Background: Only a few proteinaceous inhibitors of metallocarboxypeptidases have been characterized structurally in depth.
Results: The structure of human carboxypeptidase A4 in complex with a Nerita versicolor inhibitor (NvCI) was derived at 1.7 Å.
Conclusion: NvCI displays a different protein fold that inhibits carboxypeptidases in a substrate-like manner.
Significance: We deciphered the structural determinants for picomolar inhibition constants for A-type carboxypeptidases, the most potent by now.
Keywords: Carboxypeptidase, Crystal Structure, Enzyme Inhibitors, Enzymes, Metalloprotease, Nerita versicolor
Abstract
NvCI is a novel exogenous proteinaceous inhibitor of metallocarboxypeptidases from the marine snail Nerita versicolor. The complex between human carboxypeptidase A4 and NvCI has been crystallized and determined at 1.7 Å resolution. The NvCI structure defines a distinctive protein fold basically composed of a two-stranded antiparallel β-sheet connected by three loops and the inhibitory C-terminal tail and stabilized by three disulfide bridges. NvCI is a tight-binding inhibitor that interacts with the active site of the enzyme in a substrate-like manner. NvCI displays an extended and novel interface with human carboxypeptidase A4, responsible for inhibitory constants in the picomolar range for some members of the M14A subfamily of carboxypeptidases. This makes NvCI the strongest inhibitor reported so far for this family. The structural homology displayed by the C-terminal tails of different carboxypeptidase inhibitors represents a relevant example of convergent evolution.
Introduction
Metallocarboxypeptidases (MCPs)4 are exopeptidase enzymes involved in a great variety of processes, from digestion to blood coagulation/fibrinolysis, inflammation, and prohormone and neuropeptide processing among others, and have also been reported to be involved in the progress of certain cancers (1–3). Their activity normally takes place outside of the cell, by removing the C-terminal residues of polypeptidic chains. The activity of MCPs can be regulated in distinct organisms through the action of exogenous carboxypeptidase inhibitors (1, 3). Proteinaceous carboxypeptidase inhibitors can be found in a wide range of biological locations and in evolutionarily distant organisms such as potato, leech, ticks, and worms (4–7).
We have recently isolated a novel MCP inhibitor (NvCI) from the marine snail Nerita versicolor. It is a 53-residue protein that tightly binds to the M14A subfamily of carboxypeptidases (traditionally known as pancreatic-like carboxypeptidases). It displays an inhibition constant (Ki) in the picomolar range against several MCPs belonging to the abovementioned subfamily: bovine carboxypeptidase A1 (bCPA1), human CPA1 (hCPA1), and human carboxypeptidase A4 (hCPA4; which has been structurally characterized in this work). Such Ki values are 3 orders of magnitude lower than for other known exogenous protein inhibitors of carboxypeptidases (4–7).
Only four exogenous proteinaceous inhibitors of carboxypeptidases with a similar mode of inhibition have been studied in detail: potato carboxypeptidase inhibitor (PCI) from Solanum tuberosum (4), leech carboxypeptidase inhibitor (LCI) from Hirudo medicinalis (5), tick carboxypeptidase inhibitor (TCI) from Rhipicephalus bursa (6), and Ascaris carboxypeptidase inhibitor (ACI) from Ascaris suum (7). A common feature of all of these protein inhibitors is their small size (39–75 residues) and their stabilization by several disulfide bridges. In all cases, the inhibition relies on the interaction of the C-terminal tail with the active site groove of the carboxypeptidase in a mechanism that mimics substrate binding (8–10). Although there are neither sequence nor three-dimensional structure similarities between the main body of these inhibitors, they all have effects on the key residues for the activity of the enzyme. Remarkably, a protein carboxypeptidase inhibitor for humans, latexin, which is the only endogenous inhibitor isolated so far, displays a different mode of action, in a way reminiscent of the interaction of the prodomain with the carboxypeptidase, covering the active site of the enzyme (11).
NvCI is the first proteinaceous inhibitor of MCPs isolated and characterized from a marine organism. The marine Caribbean fauna is characterized by its richness and diversity, which make it a very attractive natural source for the identification of novel biomolecules with biological and biomedical interests. The potential of marine invertebrates as a source of these biomolecules has been reported in previous studies, particularly those focused on endoproteases such as serine and cysteine proteases and their inhibitors, some with exceptional structural and functional properties (2, 12–14).
Pro-CPA4 and its active form (CPA4), a counterpart used in this work, belong to the M14A subfamily of carboxypeptidases and have been implicated in different physiological processes (15, 16). Human pro-CPA4 was also identified as a gene product involved in prostate cancer (17).
In this work, we report the crystal structure of NvCI in complex with human CPA4 at 1.7 Å resolution. NvCI displays a different protein fold, and its interface with hCPA4 has been analyzed in detail and compared with the few reported structures of exogenous MCP inhibitors to rationally explain its exceptional ability (picomolar range) to inhibit certain MCPs.
EXPERIMENTAL PROCEDURES
Heterologous Expression and Purification of Recombinant NvCI
The NvCI amino acid sequence (UniProt ID P86912) was determined by a combination of Edman degradation and MALDI-TOF-MS. A synthetic gene encoding NvCI was designed and constructed to express this molecule in the Pichia pastoris system (GeneArt). The DNA sequence of NvCI was fused in-frame to the Saccharomyces cerevisiae prepro-α-factor signal in the XhoI site of the pPICZαA vector for secretion into the culture medium. Production of recombinant NvCI was carried out using a Zeocin hyper-resistant strain in an autoclavable bioreactor (Applikon Biotechnology). Production was monitored according to parameters such as wet cell weight, as well as by MALDI-TOF-MS, determination of the protein concentration in the supernatant by the BCA method (18), and bCPA1 inhibitory activity (19).
