Background: The significance of physical/genetic interactions between Dna2 and Fen1 is poorly understood.
Results: Fen1 stimulated the endonuclease activity of both Dna2 and Fen1 itself.
Conclusion: Fen1 is a bona fide trans-autostimulatory enzyme, and this activity resides within the C-terminal 16 amino acids of Rad27.
Significance: Our finding provides a novel insight into how Dna2 and Rad27 jointly contribute to processing of Okazaki fragments.
Keywords: DNA Enzymes, DNA Repair, DNA Replication, Enzyme Catalysis, Protein-Protein Interactions, Dna2, Fen1, Okazaki Fragment Processing, Enzymatic Stimulation, Lagging Strand DNA Synthesis
Abstract
Dna2 and Rad27 (yeast Fen1) are the two endonucleases critical for Okazaki fragment processing during lagging strand DNA synthesis that have been shown to interact genetically and physically. In this study, we addressed the functional consequences of these interactions by examining whether purified Rad27 of Saccharomyces cerevisiae affects the enzymatic activity of Dna2 and vice versa. For this purpose, we constructed Rad27DA (catalytically defective enzyme with an Asp to Ala substitution at amino acid 179) and found that it significantly stimulated the endonuclease activity of wild type Dna2, but failed to do so with Dna2Δ405N that lacks the N-terminal 405 amino acids. This was an unexpected finding because dna2Δ405N cells were still partially suppressed by overexpression of rad27DA in vivo. Further analyses revealed that Rad27 is a trans-autostimulatory enzyme, providing an explanation why overexpression of Rad27, regardless of its catalytic activity, suppressed dna2 mutants as long as an endogenous wild type Rad27 is available. We found that the C-terminal 16-amino acid fragment of Rad27, a highly polybasic region due to the presence of multiple positively charged lysine and arginine residues, was sufficient and necessary for the stimulation of both Rad27 and Dna2. Our findings provide further insight into how Dna2 and Rad27 jointly affect the processing of Okazaki fragments in eukaryotes.
Introduction
DNA replication plays an important role in maintaining the integrity of genome due to its intrinsically accurate manner in duplicating DNA (1–3). DNA repair and recombination can be regarded as backup systems that further ensure genome integrity by correcting errors introduced during or after DNA replication (4–7). Thus, the faithful maintenance of genetic information requires the cooperative action of many enzymes and protein factors involved in the major DNA transactions (7).
In DNA replication, lagging strand DNA synthesis is more complex than leading strand because it requires discontinuous DNA synthesis and joining of short pieces of DNA called Okazaki fragments (8, 9). In brief, polymerase (pol)2 α-primase synthesizes an initial RNA-DNA primer on the lagging strand template that is extended by pol δ to generate a new Okazaki fragment. Pol δ, whereas engaging in the extension, displaces the downstream Okazaki fragment and creates a 5′-flap structure (10, 11) that can be efficiently removed by the combined action of the two endonucleases, Dna2 and Rad27 in yeast. Yeast Dna2 possesses both endonuclease and helicase activities (10, 12). The endonuclease activity of Dna2 is highly specific for single-stranded DNA but unable to completely remove the 5′ single-stranded DNA flap, leaving short flaps of ∼6 nucleotides. Because of this limitation, an additional nuclease such as Fen1 is required to create ligatable nicks from 5′-flap structures. Thus, Dna2 appears to be essential only when extensive strand displacement synthesis by pol δ results in the formation of flaps long enough to be bound by replication protein A (RPA) (13, 14). RPA-bound long flaps are resistant to Fen1 cleavage but sensitive to Dna2-catalyzed cleavage. As a result, RPA-bound flaps are first cleaved by Dna2 and the shortened flaps are subsequently cleaved by Fen1 to create nicks (13). Thus, RPA acts as a molecular switch for the sequential action of Dna2 and Fen1. In addition, secondary-structured flaps, which are poor substrates for Fen1, can be resolved by Dna2 helicase activity and then cleaved by the Dna2 endonuclease activity in a coupled manner (11).
Fen1, due to its three distinct activities that include flap endonuclease, gap endonuclease, and exonuclease, participates in many DNA transactions (3, 15–22). It was reported that a double flap substrate (with a 5′-flap of varying length and a 1-nt 3′-flap) is an excellent Fen1 substrate and possibly the physiological substrate that is efficiently processed to a ligatable nick by Fen1 (23–25). Recently, the crystal structure of flap-bound human Fen1 was determined, revealing why Fen1 efficiently cleaves such structures (26).
Genetic and biochemical properties of Dna2 and Fen1 suggest that the joint action of these nucleases are critical for the efficient and seamless processing of Okazaki fragments during lagging strand DNA replication in eukaryotes (10, 13–16, 27–30). Dna2 was shown to physically interact with Rad27, and overexpression of Dna2 suppressed temperature-sensitive (ts) growth defects of rad27Δ (31). In addition, overexpression of Rad27 suppressed growth defects observed with several dna2 mutants including dna2-1 (32) and dna2Δ405N lacking the N-terminal 405-amino acid region (11). Yeast cells expressing the dna2Δ405N mutant allele devoid of the N-terminal 405 amino acid residues displayed ts growth defects (being unable to grow at 37 °C). Although the dna2Δ405N mutant allele confers this ts phenotype, the mutant enzyme (Dna2Δ405N) encoded by this allele contains endonuclease and ATPase activities comparable with those of wild type Dna2.3 Despite several reports that Fen1 and Dna2 interact, the significance of these findings is poorly understood. Recently, it was shown that Dna2 stimulated the activity of Fen1, most likely through specific protein-protein interactions (33). RPA, which stimulates the Dna2-catalyzed cleavage of long flaps (13), increased the Fen1-catalyzed cleavage of short flaps that do not bind RPA (33). This indicates that interactions between the major proteins involved in Okazaki fragment processing are important for the efficient removal of flaps.
In this study, we found that Rad27 and Dna2 are mutually stimulatory in keeping with their ability to process the same flap structures. We found that Rad27 is a trans-autostimulatory protein, and we were able to map the motif responsible for both trans-autostimulation and increasing the activity of Dna2. The mutual stimulation observed between Rad27 and Dna2 and the trans-autostimulation of Rad27 or human FEN1 (hFEN1) leads to the efficient processing of Okazaki fragments in eukaryotes.
MATERIALS AND METHODS
Enzyme, Nucleotide, and Peptides
The oligonucleotides used in this study were commercially synthesized from Genotech (Daejeon, Korea), and their sequences are as listed in Table 1. [γ-32P]ATP (3,000 Ci/mmol) was purchased from Izotop (Budapest, Hungary). Restriction enzymes and T4 polynucleotide kinase were purchased from EnzynomicsTM (Daejeon, Korea). Peptides, such as the C-terminal 16-amino acid fragment of Rad27 (Rad27(367–382)) and its mutant derivatives, were commercially synthesized by Peptron (Daejeon, Korea).
TABLE 1.
Oligonucleotides used to construct DNA substrates in this study
| No. | Oligonucleotide sequences (length in nucleotides) |
|---|---|
| 1 | GGAAAACATTATTAATGGCGTCGAGCTAGGCACAAGGCGAACTGCTAACGG (50) |
| 2 | CCGTTAGCAGTTCGCCTTGTGCCTA (25) |
| 3 | CCGTTAGCAGTTCGCCTTGTGCCTAG (26) |
| 4 | TTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCGACGCCATTAATAATGTTTTC (52) |
| 5 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC (60) |
| 6 | TTTTTTTTTTTTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC (50) |
| 7 | TTTTTTTTTTCGGACGCTCGACGCCATTAATAATGTTTTC (40) |
| 8 | CGGACGCTCGACGCCATTAATAATGTTTTC (30) |
| 9 | TGCTCGACGCCATTAATAATGTTTTC (26) |
| 10 | TTTTTTTTTTTTTGCTCGACGCCATTAATAATGTTTTC (38) |
| 11 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTCGACGCCATTAATAATGTTTTC (79) |
Construction of Expression Vectors
The pET21a-Rad27 and pET21a-Rad27DA plasmids (Novagen) were constructed to produce Rad27 enzymes (Rad27-His6 or Rad27DA-His6, respectively) with a C-terminal-tagged hexahistidine. To construct pET21a-Rad27, the open reading frame of RAD27 was first amplified using yeast genomic DNA as the template with the primer pair, 5′-CGC CAT ATG GGT ATT AAA GGT TTG-3′ and 5′-CCG CTC GAG TCT TCT TCC CTT TGT GAC-3′ (NdeI and XhoI sites are underlined, respectively). The amplified fragments were digested with NdeI and XhoI, and then cloned into pET21a cleaved with the same restriction enzymes, generating pET21a-Rad27-His6. The pET21a-Rad27DA plasmid was created using a site-directed mutagenesis kit (Enzynomics) according to the procedure recommended by the manufacturer. The substitution of Asp with Ala at amino acid 179 in Rad27 was introduced by using a pair of mutagenic primers: 5′-GCC ACA CTC TGT TAT AGA AC-3′ and 5′-CAT ATC TTC ACT TGC TGC GGC-3′ (underlined at the mutagenic base). All truncated derivatives of Rad27 were cloned into pGEX-4T-1, and the resulting expression vectors produced Rad27 as a N-terminal GST fusion protein as described previously (34).
