Background: The mechanism by which Sterile Alpha Motifs (SAMs) self-associate and polymerize to control protein function is unknown.
Results: SAM polymerization in Polyhomeotic, a Polycomb group protein, is controlled by an unstructured linker sequence in Polyhomeotic.
Conclusion: Polyhomeotic growth suppressive function is enhanced by increasing SAM polymerization.
Significance: Functions of other SAM domain-containing proteins could be manipulated through their unstructured linkers.
Keywords: Chromatin Structure, Gene Regulation, Intrinsically Disordered Proteins, Polycomb, Protein Structure
Abstract
Polyhomeotic (Ph), a member of the Polycomb Group (PcG), is a gene silencer critical for proper development. We present a previously unrecognized way of controlling Ph function through modulation of its sterile alpha motif (SAM) polymerization leading to the identification of a novel target for tuning the activities of proteins. SAM domain containing proteins have been shown to require SAM polymerization for proper function. However, the role of the Ph SAM polymer in PcG-mediated gene silencing was uncertain. Here, we first show that Ph SAM polymerization is indeed required for its gene silencing function. Interestingly, the unstructured linker sequence N-terminal to Ph SAM can shorten the length of polymers compared with when Ph SAM is individually isolated. Substituting the native linker with a random, unstructured sequence (RLink) can still limit polymerization, but not as well as the native linker. Consequently, the increased polymeric Ph RLink exhibits better gene silencing ability. In the Drosophila wing disc, Ph RLink expression suppresses growth compared with no effect for wild-type Ph, and opposite to the overgrowth phenotype observed for polymer-deficient Ph mutants. These data provide the first demonstration that the inherent activity of a protein containing a polymeric SAM can be enhanced by increasing SAM polymerization. Because the SAM linker had not been previously considered important for the function of SAM-containing proteins, our finding opens numerous opportunities to manipulate linker sequences of hundreds of polymeric SAM proteins to regulate a diverse array of intracellular functions.
Introduction
Sterile Alpha Motif (SAM)2 domains are highly prevalent in eukaryotes, found in over 3000 proteins that exist in every cell compartment. Proteins with SAM domains function as transcription factors, scaffolding proteins, nucleic acid-binding proteins, kinases and others (1). The ability to self-associate is a common feature present in a number of SAM domains. Examination of the structures of several of these SAM domains has revealed that, despite a lack of sequence identity, they all form a head-to-tail, open-ended, left-handed helical, homo-polymer architecture (2–8) (Fig. 1, A and B). A recent analysis of all SAM domain sequences upon “threading” them into known polymer structures estimates that nearly 700 SAM domains are polymeric (9). This may be a low estimate if other SAM domains also form a similar architecture utilizing different residues for polymerization than those already identified for polymer forming SAM domains.
FIGURE 1.
Ph SAM polymer. A, surface representation of nine Ph SAM units arranged in left-handed helical architecture as observed in the crystal structure (3) (PDB ID 1KW4). B, illustration of the head-to-tail arrangement of the SAM polymer. The Kd measurement was previously reported (3). C, close-up of the SAM-SAM interface of A showing the location of the residues targeted for mutagenesis. The structure shows the L1547R mutation as it was used for the crystal structure determination. Ph SAM surface mutants L1547R, H1552R, and L1565R correspond to L51R, H56R, and L69R, respectively, that were used previously (3, 4). D, domain organization of the proximal Ph of Drosophila melanogaster used in this study. HD1: homology domain 1; FCS: Phe-Cys-Ser domain; SAM: sterile alpha motif. These three domains are conserved in all Ph orthologs.
Polymerization of SAM domains has been shown to be vital to the proper function of a protein (5–8, 10) indicating a close relationship between protein function and SAM domain polymerization and suggesting protein activity could be manipulated by controlling the dynamic polymerization of the SAM domain. However, what remains largely unresolved is how SAM polymerization is regulated.
Polymeric SAM domains likely play an important role in Polycomb Group (PcG) function. The PcG is a family of gene silencing proteins that are noted for their role as developmental regulators that maintain the repressed state of Drosophila homeotic (HOX) genes (11). Of the 15 to 20 PcG proteins identified thus far, four contain SAM domains: Ph (Polyhomeotic), Scm (Sex combs on midleg), L3mbt (lethal 3 malignant brain tumor), and Sfmbt (Scm-related gene containing four mbt domains). In vitro, Ph SAM and Scm SAM can both form homo-polymers in addition to being able to hetero-polymerize with each other (3, 4, 12, 13). Binding affinity measurements suggest a model whereby the two homo-polymers come together at a single junction (4). While the polymeric status of L3mbt and Sfmbt are not known, sequence analysis of their SAM domains indicate that polymerization is likely (9). For Ph and Scm SAM (like all other SAM domains that polymerize into a helical architecture), the head-to-tail homo-polymerization is mediated through two different binding surfaces on the SAM domain: the Mid-Loop (ML) binding surface, named for the loop structure in the middle of the SAM sequence that houses key SAM-SAM-interacting residues, and the End-Helix (EH) binding surface which contains binding residues on the C-terminal helix of the SAM structure (Fig. 1B). The SAM domains of Ph and Scm are at the C terminus of both proteins (Fig. 1C). It is important to note that both the N and C termini of these and other polymeric SAM domains point outward from the polymer helix axis. Therefore, it would be feasible to both form a helical polymer structure while being able to sterically accommodate the other parts of the protein that extend away from either the N or C terminus of the SAM domain.
Ph is a core member of a multi-protein PcG complex called Polycomb Repression Complex 1 (PRC1), which also includes Posterior Sex combs (Psc), RING1, and Polycomb (Pc) (14, 15). Scm can associate with PRC1 though in less than stoichiometric amounts compared with the other PRC1 core components (16). Even though the isolated Ph SAM is able to polymerize as evidenced by its ability to form long filaments and the strong SAM-SAM self-association required for mediating polymerization demonstrated in both GST pull-down experiments and measurement of the Kd of 200 nm (3), PRC1 is not polymeric. Electron micrographs of PRC1 isolated from a baculovirus expression system does not show the presence of long, fibrous structures that would be consistent with a polymer structure (17). Scanning force microscopy analyses show limited PRC1 oligomerization (estimated in the range of four to six PRC1 units) when PRC1 binds DNA, but longer polymers were not detected (18). Interestingly, the architecture of the PRC1/DNA complex does exhibit a left-handed helical assembly, perhaps as a result of short polymers formed through Ph SAM polymerization. Because no studies to date have been reported in which Ph SAM polymerization has been directly shown to be an important element of Ph function, we sought to determine the importance of Ph SAM in Ph function. In addition, we wanted to determine why a protein complex that contains a polymeric SAM domain appears to exhibit limited polymerization in the context of PRC1. The results of our study indicate that unstructured residues adjacent to Ph SAM play an important role in controlling polymerization, perhaps facilitated by the helical architecture which is common to all SAM polymers identified thus far. Furthermore, given that the function of proteins that contain polymeric SAM domains are dependent on polymerization, our results could have consequences toward being able to modulate protein activity, and consequently cell function, through SAM linker control of the dynamic polymerization of SAM domains.
