Background: The serine deaminase CHA1 responds to heat stress in a sphingolipid-dependent manner.
Results: CHA1 requires de novo sphingoid base production for induction by serine, limiting growth-suppressing accumulation of sphingoid bases.
Conclusion: Sphingoid bases are feedback sensors of serine availability, forming a feedforward/feedback loop through CHA1.
Significance: This study defines a fundamental connection between sphingolipid and amino acid metabolic pathways with implications for disease.
Keywords: Amino Acid, Lipids, Serine, Sphingolipid, Yeast, Heat Stress, Sphingoid
Abstract
Targets of bioactive sphingolipids in Saccharomyces cerevisiae were previously identified using microarray experiments focused on sphingolipid-dependent responses to heat stress. One of these heat-induced genes is the serine deamidase/dehydratase Cha1 known to be regulated by increased serine availability. This study investigated the hypothesis that sphingolipids may mediate the induction of Cha1 in response to serine availability. The results showed that inhibition of de novo synthesis of sphingolipids, pharmacologically or genetically, prevented the induction of Cha1 in response to increased serine availability. Additional studies implicated the sphingoid bases phytosphingosine and dihydrosphingosine as the likely mediators of Cha1 up-regulation. The yeast protein kinases Pkh1 and Pkh2, known sphingoid base effectors, were found to mediate CHA1 up-regulation via the transcription factor Cha4. Because the results disclosed a role for sphingolipids in negative feedback regulation of serine metabolism, we investigated the effects of disrupting this mechanism on sphingolipid levels and on cell growth. Intriguingly, exposure of the cha1Δ strain to high serine resulted in hyperaccumulation of endogenous serine and in turn a significant accumulation of sphingoid bases and ceramides. Under these conditions, the cha1Δ strain displayed a significant growth defect that was sphingolipid-dependent. Together, this work reveals a feedforward/feedback loop whereby the sphingoid bases serve as sensors of serine availability and mediate up-regulation of Cha1 in response to serine availability, which in turn regulates sphingolipid levels by limiting serine accumulation.
Introduction
Sphingolipids constitute a unique class of lipids that have been implicated in a variety of functions in yeast, including nutrient uptake and cell cycle regulation. In particular, several responses to heat stress have been shown to require de novo synthesis of sphingolipids, and these include cell cycle arrest, proteolysis, and nutrient import (1–6).
Microarray analysis analyzing the effects of heat stress on gene expression in the lcb1-100 strain, defective in de novo synthesis, was previously used to define genes whose regulation depended on de novo synthesis of sphingolipids (7). An especially intriguing gene among these is CHA1, which encodes a serine deamidase/dehydratase, known to be up-regulated by exogenous serine (8, 9). This was of great interest because the enzyme serves to attenuate serine levels and because serine also serves as a limiting substrate in the de novo synthesis of sphingolipids at 30 °C (6). This mutual sensitivity to serine and the dependence of CHA1 up-regulation on sphingolipid synthesis suggested the intriguing possibility of a negative feedback mechanism balancing gene expresssion, serine, and sphingolipid levels.
The first step in de novo sphingolipid synthesis is the condensation of serine and palmitate by the serine palmitoyl transferase (SPT)2 complex, followed by reduction, to generate the sphingoid base dihydrosphingosine (DHS), which is then hydroxylated to form the second sphingoid base phytosphingosine (PHS). Ceramide synthases (Lac1 and Lag1 in Saccharomyces cerevisiae) further modify the sphingoid bases by addition of various fatty acids to generate dihydroceramide and phytoceramide species distinguished by the length, saturation, and/or hydroxylation of the fatty acids in amide linkage. Sphingoid bases are also modified by phosphorylation through the action of sphingoid base kinases (Lcb4 and Lcb5 in S. cerevisiae) to generate the phosphosphingoid bases. Ceramide species are further modified by addition of the sugars mannose and inositol to form the complex sphingolipids. Combinations of these covalent modifications produce a vast variety of sphingolipid species, several of which have been implicated in regulation of specific signaling pathways (1). Blocked CHA1 up-regulation could be due to blocked synthesis of any one of this multitude of lipid species or a subset thereof.
In this study, we aimed to explore the hypothesis that sphingolipid-dependent induction of CHA1 defines a feedback loop in serine metabolism mediated by sphingolipids. The results show that serine induces CHA1 through driving de novo synthesis of sphingolipids. Moreover, the results show that dysfunction of this pathway leads to hyperaccumulation of intermediate sphingolipids that results in growth suppression in response to high serine. These results support the hypotheses that sphingoid bases serve as sensors of serine availability and that CHA1 is involved in a novel negative feedback mechanism whereby Cha1 regulates sphingolipid levels by limiting available serine, and in turn sphingolipids regulate the expression of CHA1 in response to serine availability.
EXPERIMENTAL PROCEDURES
Yeast Strains and Culture Conditions
Genotypes of wild-type, temperature-sensitive, and deletion mutant strains are described in Table 1. SUR2 was deleted in JK9-3dα strains to obtain JK9-3dα sur2Δ using the following primers (forward, 5′-ACT CCG GGT CTT CGT CTT TAC TG-3′, and reverse, 5′-ACA AAC GTG GGA AGT CGG AGA C-3′) to amplify the SUR2 locus containing KanMX sequence from a genomic DNA of BY4742 sur2Δ. For the PCR, the polymerase was activated at 95 °C for 1.5 min, then cycled 35 times with at 55 °C and 72 °C elongation and annealing temperatures, respectively. The PCR product was transformed into JK9-3dα, and then G418-resistant colonies were selected for (yeast lithium acetate transformation methods). The deletion of SUR2 was verified by PCR and sequencing to confirm the replacement of SUR2 locus by KanMX sequence. YPD medium was used for the heat stress experiment. For all other experiments, synthetic complete threonine dropout (SC −Thr) medium containing 0.17% yeast nitrogen base (US Biological), 0.5% ammonium sulfate, 2 mm sodium hydroxide, and 0.07% dropout supplement, was used. The dropout supplement contains 5% of the following: adenine, uracil, tryptophan, histidine, arginine, methionine, as well as tyrosine (8%), leucine (15%), lysine (8%), phenylalanine (13%), and aspartate (26%). All dropout components were purchased from Acros Organics (Fisher Scientific), except for methionine and aspartate, purchased from Sigma-Aldrich. Serine powder was added to the medium where specified. PHS and DHS were obtained either from the Medical University of South Carolina (MUSC) Lipidomics Core or Avanti Polar Lipids. Myriocin and serine were purchased from Sigma-Aldrich, fumonisin was purchased from Enzo Clinical Labs (Farmingdale, NY), and aureobasidin was purchased from Clontech. Sphingoid bases were delivered in dimethyl sulfoxide. Myriocin and aureobasidin as in Ref. 10 were delivered in methanol, and fumonisin was delivered in water. For heat stress, cultures were grown in liquid YPD, for serine and sphingoid base induction experiments, cells were grown in liquid SC −Thr (Clontech). For all experiments, cells were grown to mid-log at 30 °C from 1 ml of SC −Thr overnight cultures. Heat stress was performed by shifting cells to a 39 °C water bath. Cultures were harvested by centrifugation at 3500 rpm for 3 min and stored at −80 °C. For spot tests, YPD and SC −Thr plates where prepared with the same composition as for liquid medium, but with 2% agar. Myriocin, aureobasidin, and fumonisin were added at 5, 0.1, and 500 μm, respectively, all plates contain 0.025% methanol vehicle. Plates were spotted from mid-log cultures grown in SC −Thr liquid medium and diluted in water.
TABLE 1.