The pRS-series of plasmids were purchased from New England Biolabs. Expression vectors for Rad27-FLAG (Rad27 with a FLAG tag fused to its C terminus) and its derivatives were constructed as follows; the open reading frame of RAD27 was first PCR-amplified using yeast genomic DNA as the template with the following pair of primers; 5′-ATA AGA ATG CGG CCG CAT GGG TAT TAA AGG TTT GAA TGC-3′ and 5′-CCG CTC GAG TCA CTT ATC GTC GTC ATC CTT GTA ATC TCT TCT TCC CTT TGT GAC TTT ATT C-3′ (NotI and XhoI sites are underlined, respectively). The amplified fragment was then cloned into NotI and XhoI sites of pRS325-ADH1, generating pRS325-ADH1-Rad27-FLAG in which expression of Rad27-FLAG is under the control of an ADH1 promoter. The pRS325-ADH1-Rad27DA-FLAG, an expression vector for Rad27DA, was constructed by amplifying the open reading frame of Rad27DA from pET21a-Rad27DA and subsequently cloning into pRS325-ADH1.
Purification of Enzymes
The pET21a-Rad27 and pET21a-Rad27DA plasmids were transformed into Escherichia coli BL21(DE3) CodonPlus-RIL (Stratagene), and expression of proteins was induced at A600 = 0.5 by addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.5 mm at 25 °C for 4 h. Cells were harvested and resuspended in lysis buffer (20 ml/g of cells, wet weight) (25 mm Tris-HCl, pH 7.8, 150 mm NaCl, 10% glycerol, 0.01% Nonidet P-40, 10 mm imidazole, 1 mm phenylmethylsulfonyl fluoride (PMSF), 0.1 mm benzamidine, 2 μg/ml of leupeptin, and 1 μg/ml of pepstatin A) and then sonicated. Crude extracts were cleared by centrifugation at 18,000 × g for 40 min using the Hanil A50S-8 rotor and the supernatant was loaded onto a 1.5-ml Ni2+-nitrilotriacetic acid column (Qiagen) at a flow rate of 0.14 ml/min, which was pre-equilibrated with >100 column volumes (CV) of lysis buffer by gravity.
After loading, beads were washed with 10 CV of lysis buffer, followed by successive washings with 40 CV of lysis buffer containing 20 and 50 mm imidazole. Bound proteins were eluted in 0.5-ml fractions with 250 mm imidazole in lysis buffer. Peak protein fractions were combined and loaded onto a heparin-Sepharose Fast Flow column (0.5 ml; GE Healthcare) by gravity, which was pre-equilibrated with 20 CV of buffer C (25 mm Tris-HCl, pH 7.5, 10% glycerol, and 0.01% Nonidet P-40) containing 150 mm NaCl. Bound proteins were washed with 40 CV of buffer C containing 200 mm NaCl and eluted with buffer C plus 500 mm NaCl. Peak protein fractions were subjected to glycerol gradient sedimentation (15–35%) containing 500 mm NaCl for 24 h at 55,000 rpm. Fractions were collected and the protein concentrations were quantified by SDS-PAGE analysis, and the amount of protein was determined using BSA as standard (Bio-Rad).
Dna2, Dna2DA (nuclease-dead mutant), Dna2Δ405N (Dna2 lacking the N-terminal 405 amino acid residues), and Dna2(1–405) (the N-terminal 405-amino acid fragment of Dna2) were purified as described previously (11, 35). Derivatives of GST-Rad27 cloned into pGEX-4T-1 vector (Amersham) were transformed to E. coli BL21(DE3) CodonPlus-RIL strain (Stratagene) and expression of proteins were induced at A600 = 0.5 by addition of 0.5 mm isopropyl β-d-thiogalactopyranoside at 25 °C for 4 h. Cells from a 0.25-liter culture were collected by centrifugation, resuspended in 15 ml of buffer T150 (50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 10% glycerol, 0.01% Nonidet P-40, 1 mm PMSF, 0.1 mm benzamidine, 1.0 μg/ml of leupeptin, and 1.0 μg/ml of pepstatin A; the subscript indicates the concentration of NaCl, in mm) and sonicated. The extracts were centrifuged at 20,000 × g for 30 min at 4 °C and the cleared lysate was loaded onto a glutathione-agarose (GSH-agarose) column, pre-equilibrated with 50 CV of T150. The column was first washed with 5 CV of buffer T1000, followed by washing with 5 CV of buffer T150. Proteins were then eluted with T150 buffer containing 20 mm reduced GSH.
Preparation of Substrates
All DNA substrates used in this study were prepared as previously reported (36). Briefly, one of the two oligonucleotides in a partial duplex DNA substrate was 5′-labeled with [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase. The 5′-labeled downstream oligonucleotides were then annealed to upstream and template oligonucleotides at a molar ratio of 1:4:2, respectively. The annealing reaction was performed using a PCR machine (95 °C, 5 min; 65 °C, 30 min; −0.5 °C/6 min). All substrates were gel-purified from 10% PAGE prior to use.
Endonuclease Assays
Standard endonuclease assays were performed in reaction mixtures (20 μl) containing 50 mm Tris-HCl, pH 7.8, 2 mm MgCl2, 2 mm dithiothreitol (DTT), 0.25 mg/ml of BSA, and 15 fmol of the flap substrate. Reactions were incubated for 30 min at 37 °C, followed by the addition of 4 μl of 6× stop solution (40% sucrose, 60 mm EDTA, 1.2% SDS, 0.05% bromphenol blue, and 0.05% xylene cyanol).
The cleavage products were separated on a 12% PAGE for 40 min at 150 V in 0.5× TBE (45 mm Tris base, 45 mm boric acid, and 1 mm EDTA). The gels were dried on a DEAE-cellulose paper (DE81, Whatman) and autoradiographed. Labeled DNA products were quantified by using a PhosphorImager (FLA-7000, Fujifilm).
Electrophoretic Mobility Shift Assay
Reaction mixtures (20 μl) containing 50 mm Tris-HCl, pH 7.8, 60 mm NaCl, 2 mm DTT, 0.25 mg/ml of BSA, 5% glycerol, 0.2 mm EDTA, and 15 fmol of DNA substrate and the indicated amounts of proteins were preincubated on ice for 10 min, followed by an additional incubation for 10 min at 37 °C. Reaction products were subjected to electrophoresis through pre-run 6% polyacrylamide gels (90 V) for 1 h in 0.5× TBE at 4 °C. Gels were dried and autoradiographed. The amount of nucleoprotein complexes formed was quantified using PhosphorImager.
Alkaline Protein Extraction from Yeast Cells
Expression levels of proteins in yeast were determined using the alkaline protein extraction as described previously (37). The dan2Δ405N mutant cells harboring pRS325-ADH1-Rad27-FLAG (or other expression vectors for mutant derivatives of Rad27) were grown to saturation. Cells (1 × 108, total) were harvested and resuspended in lysis buffer (200 μl; 1.85 m NaOH, 7.5% β-mercaptoethanol). The mixture was then incubated on ice for 10 min. TCA (20% v/v, 200 μl) was added and incubated on ice for an additional 10 min. Samples were then centrifuged at 4 °C for 5 min at 14,000 ×g in an Eppendorf centrifuge. Supernatants were discarded, and 1× SDS sample buffer was added to the precipitated materials. The samples were vigorously vortexed after 30 min of incubation at 60 °C, followed by boiling for 10 min. The samples were then subjected to electrophoresis in a 10% SDS-PAGE gel. Gels were Coomassie-stained and analyzed by Western blotting as described previously (37).