EXPERIMENTAL PROCEDURES
Luciferase Reporter Transcription Assay
The polyhomeotic-proximal (ph-p, also called ph) genes were cloned into pPAcFlag (kind gift from Dr. Albert J. Courey). The expression vectors for ph (100 ng), lacZ (7.5 ng) (kind gift from Dr. Yuzuru Shiio), both under control of the actin 5c promoter, along with the luciferase reporter gene were transfected into 1 × 105 Drosophila S2 cells using the Fugene HD transfection reagent (Roche Applied Science). For Fig. 6C, 50 ng and 3.75 ng of the ph and lacZ plasmids were used. There are three tandem repeats of the Zif268 DNA binding elements immediately upstream of a metallothionine promoter (MTp) controlling the expression of the luciferase reporter gene. Two days after transfection, the expression of the luciferase gene was induced by adding 100 μm CuSO4. The cells were harvested the following day and lysed using 100 mm potassium phosphate buffer pH 7.8, 0.2% Triton X-100, and 0.5 mm DTT. For all individual transfections, equal volumes of lysate were used for the Dual-Light Combine Reporter Gene Assay System (Applied Biosystems) to measure both luciferase and β-galactosidase activities. The data are presented as the ratio of the two enzyme activities.
FIGURE 6.
Ph RLink has enhanced growth suppressive function compared with wild-type. A, domain structures of the constructs used in the luciferase assays (B and C). Error bars are the standard deviations from three independent transfections. Insets show actual luciferase/lacZ values for Zif268-Ph and Zif268 Ph RLink. D–F, imaginal wing discs expressing exogenous wild-type ph, polymer-deficient ML mutant ph L1547R/H1552R and ph RLink, respectively. Flies were crossed to the en-Gal4; UAS-GFP driver lines to induce transgene expression in the wing disc. Hoechst dye stains DNA which shows the shape of the wing disc (blue); GFP expression defines the expression domain (green); Flag-tagged expressed transgene (red); and the last column shows the merge of all three. The bar in D measures 20 μm.
Transgenic Drosophila
All ph transgenes were cloned into pattB-UASp, a vector required for PhiC31 integration (19). The DNA was injected into early syncytial-stage blastoderm embryos (Rainbow Transgenics) that carries both a source of the PhiC31 integrase on the X chromosome and attP target site at chromosomal position 58A (yw; M{eGFP.vas-int.Dm}2A;M{RFP.attP}58A). Successful transformants were selected on the basis of the stable integration of the white gene into white mutant background. All transgenes include a Flag-epitope tag and are under control of the UAS promoter whose expression was driven by mating the transgenic flies with either the engrailed Gal4 (en-Gal4) or actin-Gal4 lines (see FlyBase database for descriptions of the Gal4 driver lines).
Protein Preparation
Supplemental Table S1 summarizes all the protein constructs used in the in vitro experiments. Except for those fused to MBP, ph genes were subcloned into a modified pET-3c vector (Novagen), transformed into BL21-pLysS cells and expression induced with 1 mm IPTG. For the MBP fused constructs, ph genes were subcloned into pBADM-41+ (EMBL), transformed into ARI814 cells (20), and expression induced with 0.2% arabinose. Typically, harvested bacterial cells from 1-liter cultures were resuspended with 10 ml of 50 mm Tris, pH 8.0, 100 mm NaCl, 25 mm imidazole, pH 7.5, 1 mm PMSF, and 10 mm βME. Cells were lysed by sonication and initial purification of the proteins was performed using Ni affinity chromatography followed by ion exchange chromatography. Proteins that required cleavage of tag sequences were purified by Ni affinity chromatography, cleaved with either TEV or Senp2 (a kind gift from Dr. Christopher D. Lima) followed by a second Ni affinity chromatography where the non-binding fractions were collected. Further purifications were performed using ion exchange chromatography. Ph sc1398–1484 1485–1577 expressed insolubly in bacteria. For this protein, bacterial cells were first resuspended in 50 mm Tris, pH 8, 200 mm NaCl, 10 mm imidazole, pH 7.5, 1 mm PMSF, 10 mm β-ME, and lysed by sonication. The soluble lysate was separated, and the pellet solubilized in the same buffer containing 6 m urea. Following non-native Ni affinity purification, the protein was refolded through dialysis into native buffer then further purified as described above for the proteins expressed in the soluble lysate.
Analytical Ultracentrifugation
All samples for AUC were placed in 10 mm Tris pH 8.0, 50 mm NaCl, and 1 mm TCEP. The SV data were analyzed with Ultrascan (21) (version 9.9 release 1282). All experiments were scanned in intensity mode at 280 nm, 20 °C, ranging between 25–40 krpm in standard 2-chanel epon centerpieces (Beckman-Coulter). All data were initially analyzed with the 2-dimensional spectrum analysis to remove time and radially invariant noise from the data (22), followed by van Holde -Weischet analysis (23), and Genetic Algorithm-Monte Carlo analysis (24, 25). All sedimentation velocity experiments were performed at the Center for Analytical Ultracentrifugation of Macromolecular Assemblies at the University of Texas Health Science Center in San Antonio using a Beckman XLI ultracentrifuge. All computations were carried out using the TeraGrid infrastructure (26).
