Background: Urea carboxylase (UC) plays an essential role in urea utilization and belongs to the biotin-dependent carboxylase superfamily.
Results: We report the crystal structure of the Kluyveromyces lactis UC.
Conclusion: The structure provides insights into the UC carboxyltransfer reaction.
Significance: Our study sheds light on the carboxyltransfer catalysis of biotin-dependent carboxylases in general and the function of the KipA-KipI complex involved in sporulation regulation.
Keywords: Biotin, Carboxylation, Enzyme Kinetics, Enzyme Structure, X-ray Crystallography, Biotin-dependent Carboxylase, KipA-KipI Complex, Urea Amidolyase, Urea Carboxylase
Abstract
Urea carboxylase (UC) is conserved in many bacteria, algae, and fungi and catalyzes the conversion of urea to allophanate, an essential step in the utilization of urea as a nitrogen source in these organisms. UC belongs to the biotin-dependent carboxylase superfamily and shares the biotin carboxylase (BC) and biotin carboxyl carrier protein (BCCP) domains with these other enzymes, but its carboxyltransferase (CT) domain is distinct. Currently, there is no information on the molecular basis of catalysis by UC. We report here the crystal structure of the Kluyveromyces lactis UC and biochemical studies to assess the structural information. Structural and sequence analyses indicate the CT domain of UC belongs to a large family of proteins with diverse functions, including the Bacillus subtilis KipA-KipI complex, which has important functions in sporulation regulation. A structure of the KipA-KipI complex is not currently available, and our structure provides a framework to understand the function of this complex. Most interestingly, in the structure the CT domain interacts with the BCCP domain, with biotin and a urea molecule bound at its active site. This structural information and our follow-up biochemical experiments provided molecular insights into the UC carboxyltransfer reaction. Several structural elements important for the UC carboxyltransfer reaction are found in other biotin-dependent carboxylases and might be conserved within this family, and our data could shed light on the mechanism of catalysis of these enzymes.
Introduction
In most living organisms, urea is generated in the degradation of nitrogen-containing molecules. In mammals, urea is excreted and is the main nitrogen-containing compound in the urine. Plants, fungi, algae, and bacteria possess an enzymatic activity that converts urea to ammonium, enabling them to utilize urea as a nitrogen source (1–3). In agriculture, urea is widely used as a fertilizer. In the digestive tracts of many animals, the residing microbes utilize urea from the host to produce biomass, which contributes to the host's nutrient intake (1, 4). Two distinct enzymes catalyze the conversion of urea to ammonium: the nickel-containing enzyme urease and the biotin-dependent enzyme urea amidolyase.
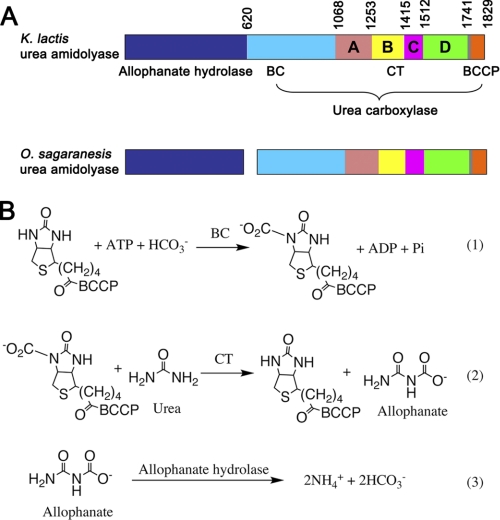
Urea amidolyase is composed of urea carboxylase (UC)2 and allophanate hydrolase (AH) activities (Fig. 1A). UC coverts urea to allophanate, and AH subsequently converts it to ammonium (Fig. 1B). In some organisms, UC and AH are encoded as separate polypeptides (5–8). Urea amidolyase has been identified in fungi (9, 10), algae (11), and bacteria (5, 6); recent studies revealed that it is widely distributed in fungi and bacteria (7, 8). In many of these organisms urease is not found. By adopting urea amidolyase for their urea utilization, they avoid the transition metal nickel, whose cellular level has to be tightly regulated (7). In addition to urea utilization, urea amidolyase is involved in a number of other processes. It is an essential component of a pyrimidine nucleic acid precursor degradation pathway (12). In the human pathogen Candida albicans it is required for the yeast-hyphal transition, an important mechanism to escape the host defense (13, 14).
FIGURE 1.
Domain organization of urea amidolyase and reactions catalyzed by it. A, domain organization of urea amidolyase. The AH component is colored in blue. The UC component is colored in cyan and orange for the BC and BCCP domains, respectively. The CT subdomains are colored in salmon (A), yellow (B), magenta (C), and green (D). The CT-BCCP linker is colored in gray. This color scheme is used throughout the paper. Domain boundaries are indicated for the K. lactis urea amidolyase. B, reactions catalyzed by urea amidolyase. Reactions 1 and 2 are catalyzed by the UC BC and CT domains, respectively. Reaction 3 is catalyzed by AH.
UC catalyzes a two-step, ATP- and biotin-dependent carboxylation reaction of urea. It is composed of biotin carboxylase (BC), carboxyltransferase (CT), and biotin carboxyl carrier protein (BCCP) domains (Fig. 1A). Biotin is covalent linked to a conserved lysine residue in the BCCP domain. The BC domain carboxylates biotin with concomitant ATP cleavage, and the CT domain subsequently transfers the carboxyl group from biotin to urea, generating allophanate (Fig. 1B). The reaction mechanism and domain organization share common features with other biotin-dependent carboxylases, including pyruvate carboxylase (PC), propionyl-CoA carboxylase (PCC), acetyl-CoA carboxylase (ACC) and 3-methylcrotonyl-CoA carboxylase. These enzymes catalyze a variety of carboxylation reactions, playing important roles in many pathways (15, 16). Like UC, they contain BC, CT, and BCCP domains and catalyze two-step carboxylation reactions (16, 17). The BC and BCCP domains of biotin-dependent carboxylases are highly homologous, but their CT domains are distinct. Recently, high resolution crystal structures of the PC (18, 19) and PCC (20) holoenzymes have been reported, which greatly advanced the understanding of biotin-dependent carboxylases. However, the molecular mechanism for how UC recognizes the urea substrate and catalyzes its carboxylation is currently not known.