RESULTS
Rad27 Stimulates the Endonuclease Activity of Dna2
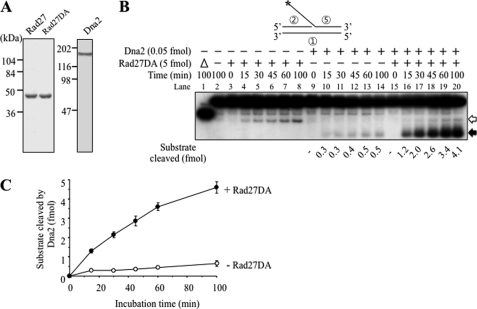
Direct physical interactions between Rad27 and Dna2 could lead to an increase in the enzymatic activities of one of the enzymes. Because Dna2 and Rad27 act on the same substrate, we prepared and isolated a nuclease-deficient Rad27 mutant enzyme (Rad27DA) by changing Asp to Ala at amino acid position 179 as well as wild type Rad27 and Dna2 (Fig. 1A). Consistent with previous reports (38, 39), the purified Rad27DA mutant enzyme displayed considerably less activity with the 5′-flap substrate than the wild type enzyme. Rad27DA (5 fmol) alone gave rise to low but detectable levels of cleavage products (Fig. 1B, lanes 3–8); the amount of cleavage products by Rad27DA did not exceed 0.5 fmol after a prolonged period (>60 min) of incubation (Fig. 1B, lanes 7 and 8). This low activity did not interfere to evaluate the influence of the Rad27 protein on the catalytic activity of Dna2 (Fig. 1B, lanes 9–20). When 5 fmol of Rad27DA was added to reactions containing a low level (0.05 fmol) of Dna2, Rad27DA markedly stimulated (up to >10-fold) the endonuclease activity of Dna2 on the 5′ 35-nt oligo(dT) flap substrate (Fig. 1B, lanes 15–20). In the absence of Rad27DA, Dna2 (0.05 fmol) alone generated low levels (up to 0.5 fmol) of cleavage products (Fig. 1B, lanes 9–14). These results showed that Rad27DA stimulates the endonuclease activity of Dna2 (Fig. 1C).
FIGURE 1.
Rad27DA stimulates the endonuclease activity of Dna2. A, SDS-PAGE analysis of the purified Rad27, Rad27D179A (Rad27DA), and Dna2 proteins. All proteins were purified as described under “Materials and Methods.” Each protein (1 μg) was subjected to 10% SDS-PAGE gel analysis following staining with Coomassie Blue R-250. The sizes of molecular mass markers are indicated in kDa. B, stimulation of Dna2 endonuclease activity by Rad27DA. Three sets of reactions (160 μl each) containing 120 fmol of the 5′ 35-nt flap substrate (standard Dna2 flap substrate, shown at the top of the figure) in the absence (−) or presence (+) of Rad27DA (5 fmol) or Dna2 (0.05 fmol) were assembled on ice and incubated at 37 °C for the indicated periods (15 to 100 min) of time. The circled numbers denote the oligonucleotides listed in Table 1. The asterisk in the substrate indicates the position of the 32P label. Aliquots (20 μl) were withdrawn from each set at the times indicated after initiation of reaction, and reactions were terminated by adding 4 μl of 6× stop solution (40% sucrose, 60 mm EDTA, 1.2% SDS, 0.05% bromphenol blue, and 0.05% xylene cyanol). Reaction products formed by each enzyme (open arrowhead, those produced by Rad27; closed, by Dna2) were separated on 12% PAGE in 0.5× TBE for 40 min at 150 V, and the amount of substrate cleaved was quantified as described under “Materials and Methods” and shown at the bottom of the figure. Lane 1 (Δ), boiled substrate; lane 2, no enzyme control. C, the amount of products formed by Dna2 in B was plotted against the time of incubation. Error bars obtained from three independent experiments are indicated.
We also determined the optimal conditions required for the stimulation of Dna2 endonuclease activity by Rad27. Rad27DA maximally stimulated Dna2 in the presence of 2–4 mm Mg2+ and 60 mm NaCl (data not shown). A further increase of Mg2+ (>10 mm) and NaCl (>100 mm) inhibited both the endonuclease activity of Dna2 and the extent of stimulation by Rad27DA. Moreover, Rad27DA efficiently stimulated the cleavage of flaps longer than 15-nt by Dna2 without altering its inability to cleave 5-nt flaps (data not shown). In contrast to the endonuclease activity, the helicase activity of Dna2 was unaffected by Rad27DA (Fig. S1B). ATP addition did not alter the Rad27DA-mediated stimulation of Dna2 (data not shown). Thus, we conclude that Rad27 stimulated only the endonuclease activity of Dna2 without affecting its helicase activity.
A Small C-terminal Fragment of Rad27 Is Sufficient to Stimulate Dna2 Endonuclease Activity
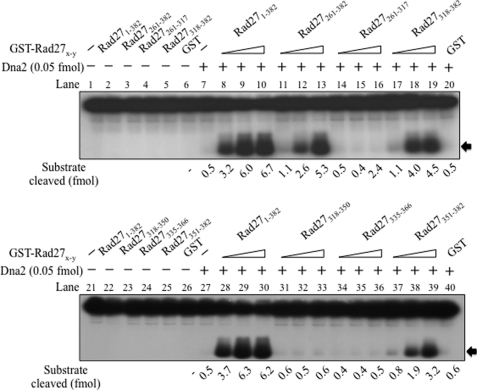
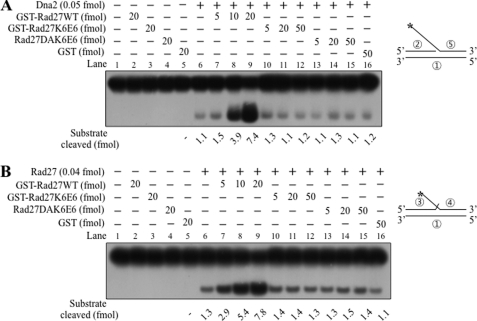
Our finding that Rad27 stimulated the endonuclease activity of Dna2 prompted us to define the region in Rad27 responsible for this effect. For this purpose, we used the N-terminal GST-tagged Rad27 derivatives (GST-Rad27x-y; where x and y indicate the amino acid residue in Rad27) that were used previously (34). We focused on the poorly conserved C-terminal region of Rad27, which is known to interact with many other proteins (34, 36, 40–42).
As shown in Fig. 2, none of the GST-Rad27x-y derivatives including GST-Rad27(1–382) (the full-length GST-Rad27, hereafter GST-Rad27) displayed flap endonuclease activity (Fig. 2, lanes 2 and 22). Thus, fusion of the GST tag to the N terminus of Rad27 inactivated the catalytic activity, in contrast to the residual activity found with Rad27DA (1% compared with wild type). Furthermore, all other isolated truncated derivatives, including GST-Rad27(261–382) (lane 3), GST-Rad27(261–317) (lane 4), GST-Rad27(318–382) (lane 5), GST-Rad27(318–350) (lane 23), GST-Rad27(335–366) (lane 24), GST-Rad27(351–382) (lane 25), and GST alone (lanes 6 and 26) did not generate cleavage products (Fig. 2), indicating that these preparations were devoid of contaminating nucleases.
FIGURE 2.
The C-terminal 32-amino acid domain of Rad27 is sufficient to stimulate endonuclease activity of Dna2. To locate the region responsible for the stimulation of Dna2, a number of the N-terminal GST-tagged Rad27 derivatives (GST-Rad27x-y; the number in subscript indicates positions of amino acid residues in Rad27 derivatives) were prepared. Standard reaction mixtures (20 μl each) containing Dna2 (0.05 fmol) and 15 fmol of the 5′ 35-nt flap substrate (see Fig. 1) were assembled, and incubated at 37 °C for 30 min in the presence of increasing concentrations (25, 50, and 100 fmol) of GST-Rad27x-y derivatives. Reaction products were analyzed as described in the legend to Fig. 1, and the amount of substrate cleaved was quantified and shown at the bottom of the figure. Additions and omissions of reaction components are indicated as (+) and (−), respectively, on top of the figure. Lanes 1 and 21, substrate only control; other controls containing GST-Rad27x-y derivatives alone include GST-Rad27(1–382) alone (lanes 2 and 22), GST-Rad27(261–382) (lane 3), GST-Rad27(261–317) (lane 4), GST-Rad27(318–382) (lane 5), GST only (lanes 6 and 26), GST-Rad27(318–350) (lane 23), GST-Rad27(335–366) (lane 24), and GST-Rad27(351–382) (lane 25). All GST-Rad27x-y only controls contained 100 fmol of the GST-Rad27x-y derivatives.