Atomic Force Microscopy
AFM imaging was performed in oscillation (tapping) mode in air, with a Multimode Nanoscope IIIa microscope (Bruker Corp.). Ph SAM, RLink Ph-(1485–1577) and Ph-(1397–1577) at 588, 620, and 620 μm stock concentration, respectively, were prepared in 10 mm Tris/HCl pH 8.0, 50 mm NaCl, and 1 mm TCEP then diluted with 5 mm Tris/HCl, pH 7.4 buffer. Between 800 and 1700 particles of at least 4 nm in length were analyzed for each sample. 3 μl of the sample was deposited on a freshly cleaved muscovite mica substrate and immediately briefly spinned on a turntable to prevent clumping of particles. After 2 min at room temperature the substrate with electrostatically attached protein molecules was washed three times with 50 μl of double distilled water and dried under a stream of nitrogen. The imaging was carried out with TESP probes (Veeco Probes) with resonant frequency tuned to 290–300 kHz, the amplitude of 100–300 mV, the setpoint between 1.5 and 1.8 V, and the scan rate of 3.05 Hz. Images of 1 × 1 μm fields were collected in a height mode with digital resolution of 512 × 512 pixels, with at least ten distinct areas imaged for each sample. The images were subjected to a standard flattening and plain-fitting in the Nanoscope software (v. 5.12). Morphometric analysis of imaged particles was performed with the SPIP software (v. 5.1.5, Image Metrology, Denmark), with the grains (particles) detected and fiber lengths approximated automatically.
CD Spectroscopy
Ph-(1397–1507) was prepared to a concentration of 0.2 mg/ml in 10 mm sodium phosphate buffer pH 7, 5 mm NaCl. The CD spectrum of was collected at room temperature using a Jasco J-815 CD spectrometer. Data were collected between 250 to 195 nm.
NMR Spectroscopy
All NMR samples (both the 15N and the doubly labeled 15N, 13C Ph-(1397–1507)) were prepared to a concentration of 1.5 mm in 10 mm NaPO4 pH 5.0, 50 mm NaCl. The concentration of 15N Ph-(1397–1507) before the addition of potential binding proteins was 1.5 mm. Ph SAM L1565R, TEL SAM A61D (2) or chicken egg white lysozyme (Sigma-Aldrich) were placed into the same buffer as Ph-(1397–1507) before adding 0.4 and 1.6 molar equivalents and measuring a {1H}-15N HSQC spectrum after each addition. Backbone assignments were made using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) on data collected on a Bruker 600 MHz spectrometer at 310 K. The backbone chemical shifts were deposited to the BMRB (accession number 17552). All other spectra were processed and analyzed with NMRPipe (27) and NMRView (28). Chemical shift perturbation experiments were performed at 310 K on a 700 MHz spectrometer. The identity of the backbone amide signals of the HSQCs collected at 300 K on the 700 MHz spectrometer were sufficiently similar to that collected at 310 K on the 600 MHz spectrometer to easily correlate the signals.
RESULTS
Ph SAM Polymerization Is Required for Ph-mediated Gene Repression
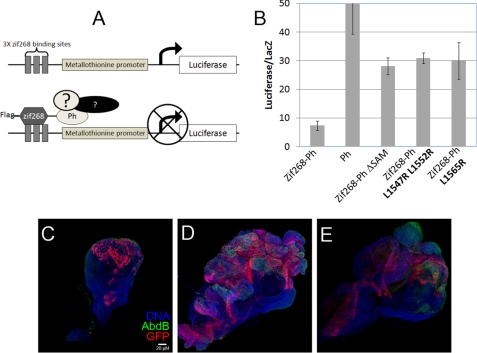
To investigate the role of Ph SAM polymerization in Ph-mediated repression, we utilized a transcription assay carried out in Drosophila S2 cells (Fig. 2A). We found that targeting wild-type Ph to the luciferase reporter gene is able to repress transcription compared with Ph that is not targeted (Fig. 2B). Moreover, a SAM domain-deleted Ph protein was unable to repress transcription showing that the SAM domain is required for the repressive function of Ph. We also tested Ph with mutations within its SAM domain that disrupt SAM-SAM interactions and are thus deficient in polymerization (3) (Fig. 1, B and C). Mutating either the ML and EH binding surface (Ph L1547R/H1552R and Ph L1565R, respectively), fails to repress luciferase compared with wild-type (Fig. 2B).
FIGURE 2.
Ph SAM polymerization is required for transcription repression. A, illustration of the transcription assay carried out in Drosophila S2 cells. B, results of the transcription assay for Ph. Error bars show the standard deviation of the results from three independent transfections. C–E, engrailed-Gal4(en-Gal4); UAS-GFP driver was used to ectopically express the Drosophila polyhomeotic-proximal (ph-p or ph) transgenes. Immunostaining of (C) WT, (D) Ph L1547R/H1552R, and (E) Ph L1565R isoforms expressed in the developing wing disc. Discs were dissected from 3rd instar larvae, fixed and stained for GFP expression (red) to define the expression domain and AbdB (green) to access derepression. Discs were counterstained Hoescht dye (blue).
We further investigated the role of Ph SAM polymerization in repression using transgenic Drosophila. We ectopically expressed Flag-tagged wild-type or either of the two SAM polymer-deficient mutant Ph proteins in the posterior compartment of the imaginal wing disc and assessed the derepression of Abdominal B (AbdB), which is normally repressed by the PcG in these cells. When exogenous wild-type Ph was expressed, there was normal wing disc development and maintenance of the repressed state of AbdB (Fig. 2C). In contrast, expression of either Ph L1547/H1552R or L1565R mutants lead to AbdB derepression, as indicated by immunostaining with anti-AbdB antibodies (Fig. 2, D and E). In addition, overexpression of the polymer-deficient Ph L1547/H1552R or L1565R mutants resulted in a dominant negative phenotype showing grossly abnormal wing discs consistent with tissue overgrowth. Taken together, these results are the first to suggest that SAM-dependent polymerization of Ph is required for transcription repression.