Purification of NvCI was performed using a combination of two ion exchange chromatographic methods: an initial weak cation exchange (AccellTM Plus CM, Waters) using 20 mm Tris-HCl (pH 7.0) and an ionic strength gradient (up to 1 m NaCl), followed by a second step of anion exchange (TSKgel® DEAE-5PW, Tosoh Bioscience LLC) using a linear gradient of 0–100% 20 mm Tris-HCl (pH 8.5) containing 1 m NaCl. The purity of NvCI was determined by its molecular mass obtained by MALDI-TOF-MS, by Tris/Tricine/SDS-PAGE, and by its functional activity against bCPA1.
Heterologous Expression and Purification of Recombinant hCPA4
Human pro-CPA4 was overexpressed and secreted into the extracellular medium using the P. pastoris heterologous system as described (11). Production of hCPA4 was carried out and monitored in the same way as described above for NvCI.
Enzyme purification was performed using a combination of hydrophobic interaction chromatography with a TOYOPEARL® butyl-650M column (Sigma-Aldrich) and weak anion exchange chromatography using a TSKgel® DEAE-5PW column according to the purification process described previously (16). The purity of hCPA4 was determined by SDS-PAGE, and its functional activity was determined by hydrolysis of the synthetic substrate N-(4-methoxyphenylazoformyl)phenylalanine (19).
Formation and Purification of NvCI-hCPA4 Complex
The formation and capture of the NvCI-hCPA4 complex were performed by preincubation of both proteins for 30 min in 50 mm Tris-HCl (pH 8.5) containing 150 mm NaCl at 37 °C. For this purpose, 16 mg of hCPA4 were incubated with 5.5 mg of recombinant NvCI in a final reaction volume of 70 ml (equivalent to an enzyme/inhibitor molar ratio of 1:2). The complex was captured on a size exclusion chromatography column (HiLoad Superdex 75 26/60, GE Healthcare) equilibrated with the same buffer used for complex formation. Elution peaks corresponding to the complex, free enzyme, and inhibitor were analyzed by PAGE. The NvCI-hCPA4 complex was concentrated to 17.6 mg/ml using an Amicon Ultra-4 centrifugal filter (Millipore).
Crystallization and Data Collection
Crystals of the complex between NvCI and hCPA4 were obtained at 18 °C by sitting-drop vapor-diffusion methods. The reservoir solution contained 0.04 m ammonium nitrate and 25% (w/v) PEG 3350. Single crystals appeared after 4 days from equal volumes of protein solution (17.6 mg/ml in 5 mm Tris (pH 8.5) and 50 mm NaCl) and reservoir solution. Crystals were cryoprotected in reservoir buffer containing 12% glycerol and flash-frozen in liquid nitrogen prior to diffraction analysis. Diffraction data were recorded from cryo-cooled crystals (100 K) at Grenoble beamline ID23-2. Data were integrated and merged using XDS (20) and scaled, reduced, and further analyzed using CCP4 (21) (Table 1).
TABLE 1.
Crystallographic data
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 69.22, 71.98, 79.84 |
| α, β, γ | 90.00°, 108.84°, 90.00° |
| Resolution (Å) | 50–1.70 (1.79–1.70)a |
| Rmergeb | 0.061 (0.51) |
| I/-σI | 13.1 (1.5) |
| Completeness (%) | 97.5 (84.5) |
| Redundancy | 3.3 (2.3) |
| Refinement | |
| Resolution (Å) | 50–1.70 |
| No. of reflections | 79,607 |
| Rwork/Rfreec | 20.54/23.46 |
| No. of atoms | 5903 |
| Protein | 702 |
| Water | 306 |
| Nitrate | 6 |
| Zinc | 2 |
| Average B-factors (Å2) | 17.25 |
| r.m.s.d.d | |
| Bond lengths (Å) | 0.007 |
| Bond angles | 1.033° |
a Statistics for the highest resolution shell are shown in parentheses.
b Rmerge = Σ|Ii − 〈I〉|/ΣIi, where Ii is the ith measurement of the intensity of an individual reflection or its symmetry-equivalent reflections, and 〈I〉 is the average intensity of that reflection and its symmetry-equivalent reflections.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo| for all reflections, and Rfree = Σ‖Fo| − |Fc‖/Σ|Fo|, calculated based on the 5% of data excluded from refinement.
d r.m.s.d., root mean square deviation.
Structure Determination and Refinement
The structure of the NvCI-hCPA4 complex was determined from the x-ray data at 1.7 Å by molecular replacement using Protein Data Bank code 2PCU for hCPA4 as a model. The quality of the diffraction data allowed automatic building of the inhibitor using wARP (22). Manual building and improvement of the model were performed using Coot (23). Refinement utilized CNS (24) and PHENIX (25). Ramachandran analysis showed that 94.70% of the residues (661) are in preferred regions, 4.58% of the residues (32) are in allowed regions, and 0.72% the of residues (5) are in outlier regions for both complexes of NvCI-hCPA4 in the asymmetric unit. Refinement and data statistics are provided in Table 1.
Determination of Inhibition Constants
Ki values were determined according to the strategy described for tight-binding inhibitors (26). The experiments were performed at 37 °C and pH 7.5 by varying the inhibitor concentration in each assay with a fixed concentration of enzyme and substrate (0.1 mm N-(4-methoxyphenylazoformyl)phenylalanine) and preincubation time. Ki values were determined for bCPA1 (Sigma-Aldrich) and for hCPA1, hCPA2, and hCPA4, which were produced by recombinant expression by our group (11, 27, 28).