In the presence of different levels of GST-Rad27 and its truncated derivatives (25, 50, and 100 fmol), we found that GST-Rad27 (Fig. 2, compare lanes 7 with 8–10 and lanes 27 with 28–30), GST-Rad27(261–382) (lanes 11–13), GST-Rad27(318–382) (lanes 17–19), and GST-Rad27(351–382) (lanes 37–39) markedly stimulated Dna2 endonuclease activity (>10-fold). In contrast, GST-Rad27(261–317) (lanes 14–16), GST-Rad27(318–350) (lanes 31–33), and GST-Rad27(335–366) (lanes 34–36) failed to do so. These findings indicate that the C-terminal 32-amino acid fragment of Rad27 (GST-Rad27(351–382)) stimulates Dna2 endonuclease activity.
Overexpression of Rad27DA Partially Suppressed Temperature-sensitive Growth Defects of dna2Δ405N Strain
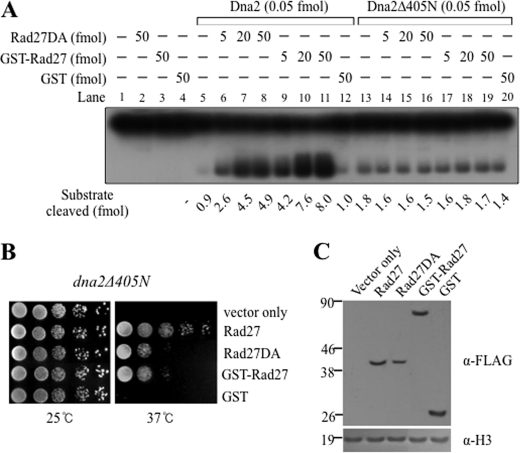
Because overexpression of RAD27 suppressed the ts growth defects of the dna2Δ405N mutant allele, we expected that Rad27 would stimulate the endonuclease activity of the Dna2Δ405N enzyme. However, neither Rad27DA nor GST-Rad27 stimulated this activity in vitro (Fig. 3A, lanes 14–16 and 17–19, respectively), suggesting that the N-terminal 405-amino acid domain of Dna2 is required for the stimulation. However, this result raised the possibility that the stimulation of Dna2Δ405N enzymatic activity by overexpression of Rad27 is not involved in suppression of dna2Δ405N mutant cells. It suggested that suppression could be due to an increase in the enzymatic activity of wild type Rad27.
FIGURE 3.
Overexpression of catalytically dead Rad27 mutant proteins suppresses the temperature-sensitive growth defects of dna2Δ405N strain. A, standard endonuclease assays of Dna2 were performed as described under “Materials and Methods” in the presence of varying amounts (5, 20, and 50 fmol) of Rad27DA (lanes 6–8 and 14–16) and GST-Rad27 (lanes 9–11 and 17–19) with Dna2 (0.05 fmol; lanes 5–12) or Dna2Δ405N (0.05 fmol; lanes 13–20). B, overexpression of Rad27 mutant proteins under control of the ADH1 promoter in the dna2Δ405N strain suppressed the ts growth defects. The dna2Δ405N cells were transformed with pRS325-ADH1 (vector only) or pRS325-AHD1-derived plasmids overexpressing Rad27-FLAG (Rad27, positive control), Rad27DA-FLAG (Rad27DA, catalytically defective), GST-Rad27-FLAG (GST-Rad27, catalytically dead), and GST-FLAG (GST, GST only control). The resulting transformants were grown in plates and a well isolated single colony was inoculated into liquid media, and cells grown were spotted in serial 10-fold dilutions (105, 104, 103, 102, and 101 cells) onto SD-L plates, followed by incubation for 5 days at the indicated temperature. C, expression levels of Rad27 proteins in each transformant used in B were examined by Western blot using anti-FLAG monoclonal antibodies (α-FLAG). Total proteins (alkaline extraction method) were prepared as described under “Materials and Methods.” As loading controls, the level of histone 3 is shown with anti-H3 monoclonal antibodies for the same blot.
Interestingly, catalytically defective Rad27DA and catalytically dead GST-Rad27 partially suppressed the ts growth defects of dna2Δ405N when they were overexpressed under control of an ADH1 promoter (Fig. 3B). As expected, overexpression of GST alone did not rescue the growth defects of dna2Δ405N cells. The difference in suppression of the dna2Δ405N mutation was not due to alteration in protein expression levels in vivo, as shown in Fig. 3C. These results suggested the following possibilities; the residual endonuclease activity of Rad27DA, even when overexpressed, is not responsible for suppression of the growth defects of dna2Δ405N. Rather, there could be unknown pathway that depends on high levels of Rad27 protein. It is worthwhile to mention that unlike the dna2Δ405N strain, the Dna2 helicase mutation (dna2K1080E) was not suppressed by overexpression of rad27DA, indicating that defects in the helicase activity are not rescued by excess levels of Rad27DA (data not shown).
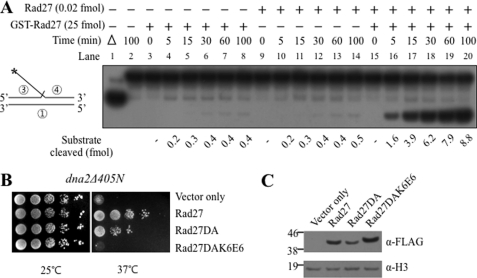
Rad27 Is a Trans-autostimulatory Protein
In light of the above finding, we investigated whether the expression of Rad27, regardless of its catalytic activity, stimulated the endonuclease activity of endogenous Rad27 present in dna2Δ405N cells to overcome the ts growth defects. We examined this notion by carrying out in vitro experiments with full-length GST-Rad27, which has negligible nuclease activity. In the presence of a double-flap substrate containing a 27-nt 5′ and a 1-nt 3′-flap, the addition of GST-Rad27 (25 fmol) or wild type Rad27 (0.02 fmol) did not lead to the formation (<0.6 fmol following a 1-h incubation) of cleavage products (Fig. 4A, lanes 3–8 and 9–14, respectively). In the presence of both proteins, a dramatic increase in substrate cleavage was observed (Fig. 4A, lanes 15–20). This observation is consistent with the notion that overexpression of the catalytically deficient GST-Rad27 or Rad27DA suppressed dna2Δ405N growth defects by stimulating the endogenous wild type Rad27 in vivo.
FIGURE 4.
Rad27 is a trans-autostimulatory protein. A, stimulation of catalytically active Rad27 enzyme by catalytically dead GST-Rad27 protein. A time course experiment was performed as shown in Fig. 1B with the double-flap substrate containing a 27-nt 5′-flap and a 1-nt 3′-flap (standard Rad27 flap substrate) as shown on the right of the figure. The circled numbers in the substrate denote the oligonucleotides listed in Table 1. The asterisk in the substrate indicates the position of 32P label. The amount of cleavage products formed by Rad27 (0.02 fmol) was determined after incubation at 37 °C for increasing time periods (5 to 100 min) in the presence (+) and absence (−) of catalytically dead GST-Rad27 (25 fmol). Aliquots (20 μl) were withdrawn from each set at the times indicated and the products were analyzed as described under “Materials and Methods.” The amount of substrate cleaved is indicated at the bottom of the figure. Lane 1 (Δ), boiled substrate; lane 2, no enzyme control. B, suppression of dna2Δ405N-associated growth defects requires the trans-autostimulatory activity of Rad27. The dna2Δ405N mutant cells were transformed with pRS325-ADH1 (Vector only) or pRS325-AHD1-derived plasmids overexpressing Rad27-FLAG (Rad27, positive control), Rad27DA-FLAG (Rad27DA, catalytically defective), and Rad27DAK6E6-FLAG (Rad27DAK6E6, defective in both catalytic and trans-autostimulatory activities). C, levels of Rad27 proteins were determined by Western blot using anti-FLAG (α-FLAG) monoclonal antibodies. α-H3, anti-histone3 monoclonal antibodies to show the level of histone 3 as loading controls.