Mini-Ph (Ph-(1291–1577)) Forms Shorter Polymers than Ph SAM Alone (Ph-(1502–1577))
The discrepancy between the evidence for Ph SAM polymerization (both in vitro (3) and gene silencing (Fig. 2) experiments) and the lack of open-ended polymers in microscopy studies (17, 18) may be due to only shorter polymers forming or polymerizing not occurring in the larger context of the entire Ph protein or when Ph is a component of PRC1. To investigate the factors that might influence Ph SAM polymerization we used sedimentation velocity (SV) to measure the level of in vitro polymerization of a series of Ph constructs (Fig. 3). We analyzed the SV data via the van Holde-Weischet (vHW) analysis (23). This approach provides sedimentation coefficient distributions from which an assessment of heterogeneity can be obtained. Ph SAM alone (Ph-(1502–1577)) shows an S-value distribution from 6–11 S (Fig. 3A), consistent with the self-association of the 10 kDa Ph SAM into high molecular weight oligomers and provides further support for the previously observed in vitro Ph SAM polymerization (3). We next characterized a Ph construct encompassing residues 1291–1577, which we refer to as mini-Ph because it contains the three predicted structured domains: HD1 (homology domain 1), the nucleic acid binding FCS (Phe-Cys-Ser) domain (29, 30), and SAM (Fig. 1D) that are conserved in all Ph homologs. In contrast to Ph SAM, the sedimentation profile of mini-Ph exhibited lower S-values ranging from 5–6 S, even though mini-Ph has a higher calculated monomeric molecular mass (32 kDa) compared with just Ph SAM (10 kDa). Thus, while the individual Ph SAM domain forms long polymers, mini-Ph appears far smaller in its oligomeric state. We next determined whether the smaller mini-Ph oligomers were smaller polymers mediated by SAM-SAM interactions. We introduced mutations intended to interfere with the SAM-SAM interaction (3) into mini-Ph and performed SV. The ML surface residue L1547 is at the edge of the SAM-SAM interaction and while the L1547R mutation does hinder polymerization, some SAM-SAM self-association ability is still retained (3). Mini-Ph L1547R shows a lower S-value distribution compared with wild-type mini-Ph indicating a smaller oligomeric state. Ph SAM with the L1565R mutations, which exhibits no ability to self-associate (3), displays an even lower S-value distribution than that of mini-Ph L1547R. These results would be predicted if SAM-SAM interactions were mediating mini-Ph oligomerization.
FIGURE 3.
Ph SAM polymerization is limited by residues N-terminal to the SAM domain. A, van Holde-Weischet (vHW) (23) combined distribution plot of the velocity sedimentation of Ph SAM alone, mini-Ph (Ph-(1291–1577)) and polymer-deficient mini-Ph mutants. The legend and concentrations of the proteins used are indicated in the plot. Rotor speed was 40 k RPM. B, molecular weight distributions determined from a genetic algorithm Monte-Carlo analysis (24, 25) from the sedimentation velocity experiments shown in A. The weighted average molecular weight of each of the distributions is shown in the inset of each plot. C–E, vHW plots of different Ph SAM N-terminal linker proteins at three different concentrations. F and G, vHW plots of MBP fused Ph SAM with different linker deletions. Rotor speeds used were 40K, 25K, and 30K RPM for C, D, and E, respectively. The rotor speed for both F and G was 30K RPM.
The wide range of S-values indicate that Ph SAM and mini-Ph are heterogeneous, existing as a range of polymer sizes that contain different number of individual Ph SAM or mini-Ph units. We used a genetic algorithm-Monte Carlo analysis (24, 25, 31, 32) to obtain molecular weight distributions for Ph SAM alone, mini-Ph and mini-Ph mutants (Fig. 3B). For Ph SAM, the polymers exhibited molecular weights as large as 5 × 105 Da with a weight average molecular mass of 220 kDa (Fig. 3B, top), while mini-Ph polymers were much smaller (Fig. 3B, top center). In agreement with the sedimentation coefficient profiles, mini-Ph L1547R (Fig. 3B, bottom center) and L1565R (Fig. 3B, bottom) mutants exhibited even lower molecular weights than wild-type mini-Ph. Interestingly, the weight average molecular mass for wild-type mini-Ph is 178 kDa, which is close to the calculated value for a hexamer. While the heterogeneous nature of the sample prevents determination of the precise molecular weight of the oligomers, the results suggest an explanation for why a 6-fold screw axis helical architecture is present for all known structures of polymeric SAM domains (see “Discussion”).
Ph SAM N-terminal Linker Can Influence Polymerization
To determine which region outside of the Ph SAM domain can influence the degree of polymerization, we prepared Ph constructs that deleted residues from the N terminus of mini-Ph and measured their level of polymerization. The sedimentation profile of Ph-(1397–1577) (Fig. 3C), which includes the linker region between the FCS and SAM domains and extends to the end of the SAM domain, exhibited much lower S-values than Ph SAM alone (compare the S-value distribution of Ph SAM alone (open diamonds) in Fig. 3A with Ph-(1397–1577) in Fig. 3C), indicating smaller polymers. We prepared additional constructs with residues deleted from the N terminus of the linker (Ph-(1427–1577) and Ph-(1461–1577)) to see if this would result in increasing the size of polymers. The S-values observed at three concentrations (37, 55, and 90 μm) for both Ph-(1397–1577) and -(1427–1577) indicated that smaller-sized polymers were still present. However, Ph-(1461–1577) shows a clear increase in S-values at 90 μm (Fig. 3, C--E). We also prepared proteins with maltose-binding protein (MBP) fused to the N terminus of these construct allowing us to perform similar SV experiments at lower protein concentrations (MBP increases the extinction coefficient) while also better mimicking the configuration of the intact protein with the additional 42 kDa to the N terminus of the protein. The MBP-fused Ph constructs exhibited increasing S-values, and thus polymerization, as more of the linker was deleted (Fig. 3, F and G). From these experiments, we conclude that the Ph SAM N-terminal linker helps control Ph SAM polymerization.
A Random, Unstructured Linker Can Hinder Polymerization but Not As Well As Native Linker
The Ph sequence that links the FCS and SAM domains is not conserved among the human Ph orthologs (Fig. 4A) and is predicted to be disordered (supplemental Fig. S1). A non-conserved, unstructured linker capable of influencing the extent of SAM polymerization may do so via conformational entropy (see “Discussion”). To test whether any unstructured sequence could produce shorter Ph SAM polymers, we engineered a randomized 87-amino acid sequence (RLink) composed of residues that have been found to be enriched in intrinsically disordered proteins (M, K, R, S, Q, P, E) (33) along with several glycines to impart additional conformational entropy to the sequence, and attached it to the N terminus of Ph-(1485–1577) (RLink Ph-(1485–1577)). The sedimentation profile of RLink Ph-(1485–1577) (Fig. 4B) exhibited a lower average S-value profile than Ph SAM, indicating that RLink can indeed control polymerization. However, compared with Ph-(1397–1577), which has the native linker, RLink Ph-(1485–1577) exhibits significantly higher S-values, indicating that there are additional features of the native linker that are involved in controlling polymerization.