We report here the crystal structure at 2.6 Å resolution of the Kluyveromyces lactis urea amidolyase UC domain (KlUC). Structural and sequence analysis indicate that its CT domain belongs to a large family of proteins with diverse functions, including the KipA-KipI complex, which has important regulatory functions in the Bacillus subtilis sporulation (21). A structure of the KipA-KipI complex is not currently available, and the UC structure provides a framework to understand the function of this complex. Most interestingly, the current UC structure captured a carboxyltransfer reaction intermediate, in which the BCCP domain interacts with the CT domain and delivers biotin into its active site. This structural information and our follow-up biochemical experiments provide molecular insights into the UC carboxyltransfer reaction. Analyses suggest that other biotin-dependent carboxylases might adopt similar modes of biotin coordination and CT-BCCP interactions in their carboxyltransfer reaction, and our data could shed light on the mechanism of the catalysis of these enzymes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The UC domain of the K. lactis urea amidolyase (KlUC, residues 617–1829) was cloned into vector pET28a (Novagen). The resulting recombinant protein contains a His6 tag and a thrombin cleavage sequence at its N terminus. The plasmid was co-transformed with a pTYB12 (New England Biolabs)-based plasmid, containing the Escherichia coli biotin ligase gene birA, into the E. coli strain BL21 Star (DE3). Cells were cultured in the Luria broth supplemented with 50 mg/liter kanamycin, 100 mg/liter ampicillin and were induced with 0.3 mm isopropyl-β-d-thiogalactopyranoside (Bio Basic Inc.), at 20 °C for 12–16 h. 30 min before the induction, 15 mg/liter d-biotin was added to the medium. Cells were harvested by centrifugation and lysed by sonification. KlUC was purified with nickel-nitrilotriacetic acid-agarose (Qiagen) and size exclusion (Sephacryl S300 HR; GE Healthcare) columns. Purified KlUC, in a buffer containing 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, 2 mm dithiothreitol (DTT), and 5% (v/v) glycerol, was concentrated to 10 mg/ml, flash-cooled in liquid nitrogen, and stored at −80 °C.
The selenomethionine (SeMet)-substituted protein was produced by growing cells in the M9 medium with specific amino acids to block methionine synthesis and supplementing with SeMet (22). Its purification was the same as the native protein, except that the DTT concentration was increased to 10 mm.
Protein Crystallization
Rod-shaped crystals of KlUC were obtained with the vapor diffusion sitting-drop method at 20 °C. The reservoir liquid contains 2.4 m ammonium sulfate and 0.1 m BisTris, pH 7.0. Before crystallization setups, urea and ADP (2 mm each) were added to the protein solution. Crystallization of the SeMet-substituted protein was the same as the native protein, except that 10 mm EDTA (Hampton Research) was added to the protein solution before crystallization setups. Crystals were cryoprotected by equilibrating in the reservoir liquid supplemented with 20% v/v glycerol, flash cooled, and stored in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Data collection was performed at 100 K. Diffraction data were collected on an ADSC Q315 CCD detector at the Shanghai Synchrotron Radiation Facility beamline BL17U. A native data set was collected at 0.979 Å. A single wavelength anomalous diffraction data set was collected at the selenium absorption edge on a SeMet-substituted crystal. Diffraction data were processed with MOSFLM (23), integrated with SCALA (24), and the intensities were converted to structure factors with CTRUNCATE (25).
Molecular replacement calculations by MOLREP (26), with the structure of the Staphyloccus aureus ACC BC subunit (Protein Data Bank (PDB) code 2VPQ, 43% sequence identity to the KlUC BC domain) (27) as the search model, identified a solution. The initial phase information was improved by incorporating the anomalous difference signal from the single wavelength anomalous diffraction data set, with Phaser (28). The electron density map was further improved by solvent flattening and histogram averaging with DM (29). The resulting electron density map allowed manual building of the atomic structure, carried out with O (30) and Coot (31). The structure was refined with CNS (32) and REFMAC (33).
Data processing and refinement statistics are summarized in Table 1. MOSFLM, SCALA, CTRUNCATE, Phaser, DM, Coot, and REFMAC are programs in the CCP4 software suite (34).
TABLE 1.
Data collection and refinement statistics
Numbers enclosed in parentheses are for the highest resolution shells.
| Crystal parameters | |
| Space group | P43212 |
| Cell dimensions | |
| a, b, c (Å) | 126.7, 126.7, 217.9 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Data collection | |
| Wavelength | 0.979 |
| Resolution (Å) | 50.0-2.6 (2.74-2.60) |
| Rmerge (%) | 9.2 (38.8) |
| I/σI | 6.1 (1.9) |
| Completeness (%) | 99.4 (99.4) |
| Redundancy | 4.7 (4.6) |
| Refinement | |
| Resolution (Å) | 50.0-2.6 (2.67-2.60) |
| No. reflections | 51,949 (3,967) |
| Rwork/Rfree (%) | 18.9 (26.8)/26.0 (33.6) |
| No. atoms | |
| Protein | 8,837 |
| Ligand/ion | 19 |
| Water | 444 |
| B-factors | |
| Protein | 40.6 |
| Ligand/ion | 27.6 |
| Water | 40.5 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.020 |
| Bond angles (°) | 1.991 |
Mutagenesis and Activity Assays
Mutagenesis was carried out with the QuikChange kit and verified by DNA sequencing. The mutants were expressed and purified the same as the wild-type protein. For activity assays, the AH domain of the K. lactis urea amidolyase (residues 1–621, KlAH) was cloned into vector pET24d (Novagen) and purified with nickel-nitrilotriacetic acid-agarose and size exclusion columns, same as KlUC.
Enzyme activity was measured by coupling ammonium generation of urea amidolyase to NADH to NAD+ conversion with glutamate dehydrogenase (35). Urea amidolyase activity was reconstituted by supplementing KlUC with KlAH. Activity assays were carried out at room temperature. Absorption change at 340 nm resulting from the NADH to NAD+ conversion was monitored on an ultraspec 2100 pro spectrophotometer (GE Healthcare). The reaction mixture contained 200 mm Tris-HCl, pH 8.0, 1 μm EGTA, 3 mm magnesium chloride, 19.5 mm potassium chloride, 200 units/ml glutamate dehydrogenase (Wako), 50 mm oxoglutaric acid, 0.3 mm NADH, 1.5 μm KlUC, 3.3 μm KlAH, 20 mm ATP, 200 mm sodium bicarbonate and variable concentrations of urea. For mutants with minimal activity, assays were repeated with higher enzyme concentrations. 15 μm KlUC and 33 μm KlAH, and 30 μm KlUC and 66 μm KlAH were used for mutants D1321A and K1605A, respectively.