To further substantiate this notion, we constructed a mutant Rad27 (Rad27DAK6E6) that was defective in both the catalytic and Rad27 stimulatory activities by combining the two separate mutations, Rad27DA and Rad27K6E6 (see below and Fig. 7). We found that overexpression of the rad27DAK6E6 mutant allele failed to suppress the ts growth defects of the dna2Δ405N (Fig. 4B), although the expression level of the Rad27DAK6E6 protein was significantly higher than that of Rad27DA (Fig. 4C). These results strongly support our suggestion that stimulation of the endogenous Rad27 nuclease activity is responsible for the observed suppression. The possibility that overexpression of Rad27DA or GST-Rad27 stimulates endonuclease activity of Dna2Δ405N in vivo indirectly through a mediator protein, however, cannot be ruled out.
FIGURE 7.
Full-length Rad27 lacking six positively charged Lys in the C-terminal region does not stimulate the endonuclease activity of Dna2 or Rad27. Standard endonuclease assays of Dna2 (A) and Rad27 (B) were performed as described in the legend to Fig. 6 in the presence of increasing amounts (5, 10, and 20 fmol) of the three Rad27 proteins (GST-Rad27WT, GST-Rad27K6E6, and Rad27DAK6E6) with flap substrates. Substrates used are schematically shown to the right of each figure. The amount of the substrate cleaved is shown at the bottom of figure.
Substrate Binding and Stimulation of Dna2 and Rad27 Depend on the Same Region of Rad27
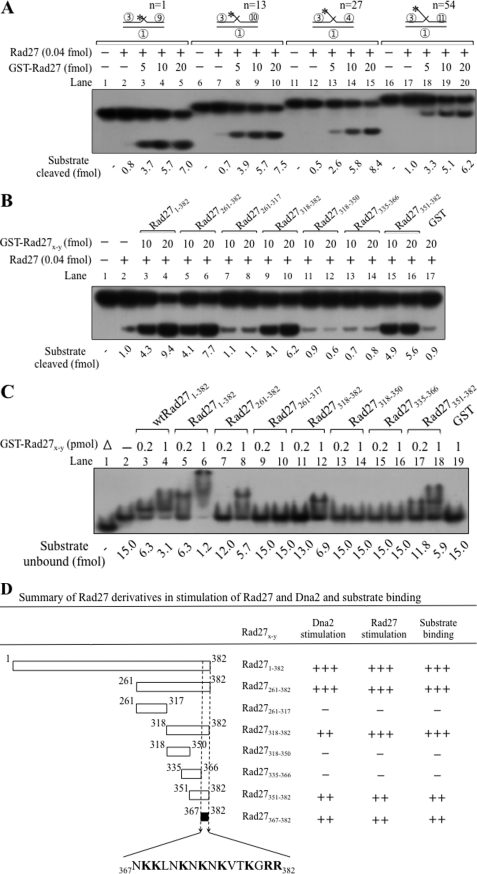
Prior to mapping of regions in Rad27 responsible for the stimulation of Rad27, we first examined the influence of GST-Rad27 on substrates containing varying 5′-flap lengths (1, 13, 27, and 54 nt). To avoid formation of secondary structures in the flap, all flaps were prepared using oligo(dT). The trans-autostimulatory effect was observed with all substrates tested, and the extent of stimulation was unaffected by the length of 5′-flaps. As shown in Fig. 5A, increasing levels (5, 10, and 20 fmol) of GST-Rad27 stimulated the activity of wild type Rad27 by 6–10-fold. Experiments with truncated forms of Rad27 revealed that the GST-Rad27(351–382) was the smallest fragment that stimulated Rad27 (Fig. 5B). We found that GST-Rad27 markedly increased (>10-fold) the endonuclease activity of Rad27 (Fig. 5B, compare lanes 2 with 3 and 4). Similar to results obtained with Dna2, GST-Rad27(261–382) (Fig. 5B, lanes 5 and 6), GST-Rad27(318–382) (lanes 9 and 10), and GST-Rad27(351–382) (lanes 15 and 16) stimulated the endonuclease activity of Rad27, whereas GST-Rad27(261–317) (lanes 7 and 8), GST-Rad27(318–350) (lanes 11 and 12), and Rad27(335–366) (lanes 13 and 14) failed to do so. These findings indicate that the same C-terminal 32-amino acid region of Rad27 enhances the endonuclease activity of both Dna2 and Rad27.
FIGURE 5.
The trans-autostimulatory activity of Rad27 resides in the same C-terminal 32-amino acid region required to stimulate the Dna2 endonuclease activity. A, standard Rad27 endonuclease assays were performed using several flap substrates as indicated schematically on top of the figure. All four substrates are double-flap substrates that differ in 5′-flap length. Length of the 5′-flap oligo(dT) is indicated by “n.” The circled numbers in the substrate denote the oligonucleotides listed in Table 1. The asterisk in the substrate indicates the position of the 32P label. B, GST-Rad27(351–382) is sufficient to stimulate Rad27 endonuclease activity. The amount of cleavage products produced by Rad27 (0.04 fmol) was measured in the presence of increasing amounts (10 and 20 fmol) of GST-Rad27x-y derivatives. C, electrophoretic mobility shift assays (EMSA) were performed as described under “Materials and Methods.” The standard double-flap substrate (15 fmol) of Rad27 was first preincubated with increasing amounts (0.2 and 1 pmol) of GST-Rad27x-y at 4 °C for 10 min, followed by an additional 10-min incubation at 37 °C. In lanes 3 and 4, the C-terminal His6-tagged wild type Rad27 (wtRad27(1–382)) was used. For all other lanes, N-terminal GST-tagged Rad27 derivatives were used. Lane 1 (Δ), boiled substrate; lane 2, no enzyme control. After incubation, samples were loaded onto a pre-run 6% polyacrylamide gel at 4 °C, and the nucleoprotein complexes formed were analyzed by autoradiography. The amount of substrate remaining was measured and the values are presented at the bottom of the gel. D, the ability of GST-Rad27x-y derivatives to bind standard flap substrate and to stimulate enzymatic activities of Rad27 and Dna2 is summarized. Positions of amino acids critical for stimulation of Dna2 and Rad27 are shown.
We also analyzed the DNA binding activity of GST-Rad27 fragments using electrophoretic mobility shift assays, and found that only those fragments that stimulated both Dna2 and Rad27 formed nucleoprotein complexes (Fig. 5C). This result, together with the above, confirms that all three activities (substrate binding, Rad27 and Dna2 stimulation) reside within the C-terminal 32-amino acid fragment of Rad27 (see Fig. 5D for summary).
Rad27(367–382) Peptide Stimulates Both Dna2 and Rad27 and Mutations within this Region Abolish Stimulatory Effect
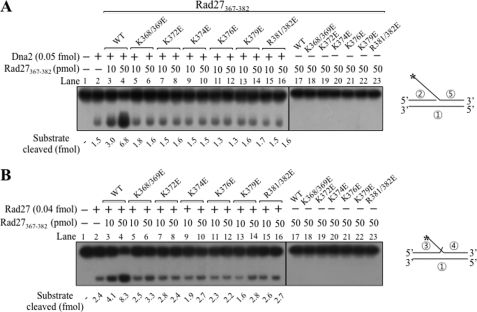
We found that boiled (10 min at 100 °C) Rad27DA and GST-Rad27 preparations retained their ability to stimulate endonuclease activities of Dna2 and Rad27 (data not shown). As shown in Fig. 5D, the shortest Rad27 fragment that possessed all three activities described above was the C-terminal 16-amino acid (Rad27(367–382)) derivative. This notion was verified with the synthetic peptide containing only the C-terminal 16-amino acid sequence. The synthetic peptide was highly soluble due to the presence of eight positively charged amino acid residues (Lys and Arg) that may be critical for DNA binding. We also prepared a number of mutant derivations of the peptide in which lysine and arginine residues were replaced with glutamic acid. These mutated peptides were used to examine the role of positively charged residues.