RESULTS
Recombinant Protein Production and Complex Formation
The natural form of NvCI was detected, isolated from a crude extract of the marine snail N. versicolor, and de novo sequenced by MALDI-TOF-MS and Edman degradation, which allowed the construction of a synthetic gene with an optimized P. pastoris codon usage. A recombinant form of NvCI with the same mass and size as the natural one (5945.6 Da by MALDI-TOF-MS and 53 residues) was produced in P. pastoris, followed by chromatographic purification as summarized under “Experimental Procedures.” NvCI was overexpressed, yielding 329.7 mg of recombinant NvCI/liter of culture broth. In the case of the enzyme counterpart, hCPA4 was also overexpressed in P. pastoris with an overall yield of 17.6 mg of hCPA4/liter of culture medium. The complex was formed and purified by gel filtration at a 1:2 molar ratio of hCPA4 to NvCI (see “Experimental Procedures” for details).
Crystal Structure of NvCI in Complex with hCPA4 at 1.7 Å
The polypeptide chain of hCPA4 and NvCI can be clearly and nearly completely traced in the electron density maps (Phe-6 to Asn-307 for hCPA4 and Val-3 to Ala-53 for NvCI) (Fig. 1). The crystal structure of NvCI-hCPA4 displays two complexes in the asymmetric unit. Both complexes are almost identical, showing a root mean square deviation of 0.8 Å and displaying similar overlapping for the hCPA4 and NvCI molecules. Based on results obtained by gel filtration chromatography, the biological unit can be considered to be a monomer, formed by only one binary complex between hCPA4 and NvCI.
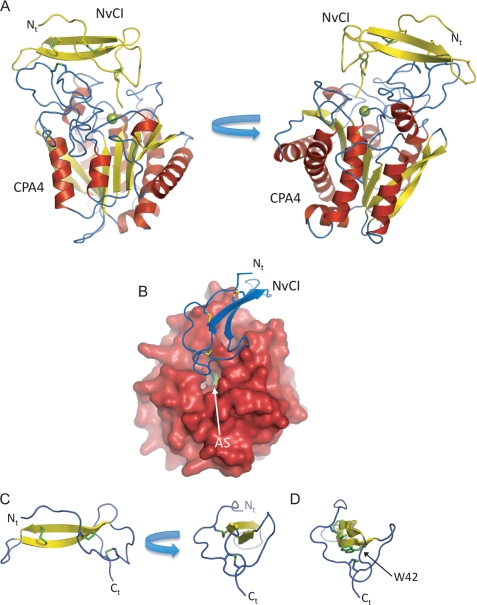
FIGURE 1.
Three-dimensional structure of NvCI in complex with human hCPA4. A, two views of the NvCI-hCPA4 complex shown in ribbon representation. The α-helix, β-strands, and coils of CPA4 are highlighted in red, yellow, and blue color, respectively. The NvCI structure is shown in yellow. The catalytic zinc atom in the metallocarboxypeptidase hCPA4 is shown in green. The three disulfide bridges formed in NvCI are shown in stick representation and visualized in green. The N terminus is labeled Nt. B, surface representation of hCPA4 in complex with NvCI shown in ribbon representation (blue). AS indicates the position of the active site groove of hCPA4. C, two views of the ribbon representation of NvCI. The β-strands and coils are colored in yellow and blue, respectively. The three disulfide bridges formed in NvCI are shown in stick representation (green). Ct, C terminus. D, same representation as in A but depicting Trp-42 in stick representation. All figures were prepared with PyMOL (32).
The structure of NvCI displays a new extended globular protein fold, which is basically formed by a central two-stranded antiparallel β-sheet connected by three major loops and two short tails that extend at the N and C termini (Fig. 1). The β-strands and the three loops are cross-connected and stabilized by three disulfide bridges formed between Cys-9 and Cys-23, Cys-15 and Cys-51, and Cys-27 and Cys-38. Despite its small size, NvCI has a small hydrophobic core located next to the C terminus of the protein formed by non-polar interactions of the side chain of Trp-42, which is sandwiched between two disulfide bridges of the inhibitor (Fig. 1D). The β-structure of the inhibitor also contains a few bulky exposed hydrophobic residues to the solvent, Phe-25, Phe-34, and Phe-44, which probably contribute to the reduced solubility of the recombinant protein. The C-terminal tail of NvCI is formed by only two residues, Tyr-52 and Ala-53, but as discussed below, this short extension is sufficient to interact with the active site residues and zinc atom of the carboxypeptidase.
The structure of the hCPA4 enzyme in complex with NvCI is related to previously reported structures of hCPA4 alone and in complex with other inhibitors (11). The structure of the enzyme displays the classical carboxypeptidase fold, with eight α-helices and a mixed eight-stranded β-sheet forming a globular α/β-motif (Fig. 1). The coordination of the zinc atom is essentially conserved compared with other MCP structures. In the absence of inhibitor, the zinc atom of MCPs is coordinated to the “catalytic” water molecule and to carboxypeptidase residues His-69 and His-196 and, in a bidentate form, to Glu-72 (1). In the NvCI-hCPA4 complex, the catalytic water is not present, and it is substituted with a bidentate coordination to the zinc atom by the C-terminal carboxylate group of NvCI, which is buried in the active site groove of the enzyme (Fig. 1B). As observed in other carboxypeptidase complexes with inhibitors, the most dramatic change observed in the active site residues is the movement of the side chain of Tyr-248, almost 180° from the “up” to the “down” position in the complex. There are other minor movements in the active site residues of hCPA4 to accommodate the C-terminal tail of the inhibitor.