FIGURE 4.
RLink only moderately limits Ph SAM polymerization. A, sequence alignment of Ph and its three human orthologs (PHC1, 2, 3) along with Ph RLink SAM. The alignment was created using multiple rounds of ClustalW (40) using the following combinations of sequences. Ph, PHC1, PHC2, and PHC3 were aligned using the sequences shown. Ph RLink SAM was separately aligned to Ph-(1397–1577). The dark-shaded residues correspond to the backbone amide signals of the two-dimensional {1H}-15N HSQC which were broadened with the 0.4 molar equivalent in trans addition of Ph SAM L1565R mutant (see Fig. 5C). The light-shaded residues are the ones that broadened with the 1.6 molar equivalent addition. B, vHW integral distribution plot for Ph RLink SAM (+). The plot for Ph SAM (open diamonds) and Ph-(1397–1577) (filled squares) are the same plots shown in Fig. 3A and 3C, respectively. They are also shown here to allow better direct comparison. All samples were at 37 μm and spun at a rotor speed of 40K RPM. C, AFM images (top) and scatter plots of polymer fiber lengths (bottom) of Ph SAM, RLink Ph-(1485–1577), and Ph-(1397–1577). All AFM fields are 200 × 300 nm, with fiber and skeleton lengths marked as red lines, as detected with the grain analysis of the SPIP software. The gray color scale in the images represents particle height, with black as a background (0 nm) and white as 10 nm. D, vHW for the scrambled linker Ph SAM chimeras.
We used AFM as an independent technique to examine the differences in polymerization between Ph SAM, Ph-(1397–1577) and RLink Ph-(1485–1577) (Fig. 4C). At 100 nm concentration, each of the three samples showed fibers of diverse lengths, suitable for particle size analysis. We used the “fiber length” parameter as a measure of the maximal linear size of a fiber. In order to include tangled polymers in our analysis, we also used the “skeleton length” parameter, which adds the length of branches to the fiber length. The scatter plots in Fig. 4C shows clear differences in polymer lengths with Ph SAM being the longest, followed by RLink Ph-(1485–1577) then Ph-(1397–1577). These differences are also discernible in the maximal fiber lengths which were 248 nm (PhSAM), 154 nm (RLink Ph-(1485–1577)), and 99 nm (Ph-(1397–1577)). Even more striking were the maximum skeleton lengths of the polymers for each protein: 468 nm (Ph SAM), 338 nm (RLink Ph-(1485–1577)), and 99 nm (Ph-(1397–1577)). The AFM results together with those from AUC SV (Fig. 4B) indicate there are longer polymers formed when RLink is attached to the SAM domain compared with the native linker.
Amino Acid Content of the SAM Linker Is Important for Controlling SAM Polymerization
We tested two additional linker sequences to further examine the role of the SAM linker in controlling SAM polymerization. We designed a construct encompassing Ph residues 1398–1577 where residues 1398–1484 were replaced with a sequence containing the same amino acids but arranged in a scrambled manner (Ph sc1398–1484 1485–1577). Likewise, the linker sequence found N-terminal to Scm SAM (Scm residues 691–777) were also scrambled and attached to Ph-(1485–1577) (Scm sc691–777 Ph-(1485–1577)). While Scm sc691–777 Ph-(1485–1577) exhibited increased polymerization compared with the Ph SAM with its native linker (Ph-(1397–1577)), Ph sc1398–1484 1485–1577, showed the identical sedimentation profile as Ph-(1397–1577) exhibiting a sedimentation coefficient of 3 s throughout the boundary fraction (Fig. 4D). These results suggest that the amino acid content of the SAM linker plays a role in controlling SAM polymerization.
Ph SAM Linker Can Interact with Ph SAM
We investigated the structural properties of the Ph SAM native linker (Ph-(1397–1507)) to try to probe why it is more effective in controlling Ph SAM polymerization better than RLink. Ph-(1397–1507) was analyzed by circular dichrosim (CD) and NMR spectroscopies. The CD spectrum of Ph-(1397–1507) (Fig. 5A) has features that would be expected for a random coil. The one-dimensional 1H NMR spectrum of Ph-(1397–1507) shows neither up- nor down-field shifted resonances, indicating the absence of any tertiary structure (Fig. 5B). Moreover, the backbone amide signals in the Ph-(1397–1507) {1H}-15N HSQC NMR spectrum (Fig. 5C, black) are all clustered within the region expected for a random coil between 7.8 and 8.6 1H ppm. Together, these results indicate that Ph-(1397–1507) lacks any structure in the absence of the SAM domain.
FIGURE 5.
The unstructured Ph SAM N-terminal linker (Ph-(1397–1507)) binds Ph SAM in trans. Ph 1397–1507 circular dichroism (A), one-dimensional 1H NMR (B), and two-dimensional {1H}-15N HSQC (C) spectra. The black, green, and red spectra in C are of Ph-(1397–1507) alone and with added 0.4 and 1.6 molar equivalents of Ph SAM L1565R, respectively. The signals that broadened with the 0.4 molar equivalent addition of the isolated Ph SAM are labeled. See also Fig. 4A.