The wild type peptide (10 and 50 pmol) stimulated the endonuclease activity of Dna2 (Fig. 6A, lanes 3 and 4) and Rad27 (Fig. 6B, lanes 3 and 4). However, changes of lysine or arginine residues, singly or in combination, to glutamic acid markedly reduced stimulation of the endonuclease activity of Dna2 or Rad27 (Fig. 6, A and B, lanes 5–16). The peptides alone did not give rise to any detectable product (Fig. 6, A and B, lanes 17–23). The wild type Rad27(367–382) peptide still stimulated Dna2 and Rad27 following boiling for 10 min (data not shown). We also prepared full-length GST-Rad27 proteins containing the identical amino acid substitutions that affected the stimulatory activity of the peptides. In contrast to the dramatic effects observed with peptides, the single or double mutated GST-Rad27 derivatives still stimulated the Dna2 and Rad27 activities (data not shown). However, when all six lysine residues (Lys-368, -369, -372, -374, -376, and -379) were changed to glutamic acid, the GST-Rad27K6E6 mutant protein failed to stimulate Dna2 (Fig. 7A, lanes 10–12) and Rad27 (Fig. 7B, lanes 10–12). Moreover, Rad27DAK6E6, the double mutant protein, failed to stimulate both Dna2 and Rad27 (Fig. 7, A and B, lanes 13–15). Collectively, these results indicate that Rad27(367–382) stimulates the endonuclease activities of Dna2 and Rad27 and the positively charged residues within this region are critical for these effects. When we compared the C-terminal 16-amino acid region of Fen1 enzymes from a number of eukaryotes, we found that the percentage of lysine and arginine residues in this region exceeded 50% (Table 2).
FIGURE 6.
The oligopeptide corresponding to the C-terminal 16-amino acid fragment of Rad27 stimulates both Dna2 and Rad27; single or double mutations in positively charged amino acid residues abolish stimulation. A, Dna2 stimulation by commercially synthesized Rad27(367–382) peptides. Dna2 endonuclease assays were performed using standard reaction conditions containing 0.05 fmol of Dna2 in the presence of increasing concentrations (10 and 50 pmol) of wild type or several mutant Rad27(367–382) peptides, as indicated. WT, wild type peptide; K372E (and other single mutation peptides), a substitution of glutamic acid for the lysine residue at amino acid position 372; K368/369E and R381/382E, substitution of glutamic acid for the two consecutive lysine and arginine residues at amino acid positions 368–369 and 381–382, respectively. The schematic structure of the substrate used is shown next to the figure. The amount of substrate cleaved is shown at the bottom of the figure. B, the same experiment shown in A was repeated with 0.04 fmol of Rad27.
TABLE 2.
Distribution of positively charged lysine and arginine residues in the C-terminal 16-amino acid region in eukaryotic Fen1 enzymes
| Organism | Total length | Total number of Lys + Arg | Sequences of the C-terminal 16-amino acid fragment | Numbera of Lys + Arg |
|---|---|---|---|---|
| aa | % | % | ||
| Homo sapiens | 380 | 60 (15.8) | KKKAKTGAAGKFKRGKb | 8 (50) |
| Canis lupus familiaris | 380 | 60 (15.8) | KKKAKTGVAGKFKRGK | 8 (50) |
| Mus musculus | 380 | 60 (15.8) | KKKAKTGGAGKFRRGK | 8 (50) |
| Gallus gallus | 381 | 59 (15.5) | KKAKTNSATAKFKKGK | 7 (43.8) |
| Drosophila melanogaster | 385 | 63 (16.4) | NKKAKTSGGGRGRRPK | 7 (43.8) |
| Caenorhabditis elegans | 382 | 64 (16.8) | AKKGAKKGGPPKKRAK | 8 (50) |
| Schizosaccharomyces pombe | 380 | 63 (16.6) | KRKRDSNKGGESKKKR | 9 (56.3) |
| Saccharomyces cerevisiae | 382 | 65 (17.0) | NKKLNKNKNKVTKGRR | 8 (50) |
| Kluyveromyces lactis | 381 | 63 (16.5) | DAKKKAAAKGKIAKRR | 8 (50) |
| Magnaporthe oryzae | 390 | 64 (16.4) | EKKEKAKAKARPRGTA | 7 (43.8) |
| Oryza sativa | 380 | 62 (16.3) | KAVANKKTKGAGGKKK | 7 (43.8) |
| Plasmodium falciparum | 672 | 112 (16.7) | PKDVTNVKKKKYTQRC | 6 (37.5) |
a The number of Lys and Arg residues in the C-terminal 16-amino acid fragment.
b Lys and Arg residus are bold.
The C-terminal Region of Both Yeast and Human Fen1 Is Essential for Autostimulation
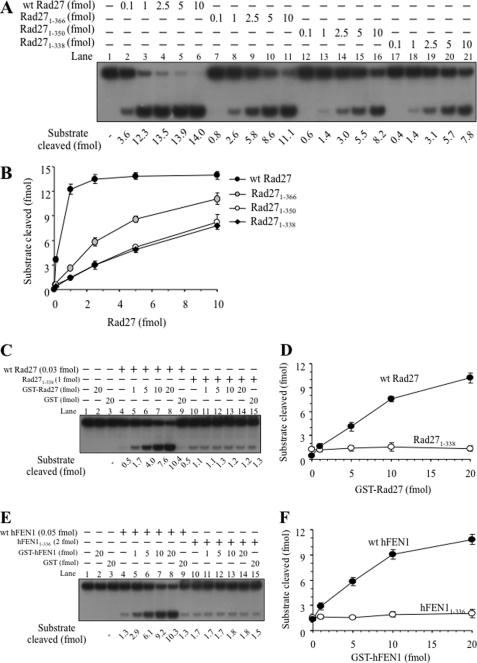
Although Fen1 is highly conserved in eukaryotes, the C-terminal regions of the yeast Rad27 and hFEN1 are less conserved compared with other regions of the enzyme. According to the three-dimensional structure of hFEN1-substrate complex (26), the C-terminal region (amino acids from 336 to 380) appears to contain a naturally unstructured extension that may participate in substrate binding, nuclear localization, and protein-protein interactions (22, 40, 42–44). Recently, we showed that human RFC stimulated the endonuclease activity of wild type hFEN1 and this activation depends on the C-terminal region of hFEN1 (36). To confirm the importance of the C terminus of Rad27 and hFEN1 for autostimulation, we constructed expression vectors for three C-terminal truncated versions of Rad27, which included Rad27(1–366), Rad27(1–350), and Rad27(1–338) (16-, 32-, and 44-amino acid deletion, respectively, from the C terminus) and purified each mutant protein as well as the wild type. We found that all three Rad27 deletion mutant proteins retained 10–20% of the endonuclease activity of the wild type protein (Fig. 8, A and B). The 16-amino acid deleted Rad27(1–366) mutant was approximately twice as active as the other two mutant enzymes that contained longer deletions (Fig. 8, A, compare lanes 7–11 with 12–21, and B). Although the wild type Rad27 was dramatically stimulated by the catalytically inactive GST-Rad27, Rad27(1–338) lacking the C-terminal 44 amino acids was hardly stimulated (Fig. 8, C, compare lanes 4–8 and 10–14, and D). GST-Rad27 also failed to stimulate the enzymatic activities of Rad27(1–350) and Rad27(1–366) prepared (data not shown). When the same experiment was repeated with the hFEN1 and its derivatives, we found that the C-terminal region of hFEN1 is also critical for autostimulation (Fig. 8, E, compare lanes 4–8 and 10–14, and F). Thus, the C-terminal regions of both yeast and human Fen1 are functionally conserved and essential for the trans-autostimulatory effects.
FIGURE 8.
The unstructured C-terminal region of both Rad27 and hFEN1 are essential for trans-autostimulation activity. A, standard endonuclease assays of Rad27 were performed with varying levels (0.1, 1, 2.5, 5, and 10 fmol) of wild type Rad27 and the three C-terminal truncated enzymes (Rad27(1–366), Rad27(1–350), Rad27(1–338)) to evaluate their relative endonuclease activities. B, the amount of substrate cleaved was plotted against the amount of Rad27 enzyme used. C, standard endonuclease assays of Rad27 were carried out in the presence of wild type Rad27 and Rad27(1–338) (lacking the C-terminal 42 amino acid residues) with increasing amounts (1, 5, 10, and 20 fmol) of GST-Rad27. D, the amount of product formed by wild type Rad27 and Rad27(1–338) was plotted against the amount of GST-Rad27 used. E, wild type hFEN1 and hFEN1(1–336) were tested for their trans-autostimulation by GST-hFEN1. F, the level of product formed in the reaction in E was plotted against the amount of GST-hFEN1 used.