Dynamic Light Scattering Experiments
Dynamic light scattering experiments were performed at 20 °C on a DynaPro Titan dynamic light scattering instrument (Wyatt Technologies). KlUC was measured at a concentration of 5 mg/ml in a buffer containing 20 mm Tris-HCl, pH 7.5, 200 mm NaCl, and 1 mm DTT. Data were collected and analyzed with the DYNAMICS V6 software (Wyatt Technologies).
RESULTS AND DISCUSSION
Structure Determination
UC from different organisms are highly conserved (supplemental Fig. S1). After screening through a number of organisms, we purified and crystallized KlUC (residues 617–1829 of the K. lactis urea amidolyase). The crystals belong to space group P43212, and contain one KlUC molecule in the asymmetric unit. The structure was determined with a combination of molecular replacement and single-wavelength anomalous scattering methods and was refined to a resolution of 2.6 Å. The refined structure agrees well with crystallographic data and expected geometric values (Table 1), with most of the residues (87.3%) in the most favored region of the Ramachandran plot and 11.4% in the additionally allowed region.
Overall Structure
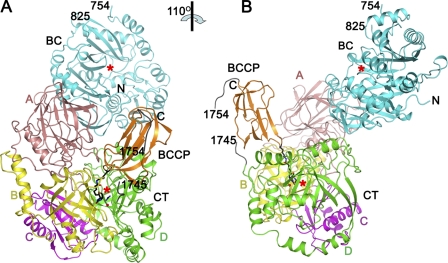
The KlUC monomer adopts an elongated structure. The CT domain (residues 1069–1741) is located at the bottom, with the BC (residues 621–1068) and BCCP (residues 1760–1829) domains on top of it. In the observed conformation interactions are found between the BC and the CT domains, and the BCCP and the CT domains, but not between the BC and the BCCP domains (Fig. 2 and supplemental Fig. S2). The BC B subdomain (36) (residues 755–824) and part of the CT-BCCP linker (residues 1742–1759) are poorly defined in the electron density map and were not included in the model. Urea and ADP (2 mm each) were included in the crystallization setups. A urea molecule was found at the CT domain active site, but no electron densities for ADP were observed.
FIGURE 2.
Overall structure of KlUC. A and B are roughly related by a 110° rotation along the vertical axis. The biotinylated lysine (Lys-1795), biotin, the urea, and water molecules found at the CT domain active site are shown in stick and ball representations. The red stars indicate active sites of the BC and the CT domains. Structural figures were produced with PyMOL.
In contrast to PC (18, 19) and PCC (20), which form oligomeric holoenzymes, the functional unit of KlUC appears to be the monomer. No substantial intermolecular interactions are observed in the crystal. In our gel filtration experiments the elution volume of KlUC corresponded to a monomeric form (supplemental Fig. S8, B and C), consistent with a molecular mass of 141 kDa measured by dynamic light scattering experiments (the molecular mass of the KlUC monomer deduced from its amino acid composition is 137 kDa). Our biochemical studies indicated that this monomeric enzyme is active. Its Km for urea is comparable with that reported for the Candida utilis urea amidolyase (9), and the kinetic parameters indicate that it is more active than the Oleomonas sagaranensis UC (5) (Table 2).
TABLE 2.
Summary of the KlUC kinetic parameters
Numbers in the parentheses are ratios to the wild-type values. S.D. values were obtained from fitting the experimental data to the Michaelis-Menten equation.
| Mutation | kcat | Km for urea | kcat/Km |
|---|---|---|---|
| s−1 | mm | s−1mm−1 | |
| Wild type | 210.8 ± 4.0 (1.0) | 0.32 ± 0.03 (1.0) | 659 ± 74 (1.0) |
| D1321A | 3.16 ± 0.20 (0.015) | 78 ± 20 (2.4×102) | 0.041 ± 0.013 (6.2×10−5) |
| Y1324F | 45.8 ± 1.2 (0.22) | 7.7 ± 1.0 (24) | 5.9 ± 0.9 (9.0×10−3) |
| D1584N | 61.8 ± 1.8 (0.29) | 11.3 ± 1.4 (35) | 5.5 ± 0.8 (8.3×10−3) |
| K1605A | 1.44 ± 0.06 (0.0068) | 493 ± 93 (1.5 ×103) | 0.0029 ± 0.0007 (4.4×10−6) |
| Y1628F | 64.9 ± 1.6 (0.31) | 15.0 ± 1.7 (47) | 4.3 ± 0.6 (6.5×10−3) |
| E1792A | 68.7 ± 1.2 (0.33) | 0.39 ± 0.03 (1.2) | 176 ± 17 (0.27) |
In the structure, the BC and CT domain active sites face each other and are separated by a distance of 60 Å (Fig. 2 and supplemental Fig. S2). This distance is comparable with that reported for PC (18, 19) and PCC (20) and is consistent with a model that the entire BCCP domain translocates during the catalysis. The CT-BCCP linker is partially disordered, suggesting it is flexible in nature, which could facilitate the translocation of the BCCP domain.
The CT Domain Belongs to a Large Family of Proteins with Diverse Functions
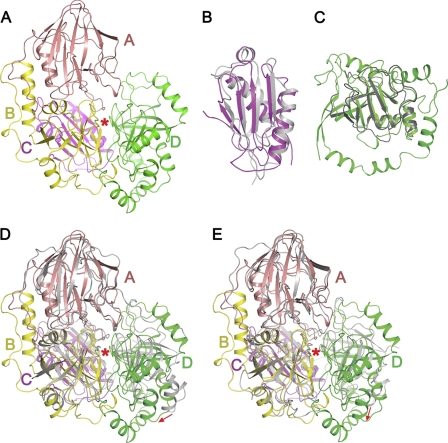
The UC CT domain does not share any sequence homology with other biotin-dependent carboxylases. Consistently, its structure is distinct from the CT domains of other biotin-dependent carboxylases (Fig. 3A). It is composed of four subdomains: A (residues 1069–1253), B (residues 1254–1415), C (residues 1416–1512), and D (residues 1513–1741). Subdomain A is a β sandwich with an α helix on one face (supplemental Fig. S3A). Subdomain C contains a five-strand antiparallel β sheet, and two α helices at one side (supplemental Fig. S3C). Both subdomains B and D adopt the cyclophilin fold (37) (supplemental Fig. S3, B and D) and can be superimposed with a r.m.s. deviation for related Cα atoms of 1.9 Å (supplemental Fig. S3E), but the sequence conservation between them is not obvious. Subdomains B, C, and D form a tight bundle. Subdomain A is located on top of the bundle, interacting mainly with subdomain B. In the structure biotin and a urea molecule are bound at the cleft between subdomains B and D (Fig. 3A).