To determine whether the linker residues directly contact the SAM domain as a way to control polymerization, we added, in trans, Ph SAM with the L1565R mutation to 15N-labeled linker (Ph-1397–1507)) and observed perturbations to its {1H}-15N HSQC spectrum amide backbone signals. Ph SAM L1565R was used for this experiment because it does not self-associate (based on GST pull-down assays (3)) thus helping to eliminate complications due to aggregation that would be seen with use of the wild-type Ph SAM polymer. The titration produced substantial perturbation of a number of signals in the Ph-(1397–1507) two-dimensional {1H}-15N HSQC spectrum (Fig. 5C). Upon addition of 0.4 equivalents of Ph SAM L1565R (Fig. 5C, green), peak broadening occurred to several residues between 1415 and 1433 to the extent they were no longer visible. Upon addition of 1.6 equivalents of Ph SAM L1565R (Fig. 5C, red), the majority of the backbone amide signals in the linker (65 residues) were no longer visible (Fig. 4A). These results indicate a direct interaction can occur, in trans, between the Ph-(1397–1507) and the Ph SAM domain. Unfortunately, we were unable to perform the complementary experiment of adding unlabeled Ph SAM linker to 15N-labeled Ph SAM L1565R because the two-dimensional {1H}-15N HSQC of the Ph SAM L1565R was not of sufficient quality to perform the experiment. Control experiments adding either a monomeric mutant SAM domain from a protein called TEL (TEL SAM) or lysozyme showed no broadening of the amide signals but did exhibit movement due to weaker binding that occurred within the fast exchange NMR timescale (supplemental Fig. S2). Interestingly, the signals that were perturbed by the addition of either the TEL SAM mutant and lysozyme are, in general, the same ones which were broadened upon addition of 0.4 molar equivalents of Ph SAM L1565R (Ph-(1415–1420) and -(1427–1433)). However, mutating these residues did not result in significant changes to polymerization or repressive ability (supplemental Fig. S3) suggesting that more extensive changes, such as with RLink (see below), are required to alter Ph SAM polymerization. We have not yet isolated 15N-labeled Ph-scrambled linker (Ph sc1398–1484) to determine whether it too can directly contact Ph SAM.
Ph RLink Has Enhanced Growth Suppressive Ability Compared with Wild-type
Given that Ph-mediated gene repression is dependent on Ph SAM polymerization (Fig. 2), it was of interest to determine whether it would be possible to produce the opposite outcome of an engineered Ph protein with a SAM domain that has a greater ability to polymerize and hence greater gene silencing ability. For example, the interaction between two Ph SAMs (Kd, 200 nm (3)) is much weaker than for the TEL SAM self-association (2 nm (2)). Thus, a Ph protein in which the SAM domain is replaced with that of TEL SAM might produce a more stable SAM polymer leading to better repression. Also, SV experiments (Fig. 3) indicated that deleting segments of the linker leads to increased S-values, i.e. polymerization, as does replacing the native linker with RLink (Fig. 4B). We incorporated these potential polymer enhancing variations and tested their repressive ability using the luciferase reporter assay (Fig. 6A). While all constructs were capable of repressing the reporter gene better than the control (Ph that is not targeted to the promoter), only the mutant Ph in which Ph-(1398–1484) was replaced with RLink (Ph RLink) showed slightly lower luciferase expression than wild-type suggesting that Ph gene silencing ability can be enhanced through increased polymerization. Ph RLink that contain polymer-deficient mutations were not capable of repressing luciferase as well as Ph RLink suggesting that Ph RLink-mediated repression is indeed dependent on Ph SAM polymerization (Fig. 6C). We have compared the repressive abilities of wild-type Ph and Ph RLink several times and in each instance we have observed Ph RLink repression better than wild-type though the differences in standard deviations from the three independent transfections were not always exceeded between Ph wild-type and Ph RLink. While these results do point toward Ph RLink being a better repressor than wild-type Ph, the lack of clear differences in repression levels did not allow us to confidently gauge whether Ph RLink is indeed a better repressor than wild-type in the context of this assay.
However, the effect of RLink was quite more evident in vivo. We engineered transgenic flies and expressed the mutant ph transgenes in the wing disc using the engrailed Gal4; UAS-GFP (en-Gal4) driver. The engrailed gene is negatively regulated by Ph (34), which we hypothesize maintains sufficiently low levels of wild-type Ph to not have an adverse effect on wing disc development unlike the more robust actin-Gal4; UAS-GFP (actin-Gal4) driver (supplemental Fig. S4). While wild-type ph expression using the en-Gal4 driver results in production of normal wing discs (Figs. 2C and 6D), expression of both the polymer-deficient Ph L1547R/H1552R and Ph L1565R mutants led to large tissue overgrowth (Figs. 2, D and E and 6E). Remarkably, Ph RLink, engineered with the expectation that it would increase polymerization, exhibited the opposite phenotype compared with the Ph SAM polymer-deficient mutants showing even smaller discs than those that expressed wild-type ph (Fig. 6F). Recent studies have shown that ph possesses growth suppressive properties where overgrowth phenotypes are observed in flies with mutant ph (35–37). Moreover, wild-type ph overexpression in the eye can induce a small eye phenotype (35). We have similarly obtained an enhanced growth suppressive function not through the overexpression of wild-type ph, but rather, by increasing the polymerization of Ph SAM. Potential molecular bases of the result shown in Fig. 6F are in the “Discussion.”
DISCUSSION
We have found that the sequence of unstructured residues adjacent to a polymeric SAM domain can control SAM polymerization. Using this knowledge, the polymerization of a SAM domain containing protein was, for the first time, able to be increased, resulting in an in vivo phenotype opposite to that produced by polymer-deficient mutants.
The measurements of Ph SAM polymerization used in our study were performed with small Ph fragments. It cannot be stated for certain whether changes in polymerization for these small fragments also occur in the context of the full-length protein. However, the function of full-length Ph does indeed correlate with the level of polymerization observed for small Ph fragments. Thus, at least in this indirect way of assessing full-length Ph polymerization, the observed changes to polymerization for small Ph fragments can be extended to full-length Ph. Future investigation of Ph and its oligomeric state will be required to determine how manipulating just the SAM domain affects the oligomeric state of the entire Ph protein.
The simplest interpretation of our cellular and in vivo data is that controlling polymerization can in turn modulate the repressive and growth suppressive functions of Ph. However, it is possible that the Ph mutants used here altered other roles of Ph. For example, the inability to repress transcription and the overgrowth phenotypes of the SAM polymer-deficient mutants (Figs. 2, D and E and 6F) may be the result of disrupting Ph binding to a positive regulator. Ph SAM uses the same binding surfaces that mediate its own SAM-SAM interaction to bind directly to Scm SAM (4). The Ph SAM ML binding surface binds to the EH surface of Scm SAM with much stronger affinity (Kd, 54 nm) than the Ph SAM EH binds to the Scm SAM ML surface (Kd ∼1 mm, all other Ph SAM/Scm SAM combinations do not result in detectable binding (4)). Thus, the Ph SAM EH mutant still possesses a viable Scm SAM binding surface, yet both ML and EH mutants allow the same level of luciferase expression (Fig. 2B) and show the same level of wing disc overgrowth (Fig. 2, D and E). For the results obtained with Ph RLink, replacing Ph residue 1398–1484 may have disrupted an interaction with a negative regulator. While possible, no such protein has yet to be identified. Furthermore, we have shown that Ph RLink-mediated repression is lost when polymer-deficient mutations are introduced in Ph RLink showing that the repression is dependent on SAM polymerization (Fig. 6C).