DISCUSSION
In this study, we examined the genetic and physical interactions between Dna2 and Rad27 and showed that Rad27 stimulated the endonuclease activity of Dna2 and that Rad27 is a trans-autostimulatory enzyme using two catalytically defective Rad27 proteins. These findings provide an explanation for the mechanism by which overexpression of Rad27 suppresses the Dna2 defects. In addition, we found that the C-terminal 16-amino acid fragment of Rad27 (Rad27(367–382)) is minimally required for the stimulation of Dna2 and Fen1 activities as well as the binding of these proteins to their substrates. We conclude that the dramatic increase of cleavage products formed when wild type and catalytically dead Rad27 enzymes were combined (Fig. 4) is due to the direct stimulation of the nuclease activity of the wild type Rad27 by GST-Rad27 or Rad27DA and not caused by the activation of a cryptic or dormant nuclease activity of mutant enzymes. This conclusion is based on the following. First, it is hard to envisage that the small increase (<0.5% in total enzyme) of wild type Rad27 resulted in the dramatic activation of inactive enzymes. Second, the marked increase in the level of cleavage product was not dependent on the presence of a protein or peptide containing the catalytic site. The finding that Rad27 can stimulate its endonuclease activity was surprising because both Rad27DA and GST-Rad27 mutant enzymes retained the DNA substrate binding activity observed with the wild type protein (data not shown). Their binding to the same substrate could lead to an inhibition of the endonuclease activity of the wild type Rad27. In accord with this, the addition of excess levels (>200 fmol) of Rad27DA inhibited the nuclease activity of Dna2 and Rad27 (data not shown).
We suggest that the Fen1 proteins are truly trans-autostimulatory because they do not require any prior modifications unlike some protein kinases or phosphatases. To date, only a few examples of self-stimulatory enzymes have been reported. One example is the self-stimulatory GTPase-activating activity of RhoC, Cdc42Hs, and Rac2, the Rho family GTPases (45, 46). The rate of GTP hydrolysis by RhoC was increased significantly by increasing levels of GTP-bound RhoC, whereas RhoA and RhoB, despite their high sequence homology with RhoC, have a slow constant rate of GTP hydrolysis in a dose-independent manner (46).
Sequence alignment revealed that the C-terminal polybasic sequence and one specific arginine residue known as “arginine fingers” present in GTPase activating proteins of Ras and Rho were required for their self-stimulatory GTPase-activating activity (47). This feature is shared by yeast Rad27 and hFEN1. The short C-terminal polybasic region of Rad27, which is sufficient and necessary to stimulate both Rad27 and Dna2, contains arginine residues, similar to that present in the self-stimulatory GTPase-activating proteins. However, our experiments with mutant peptides indicated that the polybasic residues collectively contributed to the homophilic stimulation (or trans-auto stimulation) of Rad27 and the heterophilic stimulation of Dna2, obscuring the importance of individual lysine and arginine residues in the stimulation. The homophilic or heterophilic interaction between these two functional elements and Rad27/Dna2 may result in conformational changes that activate the enzymatic activities. Our finding that GST-Rad27 stimulated Dna2 or Rad27 more efficiently (∼2-fold) than Rad27DA is also in keeping with the critical role of the C-terminal region of Fen1 in homo- or heterophilic stimulation. GST-Rad27 that contained the native C-terminal region could be more efficient in the stimulation than Rad27DA that contained an altered C terminus by a His6 tag. It appears that the high content (∼50%) of basic amino acid residues in the C-terminal 16-amino acid fragment of Fen1 (cf. ∼16% in the whole Fen1 protein, Table 2) is responsible for the trans-autostimulatory activity of all eukaryotic Fen1. We found that the highly polybasic nature, not the amino acid sequence per se, is well conserved in the C-terminal region of Fen1 enzymes from lower to higher eukaryotes. At present, how the short polybasic region found in the C terminus of Rad27 contributes to DNA binding and homophilic or heterophilic stimulation of nucleases is unknown. To further understand the molecular events underlying this effect will require additional structure information of the Rad27-polybasic sequence. We believe that the trans-autostimulation of Rad27 contributes importantly to the processing of Okazaki fragments or other related transactions. This also provides a rational mechanism by which overexpression of rad27DA suppressed the ts growth defects of the dna2Δ405N strain despite its inability to stimulate the Dna2Δ405N enzyme in vitro. Like Rad27DA, overexpression of the N-terminal GST-tagged Rad27(GST-Rad27) suppressed the growth defects of dna2Δ405N mutant allele.
We believe that the residual enzymatic activity of Rad27DA, upon overexpression, is not sufficient to suppress the phenotype of dna2Δ405N in vivo. First, the phenotype of cells overexpressing rad27DA was shown to be similar to that of rad27Δ; when the wild type RAD27 allele was replaced with rad27DA, the resulting mutant cells following its overexpression displayed ts growth defects like rad27Δ (data not shown). Second, neither low-copy nor multicopy expression of rad27DA rescued the ts growth defects of rad27Δ (data not shown), indicating that Rad27DA is not functional in vivo. Our findings that Rad27DAK6E6, which is catalytically inactive and incapable of stimulating the endonuclease activity of Rad27, failed to suppress the growth defects of dna2Δ405N cells indicate that the suppression of the dna2Δ405N mutant allele by overexpression of Rad27 or its mutant derivatives is due to its trans-autostimulatory activity and this effect requires an endogenous wild type Rad27 in dna2Δ405N cells.
The stimulation of Dna2 by Rad27 and Rad27 trans-autostimulation may reflect another important means that cells use to preserve their genome integrity during evolution. If flaps grow longer in vivo, Dna2 activity is required prior to the action of Rad27. In this situation, interactions between the two inter-dependent enzymes would be beneficial. For example, Rad27 functions to actively dissociate Dna2 from flap structures after cleavage of long flaps during Okazaki fragment processing (48, 49). Furthermore, the Rad27-mediated stimulation of Dna2 allows Dna2 to process the long flaps efficiently and the subsequent action of Rad27 is facilitated by trans-autostimulation (this study), as well as by Dna2-, RPA-, or Pif1-mediated stimulation (33), to create ligatable nicks.
Recently, it was shown that Dna2 stimulated the flap endonuclease activity of Fen1 (33). We also confirmed this in our study. In addition, we found that only the N-terminal 405-amino acid fragment of Dna2 was sufficient to stimulate Rad27. However, we noted that Dna2Δ405N lacking the N-terminal 405 amino acids also stimulated the Rad27 endonuclease activity (data not shown). Both fragments (Dna2–405N and Dna2Δ405N) of Dna2 stimulated Rad27 in an additive manner, indicating that there are at least two domains in Dna2 to enhance the Rad27 activity. We also attempted to determine whether stable complexes could be formed following incubation of mixtures of differentially tagged Fen1 proteins. However, we were not able to detect a complex containing two different tags in our experiments, indicating that Fen1 does not form a stable homodimer under the conditions used in our experiments (data not shown).
It appears that Rad27-mediated stimulation of Dna2 is not conserved among eukaryotes, because hFEN1 does not stimulate human Dna2 (data not shown). This may be due to the fact that human Dna2 lacks the N-terminal region that corresponds to yeast Dna2 and the C-terminal region of hFEN1 differs substantially from that of the yeast Rad27. Thus, multiple interactions including the trans-autostimulatory activity of Rad27 between processing enzymes could increase the efficiency, which allows ligatable nicks to be generated more rapidly, thus contributing importantly to the preservation of an intact genome.
Supplementary Material
Acknowledgment
We thank Dr. Jerard Hurwitz for critical reading of the manuscript.
This work was supported by National Research Foundation of Korea Grant 20100000009 from the Ministry of Education, Science and Technology.

This article contains supplemental Fig. S1.
S.-H. Bae, C.-H. Lee, and Y.-S. Seo, unpublished observations.
- pol
- polymerase
- RPA
- replication protein A
- nt
- nucleotide(s)
- CV
- column volume.