FIGURE 3.
Structure of the CT domain and its homologues. A, structure of the CT domain. Subdomains are labeled, the red star indicate the cleft between subdomains B and D. B, structural superimposition of the CT subdomain C and the KipI N-terminal domain (PDB code 2KWA, shown in gray). C, structural superimposition of the CT subdomain D and the KipI C-terminal domain (PDB code 2ZP2, shown in gray). D and E, structural superimpositions of the CT domain and TTHA0988 (D) and Msmeg0435/0436 (E). TTHA0988 (PDB codes 3OPF and 3ORE) and Msmeg0435/0436 (PDB code 3MML) are shown in gray. CT subdomains A, B, and C and related regions on TTHA0988 and Msmeg0435/0436 are aligned; the red arrows indicate positional differences between CT subdomain D and its counterparts. The red stars indicate the cleft between subdomains B and D.
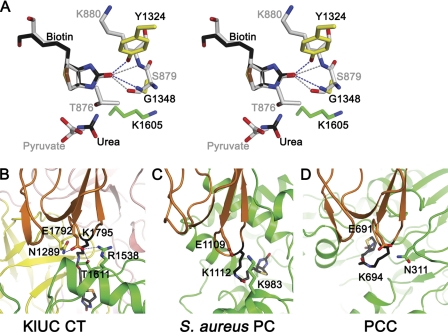
Structural and sequence analyses suggest that the CT domain of UC belongs to a large family of proteins with diverse functions. Most prominent among these homologues is the B. subtilis KipA-KipI complex, which has important regulatory functions in the sporulation process of this organism. KipI binds to the sporulation master kinase KinA and inhibits its autophosphorylation, whereas KipA interacts with KipI and prevents it from inhibiting KinA (21). KipA is equivalent to CT subdomains A and B, with sequence identity of 35%. KipI is equivalent to CT subdomains C and D, with sequence identity of 21%. Structures of the KipI N- (38) and C- (39) terminal domains have been determined. They can be superimposed onto the KlUC CT subdomains C and D, with r.m.s. deviations for related Cα atoms of 1.6 Å (Fig. 3, B and C). Interestingly, the operon encoding KipI and KipA is regulated by the availability of nitrogen, in a manner similar to that of urea amidolyase (21). However, residues that are important for the carboxyltransfer reaction in the UC CT domain are not conserved in the KipA-KipI complex (see below), and these proteins have entirely different functions.
Although structural information is currently available for the two domains of KipI, structures of KipA and the KipA-KipI complex are not known. The structure of the UC CT domain may provide a framework to understand the function of this complex. It has been proposed that KipI dissociates from KipA to bind and inhibit KinA (38), but the molecular mechanism of this process is currently unclear. In UC the cleft between CT subdomains B and D accommodates biotin and urea. These subdomains correspond to the C-terminal domains of KipA and KipI; therefore binding of small molecule (s) at the similar location in the KipA-KipI complex could modulate the KipA-KipI interaction and might induce the formation or dissociation of the complex. It remains to be seen whether the KipA-KipI complex is regulated this way. Such a process could enable the KipA-KipI complex to sense nutrient or signaling molecule levels and integrate this information into sporulation regulation.
Besides the KipA-KipI complex, the CT domain is also homologous to the Thermus thermophilus protein TTHA0988 (PDB codes 3OPF and 3ORE) (38, 40) and a heterodimeric protein complex Msmeg0435-Msmeg0436 from Mycobacterium smegmatis (PDB code 3MML, Msmeg0435/0436 hereafter). The r.m.s. distances for equivalent Cα atoms between the CT domain and these structures are 1.9 and 1.6 Å, respectively, and the sequence identities are ∼30%. Msmeg0435 is equivalent to CT subdomains C and D, whereas Msmeg0436 is equivalent to subdomains A and B. In contrast, the domain order of TTHA0988, C-D-A-B, is different compared with that of CT, suggesting distinct fusion events in the evolution of these proteins. The structural overlays also show that subdomain D can assume different positions in the three proteins (Fig. 3, D and E, and supplemental Figs. S4, A–D, and S5).
The functions of neither TTHA0988 nor Msmeg0435/0436 have been characterized. Both proteins are annotated as allophanate hydrolase but have no detectable sequence homology to the AH component of urea amidolyase. An attempt to demonstrate such an activity for TTHA0988 was unsuccessful (38, 40). UC CT domain residues important for the carboxyltransfer catalysis are not conserved in these proteins, and it is unlikely that they possess CT activities (see below). It remains to be seen whether they have functions similar to the KipA-KipI complex. It is also possible that they have yet unidentified, completely different activities.
Structure of the CT Domain Active Site Provides Insights into Carboxyltransfer Reaction
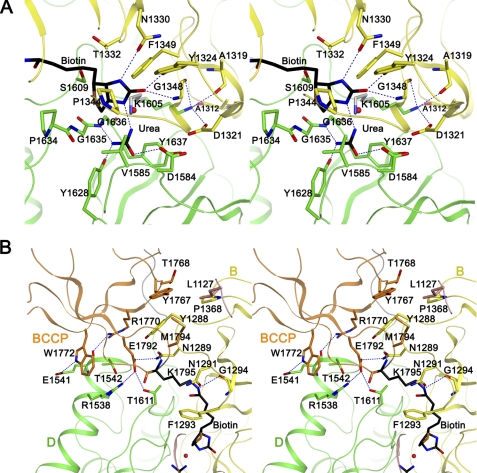
In the current KlUC structure, the biotinylated lysine (Lys-1795) side chain and the biotin valeric acid arm are fully extended, and the biotin head group (the tetrahydrothiophene and ureido rings) is bound at the cleft between subdomains B and D (Fig. 4A). The biotin group is well defined in the electron density map, a patch of electron density in the proximity of its N1′ nitrogen was modeled as urea and water molecules (supplemental Fig. S6). Conserved residues concentrate at the cleft, suggesting that it is the CT domain active site. The structural information thus provides molecular insights into the carboxyltransfer reaction in this unique CT domain.
