Background: NOTCH activation accelerates the degradation of endogenous JAK3 proteins.
Results: We show that ubiquitin-mediated JAK3 degradation involves NOTCH targets ASB2 and SKP2. The JAK3 R980W mutant exhibits impaired association with SKP2 and resists NOTCH or ASB2-induced degradation.
Conclusion: NOTCH-induced JAK3 degradation likely employs dimeric complexes consisting of Cullin1- and Cullin5-based E3 ligases.
Significance: A useful mutant is created for future in vivo studies.
Keywords: E3 Ubiquitin Ligase, Jak Kinase, Lymphocyte, NOTCH Pathway, Ubiquitination
Abstract
Although NOTCH signaling is well known to regulate lymphopoiesis, Janus kinase 3 (JAK3) also plays a critical role in promoting lymphocyte development. We have previously found that NOTCH signaling leads to the degradation of JAK3 in B lineage cells, suggesting that NOTCH signaling exerts its biological effect on lymphopoiesis through modulating JAK3 levels. Here, we delineate the biochemical mechanisms involved in NOTCH-induced JAK3 ubiquitination and degradation. NOTCH signaling is known to transcriptionally activate the genes encoding ASB2 (ankyrin-repeat SOCS box containing protein 2) and SKP2 (S-phase kinase-associated protein 2). We show that not only NOTCH but also ASB2 and SKP2 can promote the ubiquitination and degradation of JAK3. Both ASB2 and SKP2 can interact with JAK3 through different domains; the FERM and pseudo-kinase domains each had high affinities for ASB2, whereas the kinase domain primarily associated with SKP2. ASB2 and SKP2 previously have been shown to associate with each other to bridge the formation of a non-canonical Cullin1 and Cullin5-containing dimeric E3 ligase complex. Interestingly, the R980W mutant of JAK3 exhibited diminished interaction with SKP2 and resistance to NOTCH or ASB2-induced degradation. Furthermore, dominant-negative mutants of either Cullin1 or Cullin5, which lack the C terminus responsible for recruiting the E2 enzymes, were able to prevent JAK3 degradation induced by both ASB2/SKP2 and NOTCH signaling. Together, these results suggest that JAK3 ubiquitination involves the non-canonical dimeric E3 ligase complex, and the R980W mutant will serve as an excellent tool for investigating the biological significance of NOTCH-mediated JAK3 turnover.
Introduction
The NOTCH signaling pathways mediate diverse cell fate decisions in organisms ranging from Drosophila to human (1, 2). Vertebrates have four transmembrane NOTCH receptors, NOTCH1–4. Signaling through the NOTCH receptors is triggered by their interaction with Jagged or Delta-like ligands expressed on adjacent cells. Upon binding of membrane-tethered NOTCH ligands to the receptors, proteolytic cleavages occur at the trans-membrane domain of NOTCH receptors, allowing translocation of their intracellular domain (IC)2 to the nucleus. The IC domain then interacts with RBP-Jκ transcription factors to activate gene expression through displacement of repressors and recruitment of coactivators such as Mastermind (3–5). NOTCH signaling has been found to have several important functions in lymphopoiesis (6–9). For example, in the thymus, NOTCH promotes T cell development while suppressing B cell differentiation. Conditional disruption of genes encoding NOTCH1 or its DNA binding partner, RBP-Jκ, completely blocked T cell differentiation (10, 11). Instead, the thymus was populated with B cells. NOTCH also plays an important role in the formation of marginal zone B cells. Ablation of either the NOTCH2 or RBP-Jκ genes, as well as overexpression of a dominant-negative form of MASTERMIND, led to dramatic reduction in marginal zone B cell production (12–14).
Cytokine signaling also plays important roles in lymphopoiesis (15, 16). The JAK family of non-receptor tyrosine kinases, including JAK1, JAK2, JAK3, and TYK, are essential mediators of cytokine-induced signal transduction and capable of binding to various cell surface receptors. Cytokine binding to their homodimeric or heterodimeric receptors triggers the aggregation of JAKs, which leads to their phosphorylation and activation. Subsequently, JAKs phosphorylate tyrosine residues on the receptors and generate sites for interaction with cytosolic STATs (signal transducers and activators of transcription), resulting in tyrosine phosphorylation of STATs by JAKs. These phosphotyrosine residues on STATs then act as binding sites for their SH2 domains, mediating the dimerization between themselves. Activated STAT dimers then translocate into the nucleus to activate transcription of their target genes. Although Jak1, Jak2, and Tyk2 are expressed ubiquitously, Jak3 is expressed primarily in cells of hematopoietic lineages (17–19). The development of T and B lymphocytes from hematopoietic progenitor cells is strictly dependent on IL-7 signaling and JAK3 associates with γc chain-containing receptors such as IL-7R (20–24). In line with this, Jak3-deficient mice have small thymuses with deficits in thymic progenitor cells and severe blockade in B cell development (25, 26). Together, JAK3 is essential for both B and T cell differentiation. On the other hand, excess JAK3 activities have been shown to cause lymphoproliferative diseases (27).