“Primary” Interaction Site in NvCI-hCPA4
Despite its small size, NvCI interacts extensively with hCPA4, with a total contact area between both proteins of 1875.1 Å2 (Table 2). As observed with the other exogenous carboxypeptidase inhibitors (PCI, LCI, ACI, and TCI), inhibition of the enzyme can be attributed to a competitive interaction with the active site of the carboxypeptidase by occlusion of the active site subsites S1′, S1, and S2 (Fig. 2A). These sites are occupied by the C-terminal tail of NvCI (Tyr-52 and Ala-53) and constitute the primary contact region of the inhibitor. The “secondary” contact region, which is very extended and covers almost a complete face of the inhibitor, is discussed below.
TABLE 2.
Interactions between primary and secondary regions of NvCI and hCPA4
| NvCI | hCPA4 | Distance |
|---|---|---|
| Å | ||
| Primary interaction region | ||
| Cys-51 N | Glu-163 Oϵ1 | 3.06 |
| Tyr-52 N | Glu-163 Oϵ2 | 3.08 |
| Tyr-52 Cδ2 | Val-164 Cγ1 | 3.59 |
| Tyr-52 Cδ2 | Tyr-248 Cζ | 3.59 |
| Tyr-52 O | Arg-71 Nη2 | 3.02 |
| Tyr-52 O | Arg-127 Nη2 | 3.23 |
| Tyr-52 O | Phe-279 Cζ | 3.20 |
| Ala-53 N | Tyr-248 Oη | 3.78 |
| Ala-53 Cβ | Glu-270 Oϵ2 | 3.26 |
| Ala-53 O | Glu-270 Oϵ2 | 2.54 |
| Ala-53 Oτ | Arg-127 Nη2 | 2.79 |
| Secondary interaction region | ||
| Ile-10 O | Asn-159 Nδ2 | 3.30 |
| Asp-11 O | Asn-159 Nδ2 | 3.71 |
| Pro-16 Cβ | Tyr-249 Cδ2 | 3.63 |
| Leu-17 Cδ1 | Tyr-198 Cϵ1 | 3.91 |
| Leu-17 Cδ1 | Phe-279 Cζ | 3.62 |
| Phe-34 Cϵ2 | Ser-136 O | 3.43 |
| Phe-34 Cδ1 | Ser-137 Cβ | 3.67 |
| Tyr-36 Cϵ1 | Ser-137 Cβ | 3.64 |
| Glu-37 Oϵ1 | Arg-130 Nη2 | 2.81 |
| Glu-37 Oϵ2 | Arg-130 Nη1 | 2.85 |
| Cys-38 N | Ser-137 O | 3.19 |
| Cys-38 O | Cys-138 Cα | 3.24 |
| Cys-38 Sγ | Ser-137 Oγ | 3.05 |
| Gln-39 Nϵ2 | Asn-123 Oδ1 | 2.99 |
| Gln-39 Nϵ2 | Trp-126 Cζ3 | 3.58 |
| His-40 N | Cys-161 O | 3.43 |
| His-40 Cϵ1 | Asn-159 Nδ2 | 3.71 |
| Arg-48 O | Leu-125 Cδ1 | 3.86 |
| Gly-50 Cα | Arg-71 Nη1 | 3.67 |
| Gly-50 Cα | Phe-279 Cϵ2 | 3.8 |
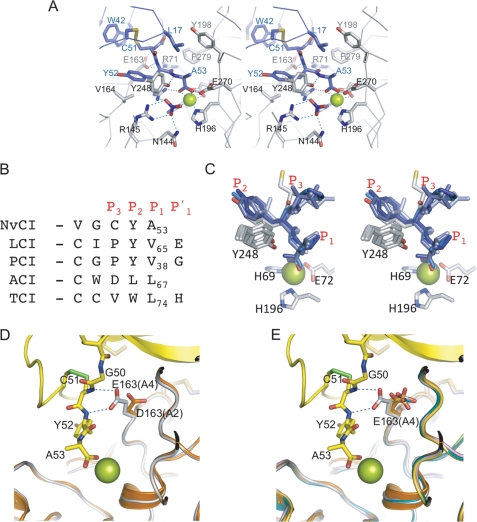
FIGURE 2.
Close-up view of NvCI primary binding region in complex with active site of hCPA4. A, close-up stereo view in stick representation of the C-terminal tail of NvCI with the active site of hCPA4. Amino acid residues corresponding to the C terminus of NvCI and the active site of hCPA4 are labeled and shown in blue and gray, respectively. The active site zinc is visualized as a green sphere. The nitrate molecule is also shown in stick representation (blue). B, sequence alignment of the C-terminal tails with other exogenous proteinaceous carboxypeptidase inhibitors, depicting the P1, P2, and P3 subsites. C, stereo representation of the structural alignment of the C-terminal tail of the inhibitor from B. Amino acid residues corresponding to the C terminus of the inhibitors are shown in blue. Zinc-binding residues and Tyr-248 are shown in gray. The active site zinc is shown as a green sphere. D, stick representation of the hydrogen bonds between Glu-163 and the C-terminal tail of NvCI. The hCPA4 structure was overlapped with hCPA2 (Protein Data Bank code 1DTD). NvCI, hCPA4, and hCPA2 chains are presented in yellow, gray, and orange, respectively. The catalytic zinc atom is presented as a green sphere. Selected amino acid residues for both carboxypeptidases and inhibitor are labeled. E, same representation as in D but with the CPA4 structure overlapped with hCPA2, hCPA1 (code 3FJU), human carboxypeptidase B (code 1ZLH), and bCPA1 (code 4CPA).