While in vitro we have been able to increase polymerization of various Ph fragments, only Ph RLink shows similar or slightly better repressive ability in our transcription assay (Fig. 6, A and B). This discrepancy may be the consequence of the precise structural features of the polymer structure unique to Ph that are required for Ph function. The affinity of the TEL SAM-TEL SAM interaction is ten times the strength of the Ph SAM-Ph SAM interaction and would lead to greater polymerization for the isolated TEL SAM. However, the TEL SAM polymer repeat distance is 53 Å (2) compared with 45 Å for the Ph SAM polymer (3) and may not ultimately assemble the precise chromatin architecture required for Ph-mediated repression. For the linker deletion constructs, it is possible that a certain length of linker sequence is required to create a proper repressive structure. It is worth noting that the number of residues measured between the end of the FCS domain and beginning of the SAM domain in Ph is 118. The three human Ph orthologs all have similar number of linker residues ranging from 103 to 121 amino acids (Fig. 4A). Thus, shortening the linker, while resulting in longer polymers for the small Ph fragments, may have altered the polymer structure sufficiently enough to negatively affect transcription repression ability. Ph RLink has the identical number of amino acids as wild-type Ph which may allow the fully repressive competent polymer structure to form.
Like many proteins that contain a SAM domain that can polymerize, Ph functions as a transcriptional repressor. However, neither the molecular mechanisms of repression mediated by Ph nor the role played by the SAM domain is known. While SAM polymerization provided an attractive model to explain how a repressive PcG complex might spread along chromatin, the lack of open-ended polymer structures suggests SAM polymers play an alternative repressive role. Given the results presented here, it is possible that polymeric SAM-containing proteins exist in a limited number of dynamic oligomeric states, instead of participating as an open-ended polymer. That is, they are able to take on multiple, but limited, stoichiometries to accommodate specific repressive chromatin architectures associated with different genetic loci. For Ph SAM, one possible role for the SAM domain is in mediating localized, higher order repressive chromatin structures. Scanning force microscopy studies of PRC1 bound to DNA revealed the presence of a left-handed helical architecture where the DNA is wrapped around several PRC1 units (18). We speculate that the structure of this PRC1 oligomer is mediated by short (four to six) PRC1 units. Alternatively, it has been suggested (38) that PcG mediated long range interactions (39) may occur through in trans PRC1 recruitment of different Polycomb response elements, perhaps through Ph SAM-Ph SAM interactions. Either mechanism of gene silencing would be enhanced with increased Ph SAM polymerization with RLink. The increased polymerization may result in greater difficulty in disassembling these complexes required during gene replication thus resulting in the reduced tissue growth phenotype observed for Ph RLink (Fig. 6D).
What remains to be determined is the precise mechanism or mechanisms by which the unstructured residues influence polymerization. Conformational entropy of the linker appears to play a key role in controlling SAM polymerization as attaching RLink to Ph SAM results in smaller polymers, though not as small as Ph SAM with its native linker. This result implies that any unstructured linker attached to a polymeric SAM domain could hinder polymerization. To this point, we have been able to obtain soluble forms of polymeric SAM domains, which by themselves are insoluble due to polymerization, by attaching RLink to them.3 The reason why an unstructured stretch of amino acids results in making shorter polymer units could stem from the helical architecture of SAM polymers. Such an architecture would make it favorable for unstructured conformations of the linker attached to the first SAM unit in the polymer to occupy conformations that would be in place to sterically hinder the addition of SAM polymer units as the extending polymer returns near the same side of the helical structure as the first SAM unit. This polymer limiting mechanism would only require conservation of the helical architecture, which is the only consistent characteristic among the known SAM polymer structures (2–8). Additional factors specific to the amino acids present in the linker also appears to play a role as a linker with the same amino acid content as the native linker but arranged in a scrambled order can still hinder polymerization as well as the native linker (Fig. 4D). We have also observed direct contacts between the native linker and SAM domain in trans (Fig. 5C), though the exact consequence of this interaction has not yet been determined. Further investigation of the role of the linker will be required to determine the mechanism and precise role of the Ph SAM linker and those of other SAM polymers.
Supplementary Material
Acknowledgments
We thank Dr. Susan T. Weintraub for comments on the manuscript. The UltraScan development is supported by NIH Grant RR022200/RGM103662B (to B. D.). Computational analysis was performed at the Texas Advanced Computing Center on Lonestar and Ranger using NSF TeraGrid allocation TG-MCB070039 (to B. D.).
This work was supported by the American Cancer Society (RSG-08-285-01-GMC), the American Heart Association (0830111N), and the Department of Defense Breast Cancer Research Program (BC075278) (to C. A. K.).

This article contains supplemental Figs. S1–S4 and Table S1.
C. A. Kim, unpublished data.
- SAM
- sterile alpha motif
- Ph
- polyhomeotic
- PcG
- polycomb group
- PRC
- polycomb repression complex
- AFM
- atomic force microscopy
- vHW
- van Holde-Weischet
- RLink
- random linker.