Yeast strains used in this work
| Strain | Genotype | Source |
|---|---|---|
| JK9–3dα | MATα leu2-3,112 ura3-52 rme1 trp1 his4 | Laboratory collection |
| lcb4Δ,lcb5Δ | MATα leu2-3, 112 ura3-52 rme1 trp1 his4 lcb4::G418 lcb5::G418 | Robert Dickson |
| cha1Δ | MATα leu2-3,112 ura3-52 rme1 trp1 his4 cha1::G418 | This study |
| BY4742 | MATα his3 leu2 lys2 ura3 | Euroscarf |
| cha4Δ | MATα his3 leu2 lys2 ura3 cha4::G418 | Euroscarf |
| Dau15 | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ | Robert Dickson |
| pkh1-ts,pkh2Δ | MATa ura3Δ0 leu2Δ0 trp1Δ his2Δ ade1Δ pkh1D398G pkh2::LEU2 | Robert Dickson |
RNA and cDNA Preparation
RNA was extracted from 2 × 107 cells using the hot acid phenol method (11). Cells were resuspended in 400 μl of TES (Tris-HCl, pH 7.5, 10 mm EDTA, 0.5% SDS) and treated with 400 μl of hot phenol. RNA was further purified using the Qiagen RNeasy RNA cleanup protocol, and the concentration was determined using the A260 nm after confirming a 260 nm/280 nm absorbance unit ratio of 1.9–2.1. Synthesis of cDNA was carried out with 0.5 μg of total RNA using the Invitrogen SSII first strand cDNA synthesis protocol.
Quantitative Real Time PCR
Real-time PCR was performed in triplicate in 25-μl reactions using the Bio-Rad SYBR Green solution on a Bio-Rad iCycler according to suggested protocols. Cycles were (i) 95 °C for 3 min; (ii) 95 °C for 10 s, 55 °C for 1 min × 40 cycles; (iii) 95 °C for 1 min; and (iv) 55 °C for 1 min. Mean normalized expression was obtained using the Q-gene formulae (12). For serine induction experiments, expression values were normalized relative to the reference gene RDN18. Expression for sphingoid base induction experiments were normalized to ACT1, due to confounding effects of sphingoid bases on RDN18. Primers used are as follows: CHA1 forward, GAT CTC CTC AGG TTT TCG CTA GTT C and reverse, ACC ATT CTC TTC TTT GTC GCT GTA G; RDN18 forward, CCA TGG TTT CAA CGG GTA ACG and reverse, GCC TTC CTT GGA TGT GGT AGC C; ACT1 forward, ATC ACC GCT TTG GCT CCA T and reverse, CCA ATC CAG ACG GAG TAC TTT CTT.
Measurement of Sphingoid Bases and Ceramides
Cells were grown to mid-log, treated, and harvested at 3500 rpm for 3 min, and then pellets were flash frozen in a methanol/dry ice bath. 1.19 × 109 cells were resuspended in 1 ml of the extraction solvent (ethanol, diethyl ether, pyridine, ammonium hydroxide, water (50:10:2:25:15)), placed in screw-cap vials containing 0.5 ml of glass beads (Sigma), and vortexed at 4 °C at maximum speed for 12 min to homogenize. The resulting suspension was diluted by adding an additional 1 ml of extraction solvent, incubated at 60 °C for 15 min, and centrifuged at 3500 rpm for 10 min at room temperature, and then the supernatant was transferred to a fresh tube. The pellet was resuspended in a fresh 2 ml of extraction solvent and then incubated and centrifuged as above. The two supernatants were combined, and the extract was dried down. Lipids were quantified according to the method described in Ref. 13. Lipid levels were normalized to total protein.
Measurement of Total Protein
Culture aliquots containing 1.19 × 107 cells were harvested in 1.5-ml screw-cap vials. Cells were pelleted and resuspended in 500 μl of lysis buffer (25 mm Tris-HCl, pH 7.4, 5 mm EDTA, 5 mm DTT, 1 mm PMSF, and Protease Inhibitor Mixture), 250 μl of glass beads were added, and cells were homogenized by vortexing at 4 °C for 12 min total, in 3-min intervals. Lysate was centrifuged at 600 × g for 10 min, and total protein in the supernatant was measured by Bio-Rad BCA protein assay using BSA as a calibration standard.
Analysis of Complex Sphingolipids
Cells were grown to mid-log in SC −Thr medium and treated with medium containing 10 or 50 mm serine and 20 μCi/ml [3H]inositol (American Radiochemicals). 6.5 × 107 cells were spun and extracted by the Mandala method (14), then resuspended in 30 μl of 2:1 chloroform and methanol, and the total volume was spotted on a TLC plate (Whatman 60 Å, 250 μm, 20-cm LK6D). Lipids were separated in (9:7:2) chloroform, methanol, and ammonium hydroxide (4.2 n). Plates were developed on x-ray film (Thermo Scientific) at −80 °C for 48 h. Spots were identified by comparing samples labeled with [3H]inositol versus those labeled with [3H]serine (supplemental Fig. S3) and by comparison with published data (15–17). Lipids were quantified by densitometry using ImageJ software. Spot density was converted to moles of lipid by calibrating spot densities containing a known quantity of [3H]inositol.
Measurement of Intracellular Serine
Cells were grown to early log in SC −Thr, transferred to fresh medium containing 0, 10, or 50 mm serine, grown for 21 h, and then 2.6 × 108 cells were harvested by centrifugation at 3500 × g for 3 min. Cells were quenched by methanol chilled to −40 °C, spun for 30 s at 16,100 × g, then washed again with cold methanol to remove residual serine left from the medium (18). Serine was extracted by resuspending and boiling in 1 ml of deionized water according to Ref. 19. Intracellular serine was measured by the method described in Ref. 20 employing a Waters 717plus/1525/2487 HPLC autosampler/pump/detector, with some modifications: amino acids were separated on an ES Industries Chromega 150 × 4.6- mm, MC18, 60-Å column. Serine was identified and quantified using the Amino Acid H standard mixture (Pierce) as a calibration standard.
Statistical Analysis
AVOVA with the Bonferroni post test was used to assign significance at p < 0.01 or p < 0.05 as specified. The specific statistical method and the number of replicates for each experiment are indicated in the figure legends.
RESULTS
De Novo Sphingolipid Synthesis Mediates Induction of CHA1 in Response to Serine
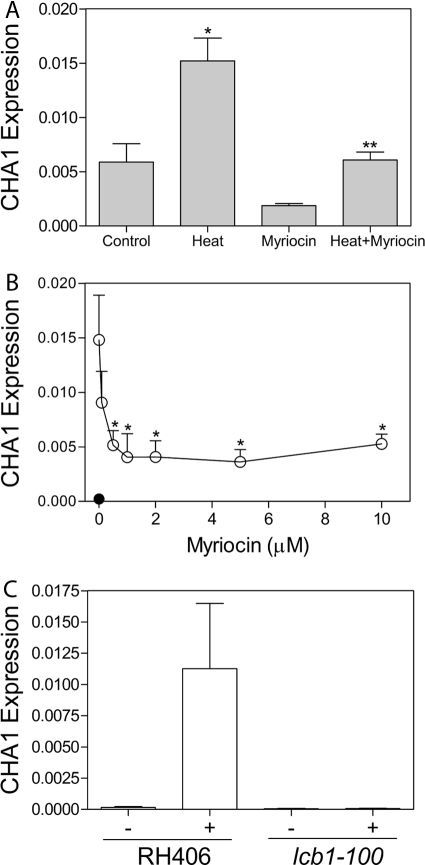
In a genomic approach, it was shown previously that heat stress up-regulates the serine deamidase/dehydratase CHA1 but that this up-regulation fails in the lcb1-100 mutant, which has a defective SPT complex and thus cannot efficiently carry out the first step of de novo sphingolipid synthesis (7). However, the lcb1-100 mutant also exhibits base-line effects on sphingolipids, and therefore, it does not fully discriminate acute effects of sphingolipids. Therefore, to evaluate the role of acute de novo synthesis directly, we employed myriocin, a pharmacological inhibitor of SPT. Yeast cells were subjected to heat stress in the presence or absence of myriocin, and the levels of CHA1 RNA were quantitated using quantitative RT-PCR (qRT-PCR). The results showed that myriocin significantly blocked the up-regulation of CHA1 in response to heat stress (Fig. 1A). Taken together, these results establish that acute de novo synthesis is required for heat induction of CHA1.