The C-terminal tail of NvCI is shorter in comparison with the other known carboxypeptidase inhibitors; it is composed of only two residues, Tyr-52 and Ala-53 (corresponding to the P2 and P1 positions), in this order when in complex with the enzyme. Three of the four reported exogenous carboxypeptidase inhibitors contain an extra C-terminal residue (which occupies the P1′ position) that is cleaved off by the carboxypeptidase and, in some cases, remains trapped in the S1′ subsite (glycine and glutamic acid are at the P1′ position in PCI and LCI, respectively) (8, 9). Interestingly, the electron density maps of the NvCI-hCPA4 complex show that part of the S1′ subsite of the carboxypeptidase is occupied by a nitrate molecule from the crystallographic buffer (Fig. 2A). The nitrate molecule is in contact with the guanidinium group of Arg-145 (at distances of 2.88 and 2.97 Å to the NH1 and NH2 atoms, respectively) and Asn-144 (at a distance of 2.79 Å to the ND2 atom). These residues belong to the S1′ subsite and interact with the carboxylate group of the cleaved residue of a carboxypeptidase substrate. In the PCI and LCI complex structures, the cleaved and trapped C-terminal residue occupies a similar position as the nitrate molecule in NvCI.
Sequence and three-dimensional structure comparisons of the C-terminal tails of NvCI with those of the different exogenous carboxypeptidase inhibitors indicated strong similarities and identical conformations of the backbone and side chains for the P1 and P2 residues (Ala-53 and Tyr-52 for NvCI, respectively) (Fig. 2, B and C, and supplemental Fig. S1). P1 and P2 residues are oriented in a substrate-like manner in all reported structures of these competitive tight-binding inhibitors. Notably, in all of them, the chemical character of the residue forming the P1 subsite is aliphatic, whereas the preference is for aromatic residues for the P2 subsite (except Leu for ACI), which establishes stacking interactions with the aromatic ring of Tyr-248 (Fig. 2C).
As mentioned above, the C-terminal carboxylate group of Ala-53 coordinates the zinc atom in a bidentate form (distances of 2.28 and 2.44 Å, respectively), whereas the amino group of Ala-53 forms a hydrogen bond with the hydroxyl side chain oxygen of Tyr-248 (2.78 Å), which is disposed in the down or “closed” conformation (when bound to substrates). The S1 subsite in the enzyme is also composed of Glu-270 and Arg-127, both of which presumably participate in the polarization of the carbonyl group of the scissile peptide bond and in proton exchange (1). In addition to coordination of the zinc atom, each of the carboxylate C-terminal oxygens of P1 Ala-53 is at hydrogen bonding distance with Glu-270 and Arg-127 (2.54 and 2.79 Å, respectively).
The S2 subsite of the enzyme, which is believed to contribute to the correct orientation of substrates during the catalytic reaction, is formed mainly by Glu-163 and Arg-71, which establish hydrogen bonds (at 3.08 and 3.02 Å, respectively) with the backbone of Tyr-52 in NvCI (Fig. 2A). In addition, the side chain of Tyr-52 is placed in a hydrophobic pocket created by Tyr-248 and Val-164 from hCPA4 and internally with Pro-16 from NvCI.
Interestingly, the side chain of Glu-163 in hCPA4 forms favorable hydrogen bond contacts with the amino groups of Cys-51 and Tyr-52 of NvCI (3.08 and 3.06 Å, respectively). Such involvement of the S3 subsite through Glu-163 interaction is novel and has not been observed in the other reported structures of complexes with carboxypeptidase inhibitors; it can be ascribed to a different backbone orientation of the P3 residue (Cys-51 in NvCI) regarding other inhibitors of MCPs. In NvCI, the hydrogen bonding network created by the C-terminal tail in complex with the active site of hCPA4 is more extended and contain two extra bonds.
NvCI displays the strongest inhibitory constants (in the picomolar range) against most CPA-type forms: bCPA1, hCPA1, and hCPA4 with Ki values of 5.8, 1.2, and 4.9 pm, respectively. An exception is hCPA2, for which the Ki value is in the nanomolar range (Table 3). In such an enzyme variant, a major structural difference in the contact residues of the primary interaction site is the substitution of Glu-163 with Asp (9), which presumably is too distant to form any hydrogen bond (Fig. 2D). This fact suggests an explanation for the nanomolar value of Ki displayed by NvCI against hCPA2 (similar to the other protein carboxypeptidase inhibitors (PCI, LCI, ACI, and TCI)) instead of the picomolar range of Ki displayed against all other A-type carboxypeptidases with a glutamic acid in that position.
TABLE 3.
Ki values for recombinant NvCI against several MCPs
Data are means ± S.D. (n = 3).
| Enzyme | Ki |
|---|---|
| pm | |
| bCPA1 | 5.8 ± 0.2 |
| hCPA1 | 1.2 ± 0.1 |
| hCPA2 | 2941.0 ± 132.4 |
| hCPA4 | 4.9 ± 0.6 |
In summary, most of the active site residues in hCPA4 involved in substrate binding and catalysis are in contact with NvCI. The interaction with the enzyme and the conformation of the C-terminal tail in NvCI (composed only of Tyr-52 and Ala-53 (P2 and P1 positions, respectively)) are similar to those of other known exogenous protein carboxypeptidase inhibitors (PCI, LCI, TCI, and ACI), in which the inhibitor tail mimics substrate binding (see Fig. 2, B and C, for comparison). All of the three-dimensional protein structures of these inhibitors from evolutionarily distant organisms are unrelated and completely different (see Fig. 4 and supplemental Fig. S1). However, the only conserved motif in all of them is the structural conformation of the P1 and P2 residues of the C-terminal tail, which can be considered as a general feature, derived from a common functional mechanism for carboxypeptidase inhibition, and as a clear example of convergent evolution. As an exception, a relevant structural difference in the NvCI-hCPA4 complex is the main chain conformation of the P3 residue, which favors the formation of two extra hydrogen bonds with Glu-163 and presumably induces a reduction of the inhibition constant by stabilization of product formation (Fig. 2D).