FIGURE 4.
Molecular insights into the carboxyltransfer reaction. A, stereo view of the CT domain active site. Important residues are highlighted. Hydrogen bonds are indicated by dotted lines, the red sphere represents a water molecule found at the active site. The Lys-1605 side chain colored in gray represents a possible conformation mediating the proton transfer between urea and biotin. B, stereo view of the CT-BCCP interface. Residues contributing to the CT-BCCP interactions are highlighted.
Highly conserved residues interact with the biotin head group and the urea molecule. The biotin head group forms three hydrogen bonds with the CT domain. Its carbonyl oxygen hydrogen bonds with the Tyr-1324 side chain hydroxyl and the Gly-1348 main chain amide, its N3′ nitrogen hydrogen bonds with the Asn-1330 side chain carbonyl. The tetrahydrothiophene ring is located in a pocket formed by residues Pro-1344, Val-1585, Pro-1634, and Gly-1635, and the ureido ring forms van der Waals interactions with side chains of Thr-1332, Phe-1349, and Ser-1609. The urea molecule is sandwiched between side chains of Val-1585 and Tyr-1637. Its carbonyl oxygen hydrogen bonds with the Asp-1584 side chain carboxyl. One of the amine groups is within hydrogen bond distance with the Tyr-1628 side chain hydroxyl and the Gly-1636 main chain amide. The amine group to be carboxylated is located 3 Å away from the biotin N1′ nitrogen (Fig. 4A). An Asp-1584–Val-1585 cis-peptide bond is found at the active site.
It has been proposed that in the carboxyltransfer reaction, biotin-dependent carboxylases utilize a general base to extract a proton from the substrate atom to be carboxylated and transfer it to the biotin N1′ nitrogen (17) (Fig. S7). In PC, a conserved threonine (Thr-908 in the human PC, Thr-876 in the S. aureus PC, Thr-882 in the Rhizobium etli PC) plays such a role (41, 42). In the current UC structure, the urea amine group to be carboxylated and the biotin N1′ nitrogen are surrounded by side chains of Asp-1321, Asp-1584, Lys-1605, and Tyr-1637. Although none of these side chains is located close enough to mediate the proton transfer, our further considerations suggest that the general base is the strictly conserved Lys-1605. In the observed conformation, the Lys-1605 side chain forms an ion pair with the conserved Asp-1321 side chain; its amine group is also within hydrogen bond distances to the main chain carbonyls of Ala-1312, Ala-1319, and Gly-1348. In this conformation, its side chain amine is located 7 Å away from the urea amine group to be carboxylated and 8 Å away from the biotin N1′ nitrogen. However, a different Lys-1605 side chain rotamer could reduce these distances to 3 Å or less, facilitating the proton transfers (Fig. 4A).
To gain further insights into the carboxyltransfer reaction, we introduced point mutations in the relevant regions and compared the activities of the wild-type protein and the mutants. All of the mutants were fully biotinylated (supplemental Fig. S8A) and behaved identically to the wild-type protein on a gel filtration column (supplemental Fig. S8B), suggesting that any change in their activity is not due to insufficient biotinylation or protein misfolding. Mutating Lys-1605 to alanine greatly reduced the activity, so did substituting its ion pair partner Asp-1321 with alanine. Other mutations in the active site, Y1324F, D1584N, and Y1628F, decreased kcat 3–5-fold and increased Km for urea 20–50-fold (Table 2 and supplemental Fig. S9). These data are consistent with our mechanism of the UC carboxyltransfer catalysis. That the K1605A mutant retains minimal activity suggests that direct “unassisted” proton transfer between urea and biotin might also occur (43). Our model could also explain the importance of Asp-1321 observed in the activity assays. The observed conformation might mimic a carboxyltransfer reaction intermediate, in which Lys-1605 has extracted a proton from urea, but has not protonated biotin, and the extracted proton is stored in the Lys-1605–Asp-1321 ion pair (supplemental Fig. S7). However, having not directly observed the conformation in which the general base interacts with biotin and the urea molecule, our hypotheses for the functions of Lys-1605 and Asp-1321 remain speculative. Further experiments are required to fully elucidate the mechanism of the UC carboxyltransfer reaction.
Some of the essential residues in the CT domain active site are not conserved in its homologues TTHA0988, Msmeg0435/0436, and the KipA-KipI complex. In these proteins there is a one-residue deletion at the position corresponds to Asp-1584, Asp-1321 is not conserved (serine), neither is Tyr-1324 (threonine in TTHA0988, alanine in Msmeg0435/0436, valine in KipA) (supplemental Fig. S4E). Mutations at these locations in KlUC reduced the activity a few hundred to more than 10,000-fold (Table 2); therefore, despite the homology, these proteins are not likely to possess the CT activity.
Function of the BCCP Domain in the CT Catalysis
In the carboxyltransfer reaction the BCCP domain interacts with the CT domain to deliver biotin, our structure provided a detailed picture of the CT-BCCP interactions. Excluding the biotin group, the CT-BCCP interface buries 680 Å2 of surface area. The β4–β5 hairpin in the BCCP domain mediates a significant portion of the interactions. The amphipathic side chain of the biotinylated Lys-1795 is completely buried and is surrounded by the side chains of Asn-1291, Phe-1293, and Thr-1611. Hydrogen bonds are formed between its side chain amine and the Asn-1291 side chain carbonyl and the Gly-1294 main chain carbonyl, and between its main chain amide and the Asn-1289 side chain carbonyl. The side chain of the adjacent and strictly conserved Glu-1792 is also completely buried. Its carboxyl oxygens hydrogen bond with side chains of the conserved Arg-1538 and of Asn-1289 and Thr-1611 (Fig. 4B). Side chains of Tyr-1767, Thr-1768, Arg-1770, Trp-1772, and Met-1794 in the BCCP domain, and Leu-1127, Tyr-1288, Pro-1368, Arg-1538, Glu-1541, and Thr-1542 in the CT domain contribute to additional van de Waals interactions. Additional hydrogen bonds are formed between the Arg-1770 side chain guanidinium group and the Glu-1541 main chain carbonyl, and side chains of Trp-1772 and Glu-1541 (Fig. 4B).