FIGURE 1.
De novo sphingolipid synthesis is required to mediate CHA1 induction in response to heat stress and to serine availability. A, CHA1 expression was measured by qRT-PCR before and after induction by heat stress with and without myriocin. Cells were grown to mid-log in YPD, pretreated with 5 μm myriocin (or vehicle) for 45 min, and then shifted from 30 to 39 °C for 15 min. The difference in expression between control and heat stress is significant (*), and the difference in expression between heat and myriocin plus heat is significant (**) by one-way ANOVA, p < 0.05. B, CHA1 was induced by serine over a range of myriocin doses from 0 to 10 μm (open circles); expression without induction by serine is shown (filled circle). JK9-3dα cells were grown to mid-log phase in SC −Thr, pretreated for 45 min with the specified dose of myriocin or vehicle, and then induced with 0.5 mm serine for 15 min. * Indicates that the difference between the vehicle (zero) and the indicated data point is significant as determined by one-way ANOVA, p < 0.05. C, lcb1-100 mutant and RH406 background strain were treated with serine to determine the degree of CHA1 induction. Cells were grown to mid-log in SC −Thr medium and then induced with 0.5 mm serine for 15 min. The difference between the wild-type and mutant expression after treatment was confirmed to be significant by one-way ANOVA at p < 0.01. For all experiments data presented are the average ± S.E. (error bars) of at least three independent experiments.
CHA1 is known to be strongly up-regulated by serine (8, 21), and we have shown previously that effects of heat stress on sphingolipids are specifically mediated by heat-induced increases in serine uptake (6). Thus, it became important to determine whether CHA1 induction by serine in the absence of heat stress requires its incorporation into sphingolipids, as this would imply participation of sphingolipids in a negative feedback mechanism regulating serine metabolism. To address this hypothesis, cells were treated with serine at several doses of myriocin, and CHA1 expression was measured by qRT-PCR. As shown in Fig. 1B, myriocin blocked CHA1 up-regulation by serine in a dose-dependent manner with 50% inhibition observed at a concentration of 0.5 μm myriocin, indicating that sphingolipid synthesis is required for CHA1 up-regulation by serine in the absence of heat. To further confirm that myriocin blocks CHA1 up-regulation specifically by blocking the SPT activity, the temperature-sensitive lcb1-100 strain, which has defective SPT activity, and its wild-type background strain RH406 were treated with serine, and CHA1 expression was measured at semipermissive temperature. CHA1 was up-regulated dramatically in the background strain RH406 but completely blocked in the lcb1-100 mutant (Fig. 1C). The semipermissive temperature of 33 °C is sufficient to block lcb1-100 activity without stimulating the heat stress response. For all experiments where serine was used to induce CHA1, cells were grown and treated in SC −Thr medium to avoid high background induction of CHA1 by threonine, which also induces this gene. The results showing that CHA1 up-regulation requires de novo sphingolipid synthesis in the absence of heat stress demonstrate that sphingolipids are the key mediators of the induction of CHA1 by serine.
Sphingoid Bases PHS and DHS Mediate CHA1 Up-regulation
To determine which class of sphingolipids is required for CHA1 up-regulation by serine, the effects of serine on CHA1 were determined in several mutants deleted in key enzymes of the sphingolipid metabolic pathway. Specifically CHA1 was induced by serine in the tsc3Δ, lcb4Δ,lcb5Δ, lag1Δ, lac1Δ, and sur2Δ strains, and expression was measured by qRT-PCR (Fig. 2A). The first strain tested, tsc3Δ, is deleted in a nonessential subunit of the SPT complex, which has diminished SPT activity. The Tsc3 subunit is also implicated in formation of the less abundant C20 sphingoid bases (6), which have a 20-carbon chain. The tsc3Δ strain did not show up-regulation significantly different from wild type, disfavoring the C20 sphingoid bases as possible mediators. The lcb4Δ,lcb5Δ double deletion strain, deleted in both sphingoid base kinases LCB4 and LCB5, showed up-regulation that was not significantly different from wild type, implying that the sphingoid base phosphates are not required for mediating CHA1 expression. This is a relevant conclusion because previous work has implicated PHS-1-phosphate in up-regulation of specific genes during heat stress (4, 22, 23) and therefore distinguishes DHS/PHS versus PHS-1-phosphate-regulated pathways. The lag1Δ and lac1Δ strains deleted in the ceramide synthases showed normal up-regulation, disfavoring the possibility that ceramide synthesis is required for CHA1 up-regulation. It also argues against the possibility that complex sphingolipid production is necessary to mediate induction. Moreover, the sur2Δ strain deleted in the sphingoid base/ceramide hydroxylase that hydroxylates DHS and dihydroceramide into PHS and phytoceramide also showed complete up-regulation. Because the result on lag1Δ and lac1Δ disfavors ceramide, the sur2Δ result implies that either PHS or DHS could potentially mediate CHA1 up-regulation. Up-regulation of CHA1 in the other mutant strains tested implies that production of the sphingoid bases DHS and/or PHS is the specific reaction required for complete CHA1 induction.
FIGURE 2.