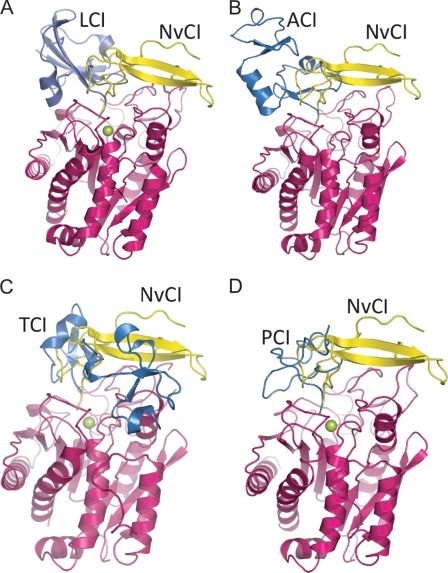
FIGURE 4.
Three-dimensional structure comparison of several complexes between exogenous proteinaceous carboxypeptidase inhibitors. The structures of different carboxypeptidases were overlapped with the NvCI-CPA4 complex. A, LCI, NvCI, and hCPA2 are shown in blue, yellow, and magenta, respectively (Protein Data Bank code 1DTD). B, ACI, NvCI, and hCPA1 are shown in blue, yellow, and magenta, respectively (code 3FJU). C, TCI, NvCI, and human carboxypeptidase B are presented in blue, yellow, and magenta, respectively (code 1ZLH). D, PCI, NvCI, and bCPA1 are shown in blue, yellow, and magenta, respectively (code 4CPA). The active site zinc is presented as a green sphere.
Secondary Interaction Site in NvCI-hCPA4
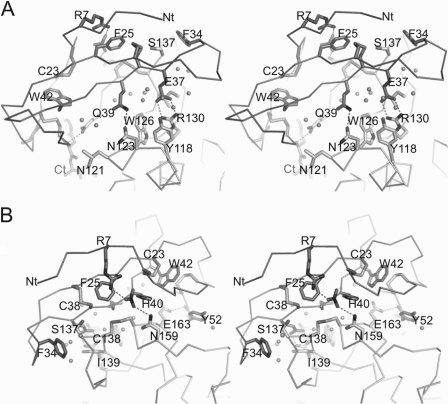
The secondary interaction site of NvCI is composed mainly of contacts between residues from the two-stranded β-sheet and regions distant from the active site groove of hCPA4 (Fig. 3). As also suggested for other carboxypeptidase inhibitors, the secondary interaction site contributes substantially to the decrease in the inhibition constant (9, 10). The contacts are in large number and varied and fit with the unusual strong inhibitory power of NvCI against most CPA-like enzymes. Besides several van der Waals interactions, there are major specific polar contacts between the backbone and side chains of NvCI with hCPA4 (Table 2). Of special interest are the specific hydrogen bonds between the side chains of Gln-39 and Asn-123 (2.99 Å), Glu-37 and Arg-130 (2.86 and 2.86 Å between O1 and O2 with the NH1 and NH2 atoms, respectively), and Arg-7 and Asn-159, with a second nitrate molecule bridging both through two hydrogen bonds (2.77 and 2.79 Å, respectively) (Fig. 3). Four hydrogen bonds involving main chain atoms are also formed: between the carbonyl oxygens of Ile-10 and Asp-11 and the side chain of Asn-159 (3.30 and 3.71 Å, respectively), between the amide hydrogen of Cys-38 and the carbonyl group of Ser-137 (3.19 A), and between the amide hydrogen of His-40 and the carbonyl group of Cys-161 (3.43 Å). A small hydrophobic core can also be distinguished in the secondary interface of NvCI with hCPA4, basically formed by Phe-25 of the inhibitor, which is buried in a pocket created by two disulfide bridges, Cys-27 and Cys-38 of NvCI and Cys-138 and Cys-161 of hCPA4.
FIGURE 3.
Close-up view of NvCI secondary binding region in complex with hCPA4. A, close-up stereo view in stick representation of the secondary binding region of NvCI with the surface of hCPA4. Amino acid residues corresponding to NvCI and hCPA4 are labeled and shown in different gray tones, respectively. The nitrate molecule is also shown in stick representation. Water molecules are shown as gray spheres. The N (Nt) and C (Ct) termini are labeled. B, same as in A but with a rotation of the complex of ∼180°, depicting the opposite face of the complex.
Structural comparisons indicated that the conformation of the residues of the enzyme (hCPA4 in this case) forming the secondary interaction surface is highly conserved in all A/B-type carboxypeptidases (M14A subfamily). This is also the case for the complex with NvCI, in particular considering the conservation and similar orientation of the specific side chain contacts described above, such as for Glu-37 and Gln-39. Thus, the lower Ki values observed for NvCI regarding other MCP inhibitors can be attributed to both the primary and secondary interaction regions, which create an extended interface with the carboxypeptidase enzyme that minimizes the product release of the catalytic reaction.