Consistent with the structural observations, substituting Glu-1792 with alanine decreased kcat 3-fold, without significantly affecting the Km for urea (Table 2 and supplemental Fig. S9A). This mutation did not cause any decrease in protein biotinylation or apparent misfolding (supplemental Fig. S8, A and B). The loss of activity is presumably due to decreased CT-BCCP affinity, which would hinder biotin delivery to the CT domain active site, slowing down the carboxyltransfer reaction and in turn the overall catalysis. The function of the interactions between the BCCP and the catalytic domains has also been demonstrated by a recent study on the R. etli PC. Introducing the T882A mutation in this enzyme, located at its CT domain active site, increased its BC activity. Residue Thr-882 contributes to the CT-BCCP interactions in the R. etli PC by forming a hydrogen bond with biotin. The authors of this study propose that the T882A mutation decreases the CT-BCCP affinity and consequently biotin occupancy at the CT domain active site, which shifts the equilibrium toward increased biotin occupancy at the BC domain active site, leading to an increase in the BC activity. Consistent with this, they found that enhancing CT-BCCP affinity with small molecules in this mutant decreased its BC activity (42).
The BCCP domain might play additional roles in the catalysis. In the observed conformation, the position of the CT subdomain D is dramatically different from its counterparts in the CT domain homologues TTHA0988 and Msmeg0435/0436. Consequently, the KlUC CT domain active site cleft between subdomains B and D is much narrower compared with the equivalent regions in its homologues (Fig. 3, D and E, and supplemental Fig. S5). Such a “closed” conformation is stabilized by the BCCP domain, which interacts simultaneously with subdomains B and D (Fig. 4B). Therefore, when the BCCP domain translocates away from the CT domain during the catalysis, the CT domain might adopt an “open” conformation resembling that of TTHA0988 and Msmeg0435/0436, with a wider active site cleft. Such open/close cycles could facilitate substrate binding and release of the reaction products.
Conserved Structural Elements in the Carboxyltransfer Catalysis of Biotin-dependent Carboxylases
Detailed structural information of biotin and substrate coordination at the CT domain active site provides insights into the carboxyltransfer reaction. To date, such information is available for PC (19), PCC (20), and for UC from the present study. In PCC the biotin group is in a partially folded, unproductive conformation (20), whereas in PC (19) and UC it is positioned for the carboxyltransfer reaction. There are no homologies between the CT domains of PC and UC, but biotin coordination at their active site shows unexpected similarities. In both enzymes, the biotin carbonyl oxygen forms two hydrogen bonds with the CT domain. In PC, it hydrogen bonds with the side chain hydroxyl of a conserved serine (Ser-911 in the human PC, Ser-879 in the S. aureus PC) and the main chain amide of a neighboring lysine (Lys-912 in the human PC, Lys-880 in the S. aureus PC). If the biotin head groups are aligned, hydrogen bond partners of the biotin carbonyl in KlUC and PC occupy similar spatial locations (Fig. 5A). Mutations affecting these hydrogen bonds in KlUC (Y1324F) and in the S. aureus PC (S879A) (41) reduced the enzyme activity. These hydrogen bonds could stabilize the enolate biotin intermediate, which explains their importance in the catalysis (supplemental Fig. S7). It remains to be seen whether similar hydrogen bonds form during the carboxyltransfer reaction of other biotin-dependent carboxylases and whether they have similar functions.
FIGURE 5.
Common structural elements shared by UC and other biotin-dependent carboxylases. A, biotin coordination at the CT domain active site. CT domain active sites of KlUC and the S. aureus PC (gray) are aligned on the biotin head group and are shown in stereo. Reaction substrates, potential general bases, and hydrogen bond partners of the biotin carbonyl are highlighted. B–D, CT-BCCP interactions. Crystal structures of KlUC (B), the S. aureus PC (C), and PCC (D) are aligned on their BCCP domains (orange) and are shown side by side. Biotin, the biotinylated lysine, the conserved glutamate in the BCCP domain, and its potential hydrogen bond partners in the CT domain (green for the S. aureus PC and PCC) are highlighted.
During the carboxyltransfer catalysis, the BCCP domain interacts with the CT domain, delivering biotin for the reaction. In KlUC, hydrogen bonds between the conserved Glu-1792 in the BCCP domain and residues in the CT domain contribute significantly to the CT-BCCP interactions (Figs. 4B and 5B), substituting it with alanine reduced the enzyme activity. CT-BCCP interactions are also observed in the crystal structures of the human and S. aureus PC (19) and PCC (20). In the human PC the glutamate is not conserved (serine) and does not form hydrogen bonds with the CT domain. In both the S. aureus PC and PCC, the glutamate is conserved, and potential hydrogen bond partners in the CT domain are found. In the S. aureus PC, the Lys-983 side chain amine is located 4.6 Å away from its carboxyl oxygen (Glu-1109, Fig. 5C) (19). In PCC, the side chain amine of Asn-311 in the β subunit is located 3.5 Å away from its carboxyl oxygen (Glu-691 in the α subunit, Fig. 5D) (20). Different side chain rotamers of these neighboring residues could reduce the distance, enabling hydrogen bond formation. The glutamate is conserved in many biotin-dependent enzymes (16), and the above analysis suggests that in these enzymes it might have a conserved function: mediating interactions between the BCCP and CT domains.
Biotin-dependent carboxylases share common mechanisms in their catalysis, homologous structural elements provide molecular basis for these mechanisms. In addition to the conserved BC and BCCP domains, our analysis suggests that biotin-dependent carboxylases share homologous structural elements in biotin coordination at their CT domain active site and in CT-BCCP interactions, which might play important roles in their carboxyltransfer reaction. Further studies are required to test these hypotheses, which will advance the understanding of this important enzyme family.
The BC Domain
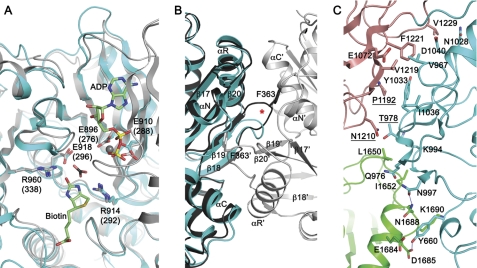
The structure of the BC domain is homologous to the BC components of other biotin-dependent enzymes. After structure alignment, the r.m.s. deviation for related Cα atoms in the KlUC BC domain and the BC subunit of E. coli ACC (36) is 0.97 Å. Essential residues at the BC domain active site (44, 45) are conserved in KlUC (Fig. 6A).