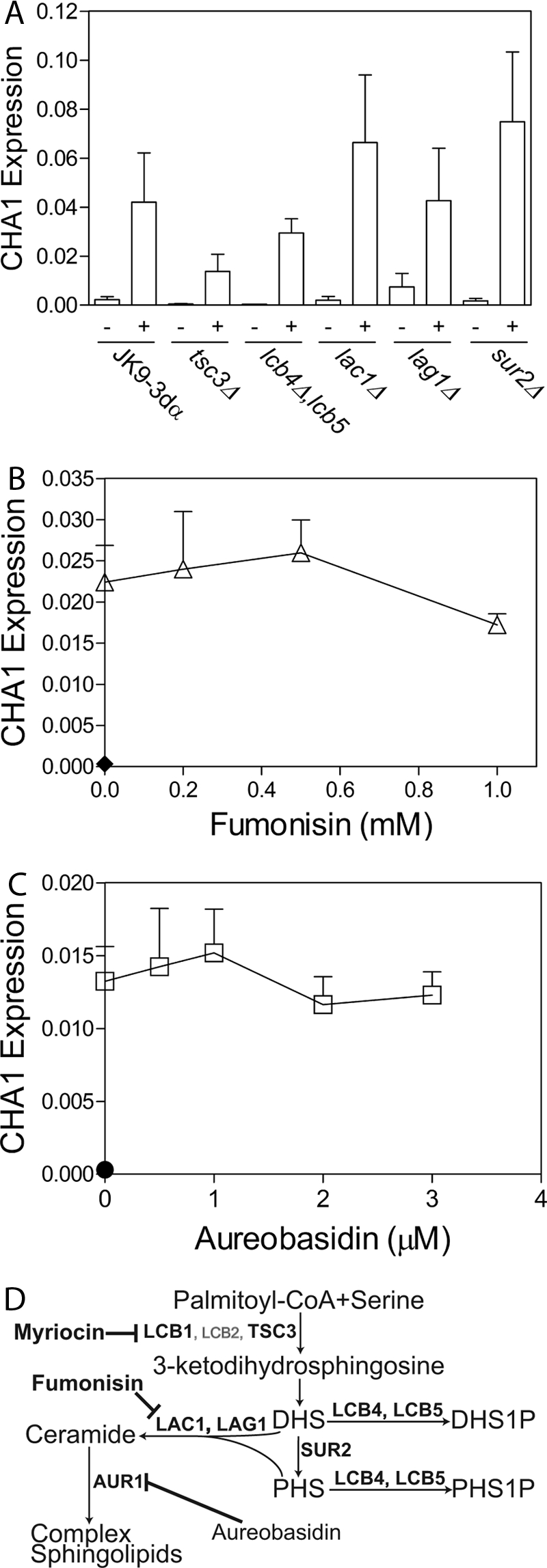
Sphingoid bases and not ceramide or phosphosphingoid bases are required for CHA1 up-regulation. CHA1 was induced by serine in key mutant strains deleted in key sphingolipid enzymes and after treatment with pharmacological inhibitors in the JK9-3dα background. A, wild-type and mutant strains were grown to mid-log in SC −Thr and induced with 0.5 mm serine for 15 min, and expression was measured by qRT-PCR. B and C, for fumonisin and aureobasidin treatment, cells were pretreated for 2 h with drug or vehicle (0.02% methanol) before induction. B, cells were not induced (filled diamond) or induced with serine after treatment with 0, 0.2, 0.5, and 1.0 mm fumonisin (open triangles), or 0, 0.5, 1, 2, and 3 μm aureobasidin (C) (open squares); expression with no induction is shown (filled circle). Induction by serine was found to be significant at p < 0.01 by one-way ANOVA in all samples. CHA1 expression was not significantly different in any of the mutant strains after induction at p < 0.05 confidence determined by two-way ANOVA. Neither pharmacological inhibitor showed significant inhibition of induction at p < 0.05 confidence by one-way ANOVA. Data presented are the average ± S.E. (error bars) of at least triplicate experiments. D, sphingolipid metabolic pathway illustrates the key points where the pathway was disrupted in this study using inhibitors or mutant strains indicated in bold font.
To confirm the conclusion derived from analysis of mutant strains, the pharmacological inhibitors fumonisin (Fig. 2B) and aureobasidin (Fig. 2C), inhibitors of ceramide (24) and complex sphingolipid synthesis (25), were employed, respectively. Neither inhibitor blocked CHA1 up-regulation (in contrast to myriocin). A small loss of up-regulation was observed at 1 mm fumonisin, but this is in excess of concentrations required to inhibit ceramide synthesis efficiently in S. cerevisiae (22, 26). Negative inhibition of up-regulation by fumonisin further consolidates the results with the lag1Δ and lac1Δ single mutants which still harbor some ceramide synthase activity; however, the lag1Δ,lac1Δ double mutant is very difficult to produce, and the only such strain available proved problematic, therefore demonstrating the effect of acute inhibition is critical to eliminating ceramide synthesis as a critical reaction for CHA1 up-regulation. Lack of inhibition of up-regulation of CHA1 by aureobasidin or by fumonisin provides strong evidence that formation of complex sphingolipids is not necessary for CHA1 up-regulation.
The above results strongly suggest that the sphingoid bases are necessary for CHA1 up-regulation. Therefore, it became important to determine whether DHS/PHS are also sufficient for up-regulation. To address this issue, the effects of the exogenous sphingoid bases DHS and PHS on CHA1 expression were evaluated by measuring dose- and time-dependent CHA1 induction by PHS/DHS. CHA1 was transiently up-regulated by both sphingoid bases, but the change in expression was variable (supplemental Fig. S1).
Yeast Protein Kinases Pkh1 and Pkh2 Mediate CHA1 Up-regulation via Transcription Factor Cha4
Previous work has implicated the yeast protein kinases Pkh1 and Pkh2 in mediating sphingoid base-dependent gene regulation (27–29), making them likely candidates as mediators of CHA1 up-regulation. Pkh1 and Pkh2 form a redundant but essential pair. A strain deleted in PKH2, but carrying a temperature-sensitive pkh1-ts allele was used to test the effects of loss of kinase function. CHA1 up-regulation in response to serine was markedly reduced in the pkh1-ts,pkh2Δ mutant strain, implying that the PKH kinases play a critical role in the up-regulation of CHA1 (Fig. 3A).
FIGURE 3.
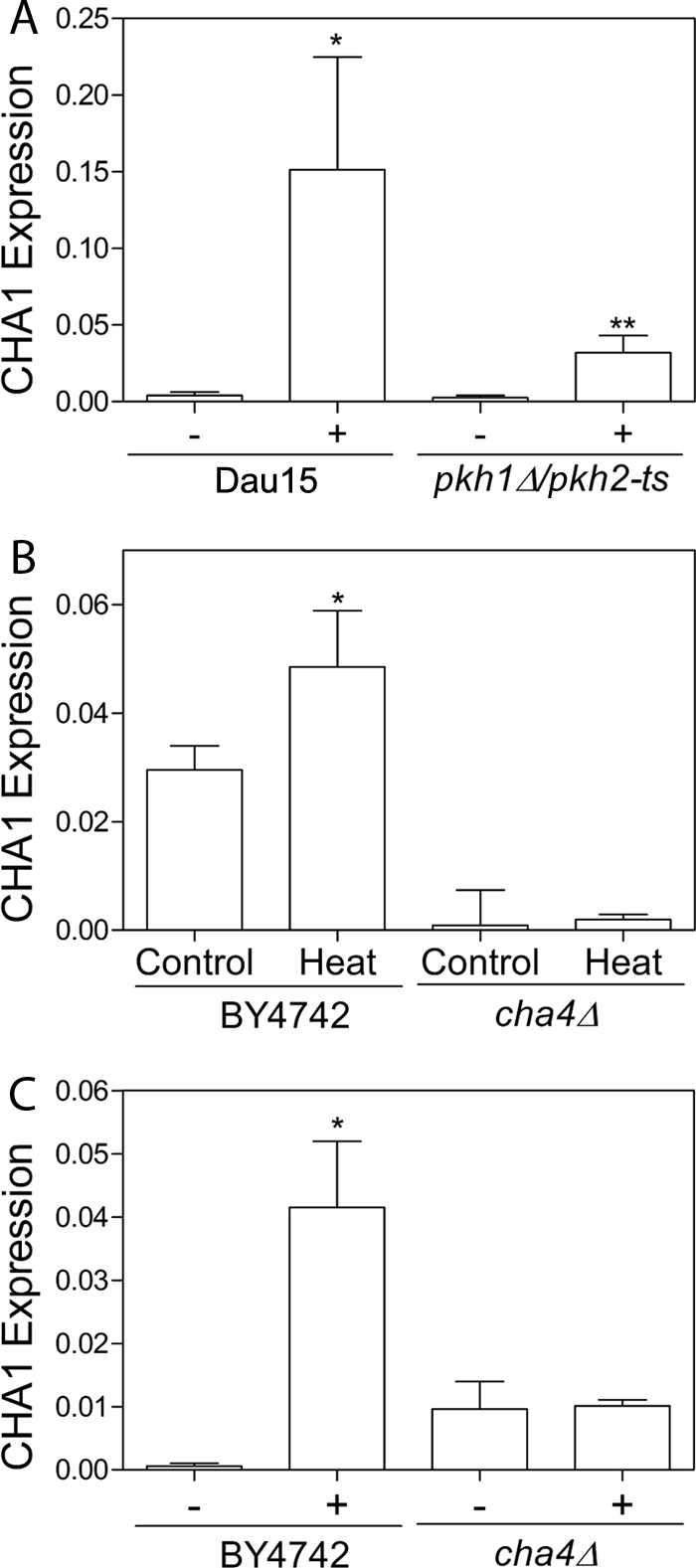
Increased sphingoid base signals CHA1 induction by transcription factor Cha4 via kinases Pkh1 and Phk2. CHA1 expression was measured by qRT-PCR after a 15-min induction with 0.5 mm serine in the pkh1-ts,pkh2Δ strain, in the cha4Δ strain and in their respective background strains Dau15 and BY4742. A, pkh1-ts,pkh2Δ strain and its background strain were shifted to 37 °C during the 15-min induction with serine. B and C, cha4Δ and its background strain were induced with 0.5 mm serine for 15 min at 30 °C in SC −Thr (C) or by 15 min at 39 °C in YPD (B). * Indicates that CHA1 was significantly up-regulated by serine or heat treatment in each background strain. ** Indicates that the difference in expression between Dau15 and pkh1-ts,pkh2Δ after treatment is significant. In both cases significance was determined by one-way ANOVA, p < 0.05. Data presented are the average ± S.E. (error bars) of triplicate experiments.