REFERENCES
- 1. Qiao F., Bowie J. U. (2005) The many faces of SAM. Sci. STKE 2005, re7. [DOI] [PubMed] [Google Scholar]
- 2. Kim C. A., Phillips M. L., Kim W., Gingery M., Tran H. H., Robinson M. A., Faham S., Bowie J. U. (2001) Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EMBO J. 20, 4173–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim C. A., Gingery M., Pilpa R. M., Bowie J. U. (2002) The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 9, 453–457 [DOI] [PubMed] [Google Scholar]
- 4. Kim C. A., Sawaya M. R., Cascio D., Kim W., Bowie J. U. (2005) Structural organization of a Sex-comb-on-midleg/polyhomeotic copolymer. J. Biol. Chem. 280, 27769–27775 [DOI] [PubMed] [Google Scholar]
- 5. Qiao F., Song H., Kim C. A., Sawaya M. R., Hunter J. B., Gingery M., Rebay I., Courey A. J., Bowie J. U. (2004) Derepression by depolymerization; structural insights into the regulation of Yan by Mae. Cell 118, 163–173 [DOI] [PubMed] [Google Scholar]
- 6. Baron M. K., Boeckers T. M., Vaida B., Faham S., Gingery M., Sawaya M. R., Salyer D., Gundelfinger E. D., Bowie J. U. (2006) An architectural framework that may lie at the core of the postsynaptic density. Science 311, 531–535 [DOI] [PubMed] [Google Scholar]
- 7. Harada B. T., Knight M. J., Imai S., Qiao F., Ramachander R., Sawaya M. R., Gingery M., Sakane F., Bowie J. U. (2008) Regulation of enzyme localization by polymerization: polymer formation by the SAM domain of diacylglycerol kinase δ1. Structure 16, 380–387 [DOI] [PubMed] [Google Scholar]
- 8. Di Pietro S. M., Cascio D., Feliciano D., Bowie J. U., Payne G. S. (2010) Regulation of clathrin adaptor function in endocytosis: novel role for the SAM domain. EMBO J. 29, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meruelo A. D., Bowie J. U. (2009) Identifying polymer-forming SAM domains. Proteins 74, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wood L. D., Irvin B. J., Nucifora G., Luce K. S., Hiebert S. W. (2003) Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwartz Y. B., Pirrotta V. (2008) Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20, 266–273 [DOI] [PubMed] [Google Scholar]
- 12. Peterson A. J., Mallin D. R., Francis N. J., Ketel C. S., Stamm J., Voeller R. K., Kingston R. E., Simon J. A. (2004) Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics 167, 1225–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyba M., Brock H. W. (1998) The SAM domain of polyhomeotic, RAE28, and scm mediates specific interactions through conserved residues. Dev. Genet. 22, 74–84 [DOI] [PubMed] [Google Scholar]
- 14. Shao Z., Raible F., Mollaaghababa R., Guyon J. R., Wu C. T., Bender W., Kingston R. E. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98, 37–46 [DOI] [PubMed] [Google Scholar]
- 15. Francis N. J., Saurin A. J., Shao Z., Kingston R. E. (2001) Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8, 545–556 [DOI] [PubMed] [Google Scholar]
- 16. Saurin A. J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R. E. (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412, 655–660 [DOI] [PubMed] [Google Scholar]
- 17. Francis N. J., Kingston R. E., Woodcock C. L. (2004) Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 18. Mohd-Sarip A., van der Knaap J. A., Wyman C., Kanaar R., Schedl P., Verrijzer C. P. (2006) Architecture of a polycomb nucleoprotein complex. Mol. Cell 24, 91–100 [DOI] [PubMed] [Google Scholar]
- 19. Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schatz P. J., Cull M. G., Martin E. L., Gates C. M. (1996) Screening of peptide libraries linked to lac repressor. Methods Enzymol. 267, 171–191 [DOI] [PubMed] [Google Scholar]
- 21. Demeler B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., Rowe A. J., eds) pp. 210–229, Royal Society of Chemistry, UK [Google Scholar]
- 22. Brookes E., Cao W., Demeler B. (2010) A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 39, 405–414 [DOI] [PubMed] [Google Scholar]
- 23. Demeler B., van Holde K. E. (2004) Sedimentation velocity analysis of highly heterogeneous systems. Anal. Biochem. 335, 279–288 [DOI] [PubMed] [Google Scholar]
- 24. Brookes E., Demeler B. (2006) in Analytical Ultracentrifugation VIII, Progr. Colloid Polym. Sci. (Wandrey C., Colfen H., eds) pp. 78, Springer [Google Scholar]
- 25. Demeler B., Brookes E. (2008) Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 286, 129–137 [Google Scholar]
- 26. Brookes E., Demeler B. (2008) Parallel computational techniques for the analysis of sedimentation velocity experiments in UltraScan. Colloid Polym. Sci. 286, 139–148 [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28. Johnson B. A., Blevins R. A. (1994) NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 29. Zhang H., Christoforou A., Aravind L., Emmons S. W., van den Heuvel S., Haber D. A. (2004) The C. elegans Polycomb gene SOP-2 encodes an RNA-binding protein. Mol. Cell 14, 841–847 [DOI] [PubMed] [Google Scholar]
- 30. Wang R., Ilangovan U., Leal B. Z., Robinson A. K., Amann B. T., Tong C. V., Berg J. M., Hinck A. P., Kim C. A. (2011) Identification of nucleic acid binding residues in the FCS domain of the polycomb group protein polyhomeotic. Biochemistry 50, 4998–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brookes E., Demeler B. (2007) Parsimonious Regularization using Genetic Algorithms Applied to the Analysis of Analytical Ultracentrifugation Experiments, GECCO Proceedings ACM 978-1-59593-69-4/07/00078 [Google Scholar]
- 32. Demeler B., Saber H., Hansen J. C. (1997) Identification and interpretation of complexity in sedimentation velocity boundaries. Biophys. J. 72, 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunker A. K., Silman I., Uversky V. N., Sussman J. L. (2008) Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18, 756–764 [DOI] [PubMed] [Google Scholar]
- 34. Randsholt N. B., Maschat F., Santamaria P. (2000) polyhomeotic controls engrailed expression and the hedgehog signaling pathway in imaginal discs. Mech. Dev. 95, 89–99 [DOI] [PubMed] [Google Scholar]
- 35. Martinez A. M., Schuettengruber B., Sakr S., Janic A., Gonzalez C., Cavalli G. (2009) Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat. Genet. 41, 1076–1082 [DOI] [PubMed] [Google Scholar]
- 36. Classen A. K., Bunker B. D., Harvey K. F., Vaccari T., Bilder D. (2009) A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat. Genet. 41, 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng S., Huang J., Wang J. (2011) Loss of the Polycomb group gene polyhomeotic induces non-autonomous cell overproliferation. EMBO Rep. 12, 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lavigne M., Francis N. J., King I. F., Kingston R. E. (2004) Propagation of silencing; recruitment and repression of naive chromatin in trans by polycomb repressed chromatin. Mol. Cell 13, 415–425 [DOI] [PubMed] [Google Scholar]
- 39. Lanzuolo C., Roure V., Dekker J., Bantignies F., Orlando V. (2007) Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 10, 1167–1174 [DOI] [PubMed] [Google Scholar]
- 40. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.