DISCUSSION
The few reports that appeared in the last decade on the structure-function relationships of MCP inhibitors of exogenous origin (4–7), after the initial one from potatoes (8, 29), indicated that they share a similar substrate-like mechanism of inhibition. Thus, they suffer a trimming of their C termini, behaving as competitive tight-binding inhibitors, mimicking the interaction of a peptide substrate with the active site of the enzyme, and requiring only the S1 and S2 subsites to be covered to fully inhibit the enzyme. Thus, even though these exogenous MCP inhibitors are isolated from evolutionarily distant organisms, this is a good example of convergent evolution dictated by the architecture of the active site of the enzyme. However, the structure of NvCI with hCPA4 indicates that the trimming action is absent (i.e. a shorter two-residue tail is sufficient) and that the S3 subsite is also implicated, in addition to S1 and S2, in promoting stronger inhibition in the picomolar range.
In addition to this primary and essential C-terminal interaction, MCP inhibitors possess a second interface (distant from active site) that covers different regions of the carboxypeptidase and confers stability to the complex formation (Fig. 4). The NvCI interface with hCPA4 is quite extended in comparison with the other known inhibitors, covering a total of 1875 Å2. In other known cases, for the protein inhibitors from leech (LCI), potato (PCI), and the Ascaris worm (ACI), the interface of CPA-like enzymes is clearly smaller: 1509, 1241, and 1426 Å2, respectively. An exception would be the two-domain (exosite-behaving) tick carboxypeptidase inhibitor (TCI), which is the largest and covers a total interface of 2108 Å2. Although the secondary interaction surface provided by all inhibitors is distinct, there is only a single way of positioning the C-terminal tail in the active site of the enzyme.
As mentioned above, the main body of each of the exogenous carboxypeptidase inhibitors has a completely different three-dimensional structure, with the C-terminal tail as the only similar structural motif (Fig. 2, B and C, and Fig. 4). The short C-terminal tail of NvCI, considered from the third disulfide bridge and formed by only two residues, Tyr-52 and Ala-53, is sufficient for a tight-binding inhibition of several types of carboxypeptidases. Interestingly, as mentioned above, in NvCI, the P3 position (Cys-51) also participates in the binding with a double main chain hydrogen bond with Glu-163 of hCPA4, presumably increasing the affinity and lowering the Ki value for NvCI, which is in the picomolar range, 3 orders of magnitude lower than the other inhibitors. This fact suggests a reason for the strongest inhibitor being NvCI in comparison with the other described proteinaceous inhibitors of MCPs.
The different ways that nature acts along evolution to inhibit carboxypeptidases can be used as a valuable tool to elucidate the determinants for a general mechanism of inhibition of MCPs. In general, these proteases are secreted and their enzymatic action takes place normally in the extracellular space, except in the case of a novel subfamily of cytosolic carboxypeptidases, which have been recently described and are presumably involved in tubulin processing (30, 31). Knowledge of the control of interference with those mechanisms, by rational structure-based or other drug design approaches, can be potentially of great interest, especially for biotechnical and biomedical industrial purposes. In addition, the expansion of this strategy to the variety of forms found in the very diverse phyla of invertebrates, including marine organisms such as N. versicolor, is revealing a rich source of discovery for novel scaffolds and lead compounds.
Supplementary Material
This work was in part supported by Grants BFU2008-0364 and BIO2010-22321-C02 from the Ministerio de Ciencia e Innovacion of Spain and by Grants MIRG-CT-2007-200346 and LIVIMODE 241919 (FP7-Health-2009) from the European Community.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (code 4A94) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MCP
- metallocarboxypeptidase
- NvCI
- N. versicolor carboxypeptidase inhibitor
- bCPA1
- bovine carboxypeptidase A1
- hCPA1
- human CPA1
- PCI
- potato carboxypeptidase inhibitor
- LCI
- leech carboxypeptidase inhibitor
- TCI
- tick carboxypeptidase inhibitor
- ACI
- Ascaris carboxypeptidase inhibitor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Vendrell J., Avilés F. X., Fricker L. D. (2004) in Handbook of Metalloproteinases (Messerschimidt A., Bode W., Cygler M., eds) Vol. 3, pp. 176–189, John Wiley & Sons, Inc., New York [Google Scholar]
- 2. Arolas J. L., Vendrell J., Avilés F. X., Fricker L. D. (2007) Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr. Pharm. Des. 13, 349–366 [DOI] [PubMed] [Google Scholar]
- 3. Fernández D., Testero S., Vendrell J., Avilés F. X., Mobashery S. (2010) The X-ray structure of carboxypeptidase A inhibited by a thiirane mechanism-based inhibitor. Chem. Biol. Drug Des. 75, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hass G. M., Nau H., Biemann K., Grahn D. T., Ericsson L. H., Neurath H. (1975) The amino acid sequence of a carboxypeptidase inhibitor from potatoes. Biochemistry 14, 1334–1342 [DOI] [PubMed] [Google Scholar]
- 5. Reverter D., Vendrell J., Canals F., Horstmann J., Avilés F. X., Fritz H., Sommerhoff C. P. (1998) A carboxypeptidase inhibitor from the medical leech Hirudo medicinalis. Isolation, sequence analysis, cDNA cloning, recombinant expression, and characterization. J. Biol. Chem. 273, 32927–32933 [DOI] [PubMed] [Google Scholar]