The transcription factor Cha4 is known to regulate CHA1 up-regulation (30, 31). Therefore, it was of interest to determine whether Cha4 is the key transcription factor mediating induction by heat as well as serine. CHA1 up-regulation by serine was measured in the cha4Δ deletion strain and in its BY4742 background strain, and it was found that up-regulation was blocked in cha4Δ, confirming its role in transcriptional activation (Fig. 3B). CHA1 was also induced in BY4742 by shifting to 39 °C for 15 min in YPD, and this induction was dependent on Cha4 (Fig. 3C). Thus, these results show that Cha4 is a key transcription factor in the induction of CHA1 in response to both heat stress and serine availability.
Serine Hyperaccumulation Leads to Hyperaccumulation of Sphingoid Bases and Ceramide in cha1Δ Strain under High Serine Load
The demonstration that serine is important for sphingolipid regulation coupled with the fact that Cha1 acts on serine led us to the hypothesis that induction of CHA1 serves not only to attenuate serine levels (thus defining a feedback pathway in serine metabolism), but also that CHA1 may then indirectly feed back on sphingolipid levels (see Fig. 6 for scheme). This speculation raises the more direct question of how the presence or absence of Cha1 might affect sphingolipid levels in the presence of high serine. To address this question, cha1Δ and its background strain JK9-3dα were grown in media with either a zero, low (10), or high (50 mm) serine concentration. The low serine condition corresponds to a concentration of serine typically found in rich media. Sphingolipids were extracted and measured by HPLC-tandem MS, and serine was extracted and measured by HPLC. It was found that after 21 h of growth, the mutant strain had accumulated 2–4-fold higher levels of both DHS, PHS as well as dihydroceramide and phytoceramide (Table 2), and 2-fold higher levels of serine. These results demonstrate that sphingolipid levels are sensitive to intracellular serine and that Cha1 is vital to regulating sphingolipid levels by way of regulating intracellular serine levels.
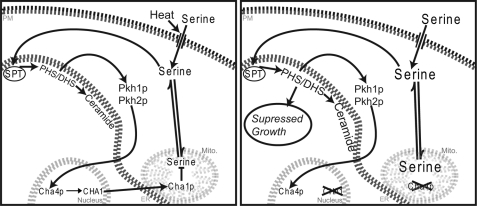
FIGURE 6.
Feedforward/feedback model for co-regulation of sphingolipid synthesis and CHA1 expression. Left, feedforward/feedback model maintains serine and sphingolipid homeostasis in the wild-type cell. Serine is taken up through the plasma membrane (PM), stimulated by heat stress or by increased serine in the medium. Intracellular serine passes into the endoplasmic reticulum (ER) and interacts with the SPT complex. The resulting increase in sphingoid bases stimulates increased CHA1 expression via Pkh1/Pkh2 and Cha4. Cha1 increases in the mitochondria (Mito.) and catabolizes serine, affecting total intracellular serine. Right, homeostasis breaks down in the cha1Δ strain under high serine load. Loss of Cha1 results in serine hyperaccumulation leading to hyperaccumulation of the sphingoid bases and ceramide and leading to suppressed growth.
TABLE 2.
Sphingoid bases and ceramides accumulate in cha1Δ with high serine load
The cha1Δ and JK9–3dα strains were grown in SC −Thr medium containing no serine or supplemented with 10 or 50 mm serine for 21 h. Equal numbers of cells for each treatment were harvested, extracted, quantified by HPLC-tandem MS, and normalized to total protein. Before treatment, cells were diluted from overnight cultures and grown to early log phase. Bold type indicates where the difference between cha1Δ and wild-type values is significant at p < 0.05 by one-way ANOVA. Data presented are the average ± S.E. of at least triplicate experiments.
| Metabolite | −Thr D.O. media |
10 mm serine |
50 mm serine |
|||
|---|---|---|---|---|---|---|
| JK9-3dα | cha1Δ | JK9-3dα | cha1Δ | JK9-3dα | cha1Δ | |
| pmol/μg protein | pmol/μg protein | pmol/μg protein | pmol/μg protein | pmol/μg protein | pmol/μg protein | |
| Phytoceramide | 10.5 ± 0.6 | 12.2 ± 0.3 | 9.1 ± 0.5 | 6.1 ± 0.2 | 8.95 ± 0.27 | 30.0 ± 1.6 |
| Dihydroceramide | 42.1 ± 0.9 | 41.1 ± 2.0 | 29.5 ± 1.7 | 27.9 ± 2.2 | 18.3 ± 1.1 | 45.2 ± 2.4 |
| Phytosphingosine | 3.35 ± 0.44 | 3.02 ± 0.4 | 5.6 ± 1.0 | 6.6 ± 1.0 | 3.63 ± 0.9 | 12.0 ± 3.1 |
| Dihydrosphingosine | 12.7 ± 0.75 | 13.0 ± 1.3 | 6.9 ± 0.7 | 6.9 ± 0.7 | 5.02 ± 1.2 | 21.5 ± 5.3 |
| Serine (×10−3) | 0.60 ± 0.01 | 0.57 ± 0.01 | 0.92 ± 0.02 | 1.43 ± 0.19 | 6.71 ± 1.09 | 13.1 ± 1.7 |
Next, the effects of Cha1 on regulating complex sphingolipid levels were evaluated using tritiated inositol to label the inositol-containing complex sphingolipids. The identified and labeled spots are phosphatidylinositol, inositol phosphoceramide, mannose-(inositol phospho)-ceramide (MIPC), and mannose-(inositol phospho)2-ceramide, and the results of three replicate experiments are shown (Fig. 4). No significant difference was observed between WT and cha1Δ (Table 3).
FIGURE 4.
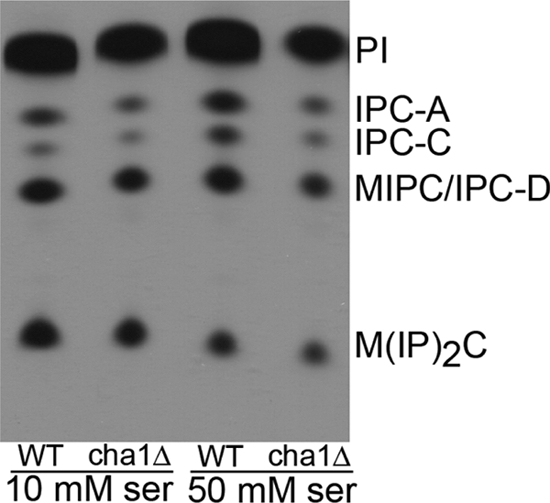
Complex sphingolipids do not accumulate in cha1Δ with high serine. The cha1Δ and JK9-3dα (WT) strains were grown in high and low serine media with 20 μCi of [3H]inositol for 21 h. Equal cells were extracted, separated by TLC, and visualized on x-ray film. The film shown is representative of triplicate experiments. PI, phosphatidylinositol; IPC, inositol phosphoceramide; MIPC, mannose-(inositol phosphoceramide); M(IP)2C, mannose-(inositol phospho)2-ceramide.