FIGURE 6.
The BC domain. A, the BC domain active site. Crystal structure of the E. coli ACC BC subunit (gray) (45) is superimposed for reference. ADP, Mg2+ ion (gray sphere), bicarbonate (black for the carbon atom), and biotin were modeled based on the E. coli BC structure. Important active site residues are highlighted; residue numbers for the E. coli BC are enclosed in parentheses. B, conformation differences at the BC dimer interface. Crystal structure of the E. coli BC (36, 44) dimer is aligned for reference and is shown in different shades of gray for the two monomers. Secondary structure elements and the E. coli BC residue Phe-363 are indicated; those labeled with the prime sign belong to the second monomer in the dimer. The red star indicates conformational differences at the β18–β19 hairpin. C, the BC-CT interface. Residues contributing to the BC-CT interactions are highlighted. Labels for those contributing to the conserved interactions are underlined.
In contrast to the E. coli BC, which form homodimers, the KlUC BC domain is monomeric. The protein surface mediating dimer interactions in the E. coli BC is not involved in protein-protein interactions in KlUC and faces a solvent channel in the crystal. Structural differences at this surface provide molecular basis for the monomeric form. KlUC contains two fewer residues in the β18–β19 hairpin. Phe-363, a major contributor to the E. coli BC dimer interactions (46), does not exist in KlUC (Fig. 6B). The Glu-23–Arg-401 intermolecular ion pair, another major contributor to the E. coli BC dimer interactions (46), is not conserved in KlUC, in which Glu-23 is replaced by an arginine (Arg-644). A similar case has recently been reported for PCC (20).
The BC domain interacts with CT subdomains A and D (Fig. 6C). The interface buries 780 Å2 of surface area. It is composed of 25% nonpolar residues, 42% polar ones, and 33% charged ones (47). At the interface with the CT subdomain D, two hydrogen bonds are formed between side chains of Tyr-660 and Asn-997 in the BC domain, and Asp-1685 and Asn-1688 in the CT domain, respectively. Side chains of Tyr-660, Gln-976, and Lys-994 in the BC domain, and Leu-1650, Ile-1652, Glu-1684, and Lys-1690 in the CT domain, contribute to the van der Waals interactions. No hydrogen bonds are formed between the BC domain and the CT subdomain A. Buried surface areas are contributed by side chains of Val-967, Thr-978, Asn-1028, Tyr-1033, Ile-1036, and Asp-1040 in the BC domain, and Glu-1072, Pro-1192, Asn-1210, Val-1219, Phe-1221, and Val-1229 in the CT subdomain A (Fig. 6C). An analysis with the Consurf server (48) on 159 selected UC sequences indicated that among the BC-CT interactions, those mediated by Thr-978, Pro-1192, and Asn-1210 are conserved. These residues are located in the center of the BC-CT interface (Fig. 6C). Therefore, although in different UC the BC-CT interactions appear to be somewhat variable, the general domain architecture is likely to be preserved.
In summary, our structural and biochemical studies provided molecular insights into the UC carboxyltransfer reaction. Because some structural elements important for the catalysis might be conserved in the biotin-dependent carboxylase family, our work also shed light on the mechanism of the carboxyltransfer reaction catalyzed by biotin-dependent carboxylases in general. In addition, structure and function of the CT domain and its homologues gave yet another interesting example of diverse functions for proteins with a common fold, and our studies on UC provided a framework to understand the function of these homologues, including the KipA-KipI complex involved in sporulation regulation in B. subtilis. Furthermore, analysis of the BC domain revealed a monomeric state similar to that reported for PCC; and even though the BC-CT interactions are somewhat variable, the UC quaternary structure is likely to be conserved.
Supplementary Material
Acknowledgments
We thank Prof. Jianhua He, Dr. Lin Tang, and Dr. Sheng Huang at the Shanghai Synchrotron Radiation Facility for setting up the BL17U beamline, and Prof. Jianping Ding, Dr. Manwu Zha, and Li Han at the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, for technical assistance.
This work was supported in part by National Institutes of Health Grant DK067238 (to L. T.). This work was also supported by grants from the Ministry of Science and Technology of China 973 Programs 2011CB910500 and 2010CB912502 and the One Hundred Talents Program of the Chinese Academy of Sciences (to S. X.).

This article contains supplemental Figs. S1–S9.
The atomic coordinates and structure factors (code 3VA7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UC
- urea carboxylase
- AH
- allophanate hydrolase
- ACC
- acetyl-CoA carboxylase
- BC
- biotin carboxylase
- BCCP
- biotin carboxyl carrier protein
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- CT
- carboxyltransferase
- KlAH
- Kluyveromyces lactis AH
- KlUC
- Kluyveromyces lactis UC
- PC
- pyruvate carboxylase
- PCC
- propionyl-CoA carboxylase
- PDB
- Protein Data Base
- r.m.s.
- root mean square.