- 6. Arolas J. L., Lorenzo J., Rovira A., Castellà J., Avilés F. X., Sommerhoff C. P. (2005) A carboxypeptidase inhibitor from the tick Rhipicephalus bursa: isolation, cDNA cloning, recombinant expression, and characterization. J. Biol. Chem. 280, 3441–3448 [DOI] [PubMed] [Google Scholar]
- 7. Sanglas L., Avilés F. X., Huber R., Gomis-Rüth F. X., Arolas J. L. (2009) Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite. Proc. Natl. Acad. Sci. U.S.A. 106, 1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rees D. C., Lipscomb W. N. (1982) Refined crystal structure of the potato inhibitor complex of carboxypeptidase A at 2.5 Å resolution. J. Mol. Biol. 160, 475–498 [DOI] [PubMed] [Google Scholar]
- 9. Reverter D., Fernández-Catalán C., Baumgartner R., Pfänder R., Huber R., Bode W., Vendrell J., Holak T. A., Avilés F. X. (2000) Structure of a novel leech carboxypeptidase inhibitor determined free in solution and in complex with human carboxypeptidase A2. Nat. Struct. Biol. 7, 322–328 [DOI] [PubMed] [Google Scholar]
- 10. Arolas J. L., Popowicz G. M., Lorenzo J., Sommerhoff C. P., Huber R., Avilés F. X., Holak T. A. (2005) The three-dimensional structures of tick carboxypeptidase inhibitor in complex with A/B carboxypeptidases reveal a novel double-headed binding mode. J. Mol. Biol. 350, 489–498 [DOI] [PubMed] [Google Scholar]
- 11. Pallarès I., Bonet R., García-Castellanos R., Ventura S., Avilés F. X., Vendrell J., Gomis-Rüth F. X. (2005) Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proc. Natl. Acad. Sci. U.S.A. 102, 3978–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delfín J., Martínez I., Antuch W., Morera V., González Y., Rodríguez R., Márquez M., Saroyán A., Larionova N., Díaz J., Padrón G., Chávez M. (1996) Purification, characterization, and immobilization of proteinase inhibitors from Stichodactyla helianthus. Toxicon 34, 1367–1376 [DOI] [PubMed] [Google Scholar]
- 13. Lenarcic B., Ritonja A., Strukelj B., Turk B., Turk V. (1997) Equistatin, a new inhibitor of cysteine proteinases from Actinia equina, is structurally related to thyroglobulin type 1 domain. J. Biol. Chem. 272, 13899–13903 [DOI] [PubMed] [Google Scholar]
- 14. González Y., Pons T., Gil J., Besada V., Alonso del Rivero M., Tanaka A. S., Araujo M. S., Chávez M. A. (2007) Characterization and comparative 3D modeling of CmPI-II, a novel “non-classical” Kazal-type inhibitor from the marine snail Cenchritis muricatus (Mollusca). Biol. Chem. 388, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 15. Reynolds D. S., Gurley D. S., Stevens R. L., Sugarbaker D. J., Austen K. F., Serafin W. E. (1989) Cloning of cDNAs that encode human mast cell carboxypeptidase A and comparison of the protein with mouse mast cell carboxypeptidase A and rat pancreatic carboxypeptidases. Proc. Natl. Acad. Sci. U.S.A. 86, 9480–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanco S., Zhang X., Morano C., Avilés F. X., Lorenzo J., Fricker L. D. (2010) Characterization of the substrate specificity of human carboxypeptidase A4 and implications for a role in extracellular peptide processing. J. Biol. Chem. 285, 18385–18396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H., Reed C. P., Zhang J. S., Shridhar V., Wang L., Smith D. I. (1999) Carboxypeptidase A3 (CPA3): a novel gene highly induced by histone deacetylase inhibitors during differentiation of prostate epithelial cancer cells. Cancer Res. 59, 2981–2988 [PubMed] [Google Scholar]
- 18. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 19. Mock W. L., Liu Y., Stanford D. J. (1996) Arazoformyl peptide surrogates as spectrophotometric kinetic assay substrates for carboxypeptidase A. Anal. Biochem. 239, 218–222 [DOI] [PubMed] [Google Scholar]
- 20. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 25. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morrison J. F. (1982) The slow-binding and slow tight-binding inhibition of enzyme-catalyzed reactions. The slow-binding and slow, tight-binding inhibition of enzyme-catalyzed reactions. Trends Biochem. Sci. 7, 102–105 [Google Scholar]
- 27. Pallarès I., Fernández D., Comellas-Bigler M., Fernández-Recio J., Ventura S., Avilés F. X., Bode W., Vendrell J. (2008) Direct interaction between a human digestive protease and the mucoadhesive poly(acrylic acid). Acta Crystallogr. D Biol. Crystallogr. 64, 784–791 [DOI] [PubMed] [Google Scholar]
- 28. Reverter D., Ventura S., Villegas V., Vendrell J., Avilés F. X. (1998) Overexpression of human procarboxypeptidase A2 in Pichia pastoris and detailed characterization of its activation pathway. J. Biol. Chem. 273, 3535–3541 [DOI] [PubMed] [Google Scholar]
- 29. Marino-Buslje C., Venhudová G., Molina M. A., Oliva B., Jorba X., Canals F., Avilés F. X., Querol E. (2000) Contribution of C-tail residues of potato carboxypeptidase inhibitor to the binding to carboxypeptidase A. A mutagenesis analysis. Eur. J. Biochem. 267, 1502–1509 [DOI] [PubMed] [Google Scholar]
- 30. Rodríguez de la Vega M., Sevilla R. G., Hermoso A., Lorenzo J., Tanco S., Diez A., Fricker L. D., Bautista J. M., Avilés F. X. (2007) Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB J. 21, 851–865 [DOI] [PubMed] [Google Scholar]
- 31. Kalinina E., Biswas R., Berezniuk I., Hermoso A., Avilés F. X., Fricker L. D. (2007) A novel subfamily of mouse cytosolic carboxypeptidases. FASEB J. 21, 836–850 [DOI] [PubMed] [Google Scholar]
- 32. Delano W. L. (2002) The PyMOL Molecular Graphics System, Version 0_99rc6, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.