TABLE 3.
High serine availability does not lead to accumulation of complex sphingolipids
The cha1Δ and JK9-3dα strains were grown in SC −Thr medium containing 10 or 50 mm serine and 20 μCi of [3H]inositol for 21 h. Equal cells were extracted, separated by TLC, visualized on x-ray film, and quantified by densitometry. Data presented are the average ± S.E. of triplicate experiments.
| Lipid/106 cellsa | 10 mm serine |
50 mm serine |
||
|---|---|---|---|---|
| JK9-3dα | cha1Δ | JK9-3dα | cha1Δ | |
| pmol | pmol | pmol | pmol | |
| IPC-A | 0.07 ± 0.03 | 0.09 ± 0.00 | 0.07 ± 0.03 | 0.09 ± 0.01 |
| IPC-C | 0.09 ± 0.02 | 0.12 ± 0.60 | 0.06 ± 0.02 | 0.08 ± 0.03 |
| MIPC/IPC-D | 0.06 ± 0.03 | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.06 ± 0.00 |
| M(IP)2C | 0.13 ± 0.01 | 0.11 ± 0.07 | 0.08 ± 0.06 | 0.08 ± 0.07 |
a IPC, inositol phosphoceramide; MIPC, mannose-(inositol phosphoceramide); M(IP)2C, mannose-(inositol phospho)2-ceramide.
Cha1Δ Strain Shows Sphingolipid-dependent Sensitivity to High Serine
Based on the above results, we predicted that feedback regulation of sphingolipid synthesis by CHA1 exerts a physiologic function in attenuating possible effects of sphingoid bases on growth, especially because these molecules have been implicated in regulation of cell cycle and growth in yeast (3, 32, 33). To test this prediction, a spot test was used to demonstrate sensitivity of the cha1Δ strain to high serine load. Relative to the WT, the cha1Δ strain showed almost no growth on high serine (50 mm) plates, but was able to grow normally on YPD or low serine (10 mm) plates (Fig. 5). To determine whether this sensitivity to high serine was due to accumulation of excess sphingolipids, plates were also prepared with 5 μm myriocin. The presence of myriocin almost completely rescued the serine sensitivity phenotype (Fig. 5), implying that in the absence of Cha1, high serine leads to cell death arising from unregulated sphingolipid production.
FIGURE 5.
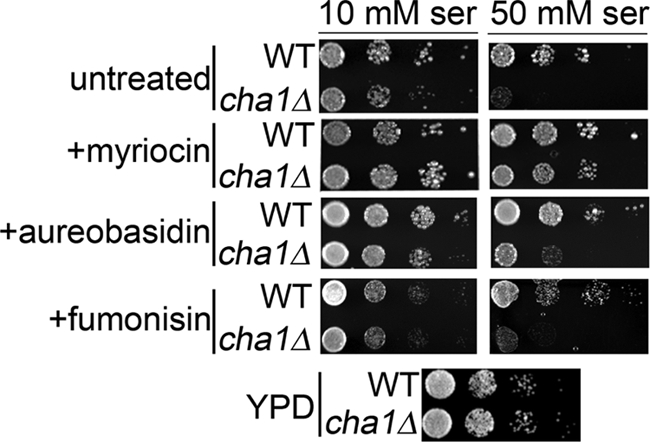
cha1Δ strain shows a sphingoid base-dependent sensitivity to high serine. The cha1Δ and JK9-3dα (WT) strains were spotted on agar plates made with SC −Thr medium supplemented with 10 and 50 mm serine. Matched plates were made with myriocin, aureobasidin, fumonisin, or vehicle. An additional YPD plate shows growth in the absence of nutrient limitation or drug treatment. Plates were incubated at 30 °C for 3 days. Cells were grown in liquid SC −Thr medium to mid-log phase and diluted in water to an A600 of 0.3, and then 5 μl of successive 10:1 dilutions were spotted with the most to least dilute cell suspensions arranged from left to right. Plates shown are representative of five independent experiments.
Although the results on CHA1 induction implicated sphingoid bases as the active species, it is not necessary that they are also the species involved in the growth suppression, which may also be mediated by the observed increase in ceramides or other sphingolipids. Therefore, in addition to myriocin, fumonisin and aureobasidin were used to determine whether synthesis of ceramide and complex sphingolipoids, respectively, are required to give rise to the serine sensitivity phenotype. However, neither fumonisin nor aureobasidin showed rescue of the phenotype to a degree similar to that of myriocin (Fig. 5) at concentrations where inhibition of lipid synthesis was confirmed (supplemental Fig. S4). Only inhibition of de novo sphingolipid synthesis at the initial step led to rescue of the high serine phenotype, thus implying that accumulation of the sphingoid bases specifically leads to the serine sensitivity phenotype.
DISCUSSION
This study reveals that sphingolipid synthesis is necessary for mediating the induction of the serine deamidase/dehydratase Cha1 in response to increased serine availability and/or uptake, which can arise from increased extracellular serine or in response to heat stress (6), respectively. The results implicate the sphingoid bases PHS/DHS as the likely key sphingolipid species required for induction illustrated in the left panel of Fig. 6. Moreover, the results show that the cha1Δ strain is unable to regulate sphingolipid synthesis, leading to hyperaccumulation of sphingoid bases and ceramides when serine availability is high. This unregulated sphingolipid synthesis leads to growth inhibition under the same conditions, as illustrated in the right panel of Fig. 6.
These results indicate the presence of a feedforward/feedback mechanism tying together the regulation of serine and sphingolipid metabolism. In turn, the results carry implications for human disease, including cancer, central nervous system (CNS) development, and neurological disease.
A major conclusion from this study is that sphingoid bases function as key sensors and signaling mediators of serine availability. It was previously shown that CHA1 is up-regulated by increased availability of serine (8), which was confirmed in this work. The results from this study demonstrate, by blocking de novo sphingolipid synthesis using myriocin and the lcb1-100 mutant, that sphingolipid synthesis is necessary for induction of CHA1 in response to increased serine availability. Moreover, previous results from our group showed that exogenous serine drives de novo synthesis of sphingolipids (6). Taken together, these results show that endogenous sphingolipids respond directly to exogenous serine levels and then mediate the effects of serine on CHA1 induction; thus fulfilling criteria as “sensors” of serine availability.
Interestingly, previous work has shown that sphingolipid synthesis is up-regulated by heat stress (23), and previous work from our group has shown that this effect requires a heat-mediated increase in serine uptake (6, 22). Moreover, CHA1 was one of the genes that were highly induced in response to acute heat stress (7). Therefore, this proposed role of sphingolipids in sensing serine extends to heat stress and possibly other mechanisms that may regulate serine availability. Such mechanisms now appear to employ this intrinsic mechanism of coupling serine to sphingolipids and then to downstream targets.
The sphingoid bases are the most likely mediators of CHA1 induction based on the genetic and pharmacological data presented here. Specifically, the inhibitor myriocin as well as the lcb1-100 mutant, both of which act upstream to block sphingoid base production, block or partially block CHA1 induction. Other mutants and inhibitors, which block lipid production downstream of the sphingoid bases, have no effect (Fig. 2, A–C). Thus, this combined genetic and pharmacologic approach “isolates” sphingoid bases as the likely mediators. Moreover, PHS/DHS were sufficient for CHA1 induction. Thus, sphingoid bases are necessary and sufficient for induction of Cha1 in response to increased serine availability.