REFERENCES
- 1. Waga S., Stillman B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 2. Johnson A., O'Donnell M. (2005) Cellular DNA replicases. Components and dynamics at the replication fork. Annu. Rev. Biochem. 74, 283–315 [DOI] [PubMed] [Google Scholar]
- 3. Burgers P. M. (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flores-Rozas H., Kolodner R. D. (2000) Links between replication, recombination, and genome instability in eukaryotes. Trends Biochem. Sci. 25, 196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McGlynn P., Lloyd R. G. (2002) Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3, 859–870 [DOI] [PubMed] [Google Scholar]
- 6. Cox M. M. (2002) The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 510, 107–120 [DOI] [PubMed] [Google Scholar]
- 7. Maher R. L., Branagan A. M., Morrical S. W. (2011) Coordination of DNA replication and recombination activities in the maintenance of genome stability. J. Cell. Biochem. 112, 2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsurimoto T., Stillman B. (1991) Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J. Biol. Chem. 266, 1961–1968 [PubMed] [Google Scholar]
- 9. Denis D., Bullock P. A. (1993) Primer-DNA formation during simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 13, 2882–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bae S. H., Seo Y. S. (2000) Characterization of the enzymatic properties of the yeast dna2 helicase/endonuclease suggests a new model for Okazaki fragment processing. J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 11. Bae S. H., Kim J. A., Choi E., Lee K. H., Kang H. Y., Kim H. D., Kim J. H., Bae K. H., Cho Y., Park C., Seo Y. S. (2001) Tripartite structure of Saccharomyces cerevisiae Dna2 helicase/endonuclease. Nucleic Acids Res. 29, 3069–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bae S. H., Kim D. W., Kim J., Kim J. H., Kim D. H., Kim H. D., Kang H. Y., Seo Y. S. (2002) Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277, 26632–26641 [DOI] [PubMed] [Google Scholar]
- 13. Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
- 14. Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., Bambara R. A. (2004) On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 279, 15014–15024 [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Kao H. I., Bambara R. A. (2004) Flap endonuclease 1. A central component of DNA metabolism. Annu. Rev. Biochem. 73, 589–615 [DOI] [PubMed] [Google Scholar]
- 16. Garg P., Burgers P. M. (2005) DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 17. Reagan M. S., Pittenger C., Siede W., Friedberg E. C. (1995) Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177, 364–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tishkoff D. X., Filosi N., Gaida G. M., Kolodner R. D. (1997) A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88, 253–263 [DOI] [PubMed] [Google Scholar]
- 19. Johnson R. E., Kovvali G. K., Prakash L., Prakash S. (1998) Role of yeast Rth1 nuclease and its homologs in mutation avoidance, DNA repair, and DNA replication. Curr. Genet. 34, 21–29 [DOI] [PubMed] [Google Scholar]
- 20. Harrington J. J., Lieber M. R. (1994) The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng L., Zhou M., Chai Q., Parrish J., Xue D., Patrick S. M., Turchi J. J., Yannone S. M., Chen D., Shen B. (2005) Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6, 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen B., Singh P., Liu R., Qiu J., Zheng L., Finger L. D., Alas S. (2005) Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Biol. Essays 27, 717–729 [DOI] [PubMed] [Google Scholar]
- 23. Storici F., Henneke G., Ferrari E., Gordenin D. A., Hübscher U., Resnick M. A. (2002) The flexible loop of human FEN1 endonuclease is required for flap cleavage during DNA replication and repair. EMBO J. 21, 5930–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chapados B. R., Hosfield D. J., Han S., Qiu J., Yelent B., Shen B., Tainer J. A. (2004) Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116, 39–50 [DOI] [PubMed] [Google Scholar]
- 25. Friedrich-Heineken E., Hübscher U. (2004) The Fen1 extrahelical 3′-flap pocket is conserved from archaea to human and regulates DNA substrate specificity. Nucleic Acids Res. 32, 2520–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsutakawa S. E., Classen S., Chapados B. R., Arvai A. S., Finger L. D., Guenther G., Tomlinson C. G., Thompson P., Sarker A. H., Shen B., Cooper P. K., Grasby J. A., Tainer J. A. (2011) Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145, 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. MacNeill S. A. (2001) DNA replication. Partners in the Okazaki two-step. Curr. Biol. 11, R842–R844 [DOI] [PubMed] [Google Scholar]
- 28. Hübscher U., Seo Y. S. (2001) Replication of the lagging strand. A concert of at least 23 polypeptides. Mol. Cells 12, 149–157 [PubMed] [Google Scholar]
- 29. Rossi M. L., Purohit V., Brandt P. D., Bambara R. A. (2006) Lagging strand replication proteins in genome stability and DNA repair. Chem. Rev. 106, 453–473 [DOI] [PubMed] [Google Scholar]
- 30. Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 31. Budd M. E., Campbell J. L. (1997) A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17, 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Budd M. E., Campbell J. L. (2000) The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459, 173–186 [DOI] [PubMed] [Google Scholar]
- 33. Henry R. A., Balakrishnan L., Ying-Lin S. T., Campbell J. L., Bambara R. A. (2010) Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J. Biol. Chem. 285, 28496–28505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang M. J., Lee C. H., Kang Y. H., Cho I. T., Nguyen T. A., Seo Y. S. (2010) Genetic and functional interactions between Mus81-Mms4 and Rad27. Nucleic Acids Res. 38, 7611–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) Dna2 of Saccharomyces cerevisiae possesses a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 36. Cho I. T., Kim D. H., Kang Y. H., Lee C. H., Amangyelid T., Nguyen T. A., Hurwitz J., Seo Y. S. (2009) Human replication factor C stimulates flap endonuclease 1. J. Biol. Chem. 284, 10387–10399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee C. H., Shin Y. K., Phung T. T., Bae J. S., Kang Y. H., Nguyen T. A., Kim J. H., Kim D. H., Kang M. J., Bae S. H., Seo Y. S. (2010) Involvement of Vts1, a structure-specific RNA-binding protein, in Okazaki fragment processing in yeast. Nucleic Acids Res. 38, 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gary R., Park M. S., Nolan J. P., Cornelius H. L., Kozyreva O. G., Tran H. T., Lobachev K. S., Resnick M. A., Gordenin D. A. (1999) A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19, 5373–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shen B., Nolan J. P., Sklar L. A., Park M. S. (1997) Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 25, 3332–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo Z., Chavez V., Singh P., Finger L. D., Hang H., Hegde M. L., Shen B. (2008) Comprehensive mapping of the C terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J. Mol. Biol. 377, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karanja K. K., Livingston D. M. (2009) C-terminal flap endonuclease (rad27) mutations. Lethal interactions with a DNA ligase I mutation (cdc9-p) and suppression by proliferating cell nuclear antigen (POL30) in Saccharomyces cerevisiae. Genetics 183, 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frank G., Qiu J., Zheng L., Shen B. (2001) Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 276, 36295–36302 [DOI] [PubMed] [Google Scholar]
- 43. Qiu J., Li X., Frank G., Shen B. (2001) Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 276, 4901–4908 [DOI] [PubMed] [Google Scholar]
- 44. Stucki M., Jónsson Z. O., Hübscher U. (2001) In eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. J. Biol. Chem. 276, 7843–7849 [DOI] [PubMed] [Google Scholar]
- 45. Zhang B., Zheng Y. (1998) Negative regulation of Rho family GTPases Cdc42 and Rac2 by homodimer formation. J. Biol. Chem. 273, 25728–25733 [DOI] [PubMed] [Google Scholar]
- 46. Zhang B., Zhang Y., Collins C. C., Johnson D. I., Zheng Y. (1999) A built-in arginine finger triggers the self-stimulatory GTPase-activating activity of rho family GTPases. J. Biol. Chem. 274, 2609–2612 [DOI] [PubMed] [Google Scholar]
- 47. Bourne H. R. (1997) G proteins. The arginine finger strikes again. Nature 389, 673–674 [DOI] [PubMed] [Google Scholar]
- 48. Stewart J. A., Campbell J. L., Bambara R. A. (2006) Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 281, 38565–38572 [DOI] [PubMed] [Google Scholar]
- 49. Stewart J. A., Campbell J. L., Bambara R. A. (2009) Significance of the dissociation of Dna2 by flap endonuclease 1 to Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 284, 8283–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.