptors. Trends Pharmacol. Sci. 31, 221–228 [DOI] [PubMed] [Google Scholar]
- 46. Moore C. A., Milano S. K., Benovic J. L. (2007) Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 69, 451–482 [DOI] [PubMed] [Google Scholar]
- 47. Nguyen T., Südhof T. C. (1997) Binding properties of neuroligin 1 and neurexin 1β reveal function as heterophilic cell adhesion molecules. J. Biol. Chem. 272, 26032–26039 [DOI] [PubMed] [Google Scholar]
- 48. Rommelse N. N., Geurts H. M., Franke B., Buitelaar J. K., Hartman C. A. (2011) A review on cognitive and brain endophenotypes that may be common in autism spectrum disorder and attention-deficit/hyperactivity disorder and facilitate the search for pleiotropic genes. Neurosci. Biobehav. Rev. 35, 1363–1396 [DOI] [PubMed] [Google Scholar]
- 49. Lelianova V. G., Davletov B. A., Sterling A., Rahman M. A., Grishin E. V., Totty N. F., Ushkaryov Y. A. (1997) α-Latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J. Biol. Chem. 272, 21504–21508 [DOI] [PubMed] [Google Scholar]
- 50. Langenhan T., Prömel S., Mestek L., Esmaeili B., Waller-Evans H., Hennig C., Kohara Y., Avery L., Vakonakis I., Schnabel R., Russ A. P. (2009) Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev. Cell 17, 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng S. B., Amici S. A., Ren X. Q., McKay S. B., Treuil M. W., Lindstrom J. M., Rao J., Anand R. (2009) Presynaptic targeting of α4β2 nicotinic acetylcholine receptors is regulated by neurexin-1β. J. Biol. Chem. 284, 23251–23259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tobaben S., Südhof T. C., Stahl B. (2000) The G protein-coupled receptor CL1 interacts directly with proteins of the Shank family. J. Biol. Chem. 275, 36204–36210 [DOI] [PubMed] [Google Scholar]
- 53. Arac D., Boucard A. A., Bolliger M. F., Nbuyen J., Soltis M., Südhof T. C., Brunger A. T. (2012) A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. doi: 10.1038/emboj.2012.26 [DOI] [PMC free article] [PubMed] [Google Scholar]