Mechanistically, the results implicate the kinases Pkh1 and Pkh2 as mediators of CHA1 induction downstream of the sphingoid bases. Activation of Pkh1/Pkh2 by the sphingoid bases has been implicated in several cell functions, including protein synthesis upon recovery from heat stress (34), P-body formation (35, 36), cell wall integrity, and endocytosis (37, 38). Other studies have implicated a specific phosphorylation site on Ypk1, the primary substrate of Pkh1, in mediating sphingolipid-mediated signaling (39), and PHS was shown to activate Pkh1 in vitro (32); however, some doubt has been raised regarding the latter (40).
Because Cha1 acts to decrease the levels of serine, the results on induction of Cha1 by serine through sphingolipids suggested to us that dysfunction of this homeostatic mechanism would result not only in hyperaccumulation of serine but also in derangements in sphingolipid metabolism. Indeed, serine levels were elevated in the absence of CHA1 expression in response to increased extracellular serine (Table 2). Importantly, the levels of sphingoid bases and ceramides were also significantly elevated under the same conditions, providing further evidence for the coupling of serine metabolism and sphingolipid metabolism. Complex sphingolipids in contrast did not increase under the same conditions, consistent with a recent study (41) where hyperaccumulation of ceramides and sphingoid bases was not accompanied by accumulation of complex sphingolipids. Functionally, the loss of feedback control of serine and sphingolipids in the CHA1 mutant led to a growth defect on high serine media. This growth defect, due to hyperaccumulated intracellular serine, was mediated by sphingolipids and most likely sphingoid bases. Thus, the results reveal a fundamental role for sphingolipids, as direct downstream metabolites of serine, in sensing serine availability, and then initiating a feedback mechanism to induce Cha1. CHA1 expression then serves to attenuate serine levels and consequently sphingolipid levels. It should be noted that the action of CHA1 also provides a nitrogen source (8) and diverts serine toward other metabolites (via conversion to pyruvate) (9, 21). It is intriguing to speculate that sphingolipids may have primarily evolved to sense (metabolically) and regulate (via signaling) these fundamental metabolic feedback pathways.
The close relationship between serine and sphingolipid levels presented here suggests a fundamental connection between amino acid and sphingolipid metabolic pathways. Preliminary data from our laboratory, which led us to focus on CHA1, also identified a number of other genes involved in many aspects of amino acid metabolism/catabolism that are regulated in a sphingolipid-dependent manner. PHS in yeast and ceramide in mammalian cells have been previously shown to regulate amino acid uptake by regulating levels of amino acid transporters (2, 42). The homeostatic mechanism described here may therefore affect amino acid transporter activity and vice versa. Together, these data suggest a complex interwoven connection between amino acid and sphingolipid metabolic pathways.
The feedback mechanism revealed in this study may also have implications for cancer, CNS development, and neurological disease (43). The human CHA1 homolog hSDH is expressed in lung cancer cells as cSDH, a mutant enzyme with lower activity (44, 45). The effect of the cSDH mutation on intracellular serine has not been studied; however, defective serine metabolism has been associated with a number of cancers (46–48). The cSDH mutant enzyme also has altered amino acid substrate specificity, favoring threonine over serine (44, 45). Altered amino acid specificity of a mutant form of the human SPT subunits, favoring glycine and alanine over serine, leads to formation of deoxysphinganine and de-(oxymethyl)sphinganine, leading to hereditary sensory type I neuropathy (49, 50). Because altered availability of glycine and alanine leads to production of deoxysphinganine and de-(oxymethyl)sphinganine, disruption of their metabolism may also potentially lead to neurological disease.
In conclusion, this study reveals the existence of a novel regulatory mechanism whereby sphingoid bases indirectly regulate serine levels by controlling CHA1 expression, and Cha1 expression in turn indirectly regulates sphingolipid levels by regulating serine levels. Importantly, these mechanisms rely on the direct coupling of serine to sphingolipid metabolism. It was also concluded that this homeostatic mechanism is essential for cell growth regulation under high serine load.
Supplementary Material
Acknowledgments
We thank Dr. Robert Dickson and Dr. Jeremy Thorner for providing the strains and K. Alexa Orr Gandy for help with statistical analysis.
This work was supported, in whole or in part, by National Institutes of Health Grant 2R01GM063265 (to Y. A. H.).

This article contains supplemental Figs. S1–S4.
- SPT
- serine palmitoyl transferase
- DHS
- dihydrosphingosine
- PHS
- phytosphingosine
- qRT-PCR
- quantitative reverse transcription PCR
- SC-Thr medium
- synthetic complete threonine dropout medium
- YPD
- yeast extract peptone dextrose.
REFERENCES
- 1. Dickson R. C. (2008) Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J. Lipid Res. 49, 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chung N., Mao C., Heitman J., Hannun Y. A., Obeid L. M. (2001) Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J. Biol. Chem. 276, 35614–35621 [DOI] [PubMed] [Google Scholar]
- 3. Jenkins G. M., Hannun Y. A. (2001) Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 276, 8574–8581 [DOI] [PubMed] [Google Scholar]
- 4. Cowart L. A., Shotwell M., Worley M. L., Richards A. J., Montefusco D. J., Hannun Y. A., Lu X. (2010) Revealing a signaling role of phytosphingosine 1-phosphate in yeast. Mol. Syst. Biol. 6, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Edinger A. L. (2009) Starvation in the midst of plenty: making sense of ceramide-induced autophagy by analyzing nutrient transporter expression. Biochem. Soc. Trans. 37, 253–258 [DOI] [PubMed] [Google Scholar]
- 6. Cowart L. A., Hannun Y. A. (2007) Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. J. Biol. Chem. 282, 12330–12340 [DOI] [PubMed] [Google Scholar]
- 7. Cowart L. A., Okamoto Y., Lu X., Hannun Y. A. (2006) Distinct roles for de novo versus hydrolytic pathways of sphingolipid biosynthesis in Saccharomyces cerevisiae. Biochem. J. 393, 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bornaes C., Ignjatovic M. W., Schjerling P., Kielland-Brandt M. C., Holmberg S. (1993) A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol. Cell. Biol. 13, 7604–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bornaes C., Petersen J. G., Holmberg S. (1992) Serine and threonine catabolism in Saccharomyces cerevisiae: the CHA1 polypeptide is homologous with other serine and threonine dehydratases. Genetics 131, 531–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schorling S., Vallée B., Barz W. P., Riezman H., Oesterhelt D. (2001) Lag1p and Lac1p are essential for the acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisiae. Mol. Biol. Cell 12, 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collart M. A., Oliviero S. (2001) in Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., eds), pp. 13.12.1–13.12.5, John Wiley & Sons, New York [Google Scholar]
- 12. Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. (2002) Processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32, 1372–1374, 1376,, 1378–1379 [PubMed] [Google Scholar]
- 13. Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Simultaneous quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 14. Mandala S. M., Thornton R. A., Frommer B. R., Curotto J. E., Rozdilsky W., Kurtz M. B., Giacobbe R. A., Bills G. F., Cabello M. A., Martín I. (1995) The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis: producing organism, fermentation, isolation, and biological activity. J. Antibiot. 48, 349–356 [DOI] [PubMed] [Google Scholar]