REFERENCES
- 1. Mobley H. L., Hausinger R. P. (1989) Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mobley H. L., Island M. D., Hausinger R. P. (1995) Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sirko A., Brodzik R. (2000) Plant ureases: roles and regulation. Acta Biochim. Pol. 47, 1189–1195 [PubMed] [Google Scholar]
- 4. Singer M. A. (2003) Do mammals, birds, reptiles, and fish have similar nitrogen conserving systems? Comp. Biochem. Physiol. B Biochem. Mol. Biol. 134, 543–558 [DOI] [PubMed] [Google Scholar]
- 5. Kanamori T., Kanou N., Atomi H., Imanaka T. (2004) Enzymatic characterization of a prokaryotic urea carboxylase. J. Bacteriol. 186, 2532–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanamori T., Kanou N., Kusakabe S., Atomi H., Imanaka T. (2005) Allophanate hydrolase of Oleomonas sagaranensis involved in an ATP-dependent degradation pathway specific to urea. FEMS Microbiol. Lett. 245, 61–65 [DOI] [PubMed] [Google Scholar]
- 7. Navarathna D. H., Harris S. D., Roberts D. D., Nickerson K. W. (2010) Evolutionary aspects of urea utilization by fungi. FEMS Yeast Res. 10, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strope P. K., Nickerson K. W., Harris S. D., Moriyama E. N. (2011) Molecular evolution of urea amidolyase and urea carboxylase in fungi. BMC Evol. Biol. 11, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roon R. J., Levenberg B. (1972) Urea amidolyase. I. Properties of the enzyme from Candida utilis. J. Biol. Chem. 247, 4107–4113 [PubMed] [Google Scholar]
- 10. Whitney P. A., Cooper T. G. (1972) Urea carboxylase and allophanate hydrolase: two components of adenosine triphosphate:urea amidolyase in Saccharomyces cerevisiae. J. Biol. Chem. 247, 1349–1353 [PubMed] [Google Scholar]
- 11. Leftley J. W., Syrett P. J. (1973) J. Gen. Microbiol. 77, 109–115 [Google Scholar]
- 12. Andersen G., Björnberg O., Polakova S., Pynyaha Y., Rasmussen A., Møller K., Hofer A., Moritz T., Sandrini M. P., Merico A. M., Compagno C., Akerlund H. E., Gojković Z., Piskur J. (2008) A second pathway to degrade pyrimidine nucleic acid precursors in eukaryotes. J. Mol. Biol. 380, 656–666 [DOI] [PubMed] [Google Scholar]
- 13. Ghosh S., Navarathna D. H., Roberts D. D., Cooper J. T., Atkin A. L., Petro T. M., Nickerson K. W. (2009) Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect. Immun. 77, 1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vylkova S., Carman A. J., Danhof H. A., Collette J. R., Zhou H., Lorenz M. C. (2011) The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio 2, e00055–00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tong L. (2005) Acetylcoenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cell. Mol. Life Sci. 62, 1784–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jitrapakdee S., Wallace J. C. (2003) The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr. Protein Pept. Sci. 4, 217–229 [DOI] [PubMed] [Google Scholar]
- 17. Attwood P. V., Wallace J. C. (2002) Chemical and catalytic mechanisms of carboxyl transfer reactions in biotin-dependent enzymes. Acc. Chem. Res. 35, 113–120 [DOI] [PubMed] [Google Scholar]
- 18. St Maurice M., Reinhardt L., Surinya K. H., Attwood P. V., Wallace J. C., Cleland W. W., Rayment I. (2007) Domain architecture of pyruvate carboxylase, a biotin-dependent multifunctional enzyme. Science 317, 1076–1079 [DOI] [PubMed] [Google Scholar]
- 19. Xiang S., Tong L. (2008) Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction. Nat. Struct. Mol. Biol. 15, 295–302 [DOI] [PubMed] [Google Scholar]
- 20. Huang C. S., Sadre-Bazzaz K., Shen Y., Deng B., Zhou Z. H., Tong L. (2010) Crystal structure of the α(6)β(6) holoenzyme of propionyl-coenzyme A carboxylase. Nature 466, 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang L., Grau R., Perego M., Hoch J. A. (1997) A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 11, 2569–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Doublié S., Kapp U., Aberg A., Brown K., Strub K., Cusack S. (1996) Crystallization and preliminary x-ray analysis of the 9-kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 384, 219–221 [DOI] [PubMed] [Google Scholar]
- 23. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 25. Padilla J. E., Yeates T. O. (2003) A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 59, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 26. Vagin A., Teplyakov A. (2000) An approach to multicopy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 27. Mochalkin I., Miller J. R., Evdokimov A., Lightle S., Yan C., Stover C. K., Waldrop G. L. (2008) Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase. Protein Sci. 17, 1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cowtan K. (1999) Error estimation and bias correction in phase-improvement calculations. Acta Crystallogr. D Biol. Crystallogr. 55, 1555–1567 [DOI] [PubMed] [Google Scholar]
- 30. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaltwasser H., Schlegel H. G. (1966) NADH-dependent coupled enzyme assay for urease and other ammonia-producing systems. Anal. Biochem. 16, 132–138 [DOI] [PubMed] [Google Scholar]
- 36. Waldrop G. L., Rayment I., Holden H. M. (1994) Three-dimensional structure of the biotin carboxylase subunit of acetyl-CoA carboxylase. Biochemistry 33, 10249–10256 [DOI] [PubMed] [Google Scholar]
- 37. Wang P., Heitman J. (2005) The cyclophilins. Genome Biol. 6, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jacques D. A., Langley D. B., Hynson R. M., Whitten A. E., Kwan A., Guss J. M., Trewhella J. (2011) A novel structure of an antikinase and its inhibitor. J. Mol. Biol. 405, 214–226 [DOI] [PubMed] [Google Scholar]
- 39. Jacques D. A., Langley D. B., Jeffries C. M., Cunningham K. A., Burkholder W. F., Guss J. M., Trewhella J. (2008) Histidine kinase regulation by a cyclophilin-like inhibitor. J. Mol. Biol. 384, 422–435 [DOI] [PubMed] [Google Scholar]
- 40. Jacques D. A., Langley D. B., Kuramitsu S., Yokoyama S., Trewhella J., Guss J. M. (2011) The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase. Acta Crystallogr. D Biol. Crystallogr. 67, 105–111 [DOI] [PubMed] [Google Scholar]
- 41. Yu L. P., Xiang S., Lasso G., Gil D., Valle M., Tong L. (2009) A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure 17, 823–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeczycki T. N., St Maurice M., Jitrapakdee S., Wallace J. C., Attwood P. V., Cleland W. W. (2009) Insight into the carboxyl transferase domain mechanism of pyruvate carboxylase from Rhizobium etli. Biochemistry 48, 4305–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Knowles J. R. (1989) The mechanism of biotin-dependent enzymes. Annu. Rev. Biochem. 58, 195–221 [DOI] [PubMed] [Google Scholar]
- 44. Thoden J. B., Blanchard C. Z., Holden H. M., Waldrop G. L. (2000) Movement of the biotin carboxylase B domain as a result of ATP binding. J. Biol. Chem. 275, 16183–16190 [DOI] [PubMed] [Google Scholar]
- 45. Chou C. Y., Yu L. P., Tong L. (2009) Crystal structure of biotin carboxylase in complex with substrates and implications for its catalytic mechanism. J. Biol. Chem. 284, 11690–11697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen Y., Chou C. Y., Chang G. G., Tong L. (2006) Is dimerization required for the catalytic activity of bacterial biotin carboxylase? Mol. Cell 22, 807–818 [DOI] [PubMed] [Google Scholar]
- 47. Reynolds C., Damerell D., Jones S. (2009) ProtorP: a protein-protein interaction analysis server. Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]
- 48. Glaser F., Pupko T., Paz I., Bell R. E., Bechor-Shental D., Martz E., Ben-Tal N. (2003) ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.