- 15. Uemura S., Kihara A., Iwaki S., Inokuchi J., Igarashi Y. (2007) Regulation of the transport and protein levels of the inositol phosphorylceramide mannosyltransferases Csg1 and Csh1 by the Ca2+-binding protein Csg2. J. Biol. Chem. 282, 8613–8621 [DOI] [PubMed] [Google Scholar]
- 16. Reggiori F., Conzelmann A. (1998) Biosynthesis of inositol phosphoceramides and remodeling of glycosylphosphatidylinositol anchors in Saccharomyces cerevisiae are mediated by different enzymes. J. Biol. Chem. 273, 30550–30559 [DOI] [PubMed] [Google Scholar]
- 17. Cerbón J., Falcon A., Hernández-Luna C., Segura-Cobos D. (2005) Inositol phosphoceramide synthase is a regulator of intracellular levels of diacylglycerol and ceramide during the G1 to S transition in Saccharomyces cerevisiae. Biochem. J. 388, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Villas-Bôas S. G., Højer-Pedersen J., Akesson M., Smedsgaard J., Nielsen J. (2005) Global metabolite analysis of yeast: evaluation of sample preparation methods. Yeast 22, 1155–1169 [DOI] [PubMed] [Google Scholar]
- 19. Hans M. A., Heinzle E., Wittmann C. (2003) Free intracellular amino acid pools during autonomous oscillations in Saccharomyces cerevisiae. Biotechnol. Bioeng. 82, 143–151 [DOI] [PubMed] [Google Scholar]
- 20. Frank M. P., Powers R. W. (2007) Simple and rapid quantitative high-performance liquid chromatographic analysis of plasma amino acids. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 852, 646–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Petersen J. G., Kielland-Brandt M. C., Nilsson-Tillgren T., Bornaes C., Holmberg S. (1988) Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics 119, 527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jenkins G. M., Richards A., Wahl T., Mao C., Obeid L., Hannun Y. (1997) Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem. 272, 32566–32572 [DOI] [PubMed] [Google Scholar]
- 23. Dickson R. C., Nagiec E. E., Skrzypek M., Tillman P., Wells G. B., Lester R. L. (1997) Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272, 30196–30200 [DOI] [PubMed] [Google Scholar]
- 24. Merrill A. H., Jr., van Echten G., Wang E., Sandhoff K. (1993) Fumonisin B1 inhibits sphingosine (sphinganine) N-acyltransferase and de novo sphingolipid biosynthesis in cultured neurons in situ. J. Biol. Chem. 268, 27299–27306 [PubMed] [Google Scholar]
- 25. Nagiec M. M., Nagiec E. E., Baltisberger J. A., Wells G. B., Lester R. L., Dickson R. C. (1997) Sphingolipid synthesis as a target for antifungal drugs: complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem. 272, 9809–9817 [DOI] [PubMed] [Google Scholar]
- 26. Wu W. I., McDonough V. M., Nickels J. T., Jr., Ko J., Fischl A. S., Vales T. R., Merrill A. H., Jr., Carman G. M. (1995) Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J. Biol. Chem. 270, 13171–13178 [DOI] [PubMed] [Google Scholar]
- 27. Sun Y., Taniguchi R., Tanoue D., Yamaji T., Takematsu H., Mori K., Fujita T., Kawasaki T., Kozutsumi Y. (2000) Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol. Cell. Biol. 20, 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Villa N. Y., Kupchak B. R., Garitaonandia I., Smith J. L., Alonso E., Alford C., Cowart L. A., Hannun Y. A., Lyons T. J. (2009) Sphingolipids function as downstream effectors of a fungal PAQR. Mol. Pharmacol. 75, 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. deHart A. K., Schnell J. D., Allen D. A., Hicke L. (2002) The conserved Pkh-Ypk kinase cascade is required for endocytosis in yeast. J. Cell Biol. 156, 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holmberg S., Schjerling P. (1996) Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. He Q., Battistella L., Morse R. H. (2008) Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J. Biol. Chem. 283, 5276–5286 [DOI] [PubMed] [Google Scholar]
- 32. Liu K., Zhang X., Lester R. L., Dickson R. C. (2005) The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280, 22679–22687 [DOI] [PubMed] [Google Scholar]
- 33. Kim S., Fyrst H., Saba J. (2000) Accumulation of phosphorylated sphingoid long chain bases results in cell growth inhibition in Saccharomyces cerevisiae. Genetics 156, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meier K. D., Deloche O., Kajiwara K., Funato K., Riezman H. (2006) Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol. Biol. Cell 17, 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo G., Costanzo M., Boone C., Dickson R. C. (2011) Nutrients and the Pkh1/2 and Pkc1 protein kinases control mRNA decay and P-body assembly in yeast. J. Biol. Chem. 286, 8759–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowart L. A., Gandy J. L., Tholanikunnel B., Hannun Y. A. (2010) Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 431, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roelants F. M., Torrance P. D., Bezman N., Thorner J. (2002) Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13, 3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo G., Gruhler A., Liu Y., Jensen O. N., Dickson R. C. (2008) The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283, 10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roelants F. M., Torrance P. D., Thorner J. (2004) Differential roles of PDK1- and PDK2-phosphorylation sites in the yeast AGC kinases Ypk1, Pkc1, and Sch9. Microbiology 150, 3289–3304 [DOI] [PubMed] [Google Scholar]
- 40. Roelants F. M., Baltz A. G., Trott A. E., Fereres S., Thorner J. (2010) A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. U.S.A. 107, 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guenther G. G., Peralta E. R., Rosales K. R., Wong S. Y., Siskind L. J., Edinger A. L. (2008) Ceramide starves cells to death by down-regulating nutrient transporter proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Koning T. J., Snell K., Duran M., Berger R., Poll-The B. T., Surtees R. (2003) l-Serine in disease and development. Biochem. J. 371, 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ogawa H., Gomi T., Nishizawa M., Hayakawa Y., Endo S., Hayashi K., Ochiai H., Takusagawa F., Pitot H. C., Mori H., Sakurai H., Koizumi K., Saiki I., Oda H., Fujishita T., Miwa T., Maruyama M., Kobayashi M. (2006) Enzymatic and biochemical properties of a novel human serine dehydratase isoform. Biochim. Biophys. Acta 1764, 961–971 [DOI] [PubMed] [Google Scholar]
- 45. Yamada T., Komoto J., Kasuya T., Takata Y., Ogawa H., Mori H., Takusagawa F. (2008) A catalytic mechanism that explains a low catalytic activity of serine dehydratase like-1 from human cancer cells: crystal structure and site-directed mutagenesis studies. Biochim. Biophys. Acta 1780, 809–818 [DOI] [PubMed] [Google Scholar]
- 46. Bachelor M. A., Lu Y., Owens D. M. (2011) l-3-Phosphoserine phosphatase (PSPH) regulates cutaneous squamous cell carcinoma proliferation independent of l-serine biosynthesis. J. Dermatol. Sci. 63, 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pollari S., Käkönen S. M., Edgren H., Wolf M., Kohonen P., Sara H., Guise T., Nees M., Kallioniemi O. (2011) Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 125, 421–430 [DOI] [PubMed] [Google Scholar]
- 48. Tan E. H., Ramlau R., Pluzanska A., Kuo H. P., Reck M., Milanowski J., Au J. S., Felip E., Yang P. C., Damyanov D., Orlov S., Akimov M., Delmar P., Essioux L., Hillenbach C., Klughammer B., McLoughlin P., Baselga J. (2010) A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non-small-cell lung cancer. Ann. Oncol. 21, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Penno A., Reilly M. M., Houlden H., Laurá M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H., Jr., von Eckardstein A., Hornemann T. (2010) Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285, 11178–11187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rotthier A., Auer-Grumbach M., Janssens K., Baets J., Penno A., Almeida-Souza L., Van Hoof K., Jacobs A., De Vriendt E., Schlotter-Weigel B., Löscher W., Vondráček P., Seeman P., De Jonghe P., Van Dijck P., Jordanova A., Hornemann T., Timmerman V. (2010) Mutations in the SPTLC2 subunit of serine palmitoyltransferase cause hereditary sensory and autonomic neuropathy type I. Am. J. Hum. Genet. 87, 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.