To precisely modulate cellular responses, JAK proteins are subjected to multiple tiers of regulation through phosphorylation, de-phosphorylation, inhibitor binding, and ubiquitin-mediated degradation (28, 29). Several well known regulatory mediators of JAK signaling belong to a family of cytoplasmic suppressors of cytokine signaling (SOCS)-containing proteins, including SOCS1–7 proteins and CIS (cytokine inducible SH2-domain-containing protein) (30). Expression of these SOCS proteins is transcriptionally activated by signaling from cytokine receptors via the JAK-STAT pathway. These proteins in turn down-regulate cytokine signaling in a negative feedback loop by three distinct mechanisms: inhibition of JAK activities, competition with STATs for binding sites, and ubiquitin-mediated degradation of cytokine receptors and JAKs. SOCS proteins have been shown to serve as substrate-binding subunits of the ElonginB/C/Cullin5/SOCS (ECS) E3 ubiquitin ligase complexes and facilitate the ubiquitination of JAK2 (31–33). Whether JAK3 is regulated in a similar manner has not been investigated.
Our group has previously shown that activation of endogenous NOTCH receptors accelerates the degradation of endogenous JAK2 and JAK3 in B but not in T lineage cells in a MAPK-dependent manner, demonstrating a link between NOTCH and JAK-STAT signaling pathways (33, 34). It is thus possible that NOTCH signaling exerts its effect on lymphopoiesis, at least in part, by influencing the stability of JAK3. Indeed, we have shown that constitutive activation of STAT5 cooperates with a NOTCH-resistant E2A mutant to drive B cell differentiation in the thymus (34). NOTCH-induced protein ubiquitination depends on its ability to activate transcription, which is consistent with the findings that NOTCH signaling stimulates the transcription of genes encoding SKP2 and ASB2 (33, 35, 36). Although SKP2 is a substrate-binding component of the SKP1/Cullin1/F-box protein (SCF) E3 ligase complexes, ASB2 represents a subgroup of the SOCS superfamily (37, 38). ASB2 has been shown to compete with SOCS1 to interact with Elongins and Cullin5 (33). Furthermore, we have recently provided evidence to suggest that ASB2 and SKP2 associate with each other to bridge the formation of non-canonical E3 ligase complexes composed of ElonginB/C/Cullin5 and SKP1-Cullin1 (33). Although we showed that such complexes are likely responsible for the ubiquitination of JAK2, the molecular details involved in the relationship between JAK2 and the E3 ligase complexes has not been elucidated.
The purposes of the current study are to test whether NOTCH-induced JAK3 degradation employs similar non-canonical E3 ligase complexes and to understand the underlying molecular mechanisms, which will help develop critical reagents for future in vivo studies. We showed that like activated NOTCH1, ASB2 and SKP2 promoted the ubiquitination and degradation of JAK3. Interestingly, the R980W mutant of JAK3, which displayed diminished affinity for SKP2, was resistant to NOTCH or ASB2/SKP2-induced degradation. Moreover, dominant-negative mutants of either Cullin1 or Cullin5 rescued JAK3 proteins in the presence of activated NOTCH or ASB2/SKP2. Therefore, these results suggest that NOTCH-induced JAK3 ubiquitination involves the non-canonical heterodimeric E3 ligase complex. The Arg-980 residue plays an important role in JAK3 ubiquitination, and thus, the mutant could serve as a valuable tool for investigating the biological function of NOTCH-mediated JAK3 turnover in lymphopoiesis.
MATERIALS AND METHODS
Plasmids
A retroviral construct expressing HPC4-tagged mouse JAK3 was created by PCR amplification of mouse cDNA with JAK3-specific primers and in-frame insertion downstream of a HPC4 tag in a modified MIGR1 retroviral vector (39). The R980W mutation in kinase domain of JAK3 was introduced by using two-step PCR to obtain a 930-bp DNA fragment flanked by XhoI and EcoRI sites. This fragment then replaced the wild type XhoI-EcoRI fragment.
In addition, HPC4-tagged wild type or mutant JAK3 cDNAs were also cloned into pcDNA3 by transferring the full-length JAK3 with HPC4 tag from the MIGR1 retroviral vector to the pcDNA3 vector. To do so, the 1.4-kb fragment containing N-terminal JAK3 was amplified by PCR using HPC4-JAK3 as a template. Amplified DNA fragments were inserted into the pGEM-T Easy vector for sequencing and then cut out with EcoRI. The EcoRI fragment was cloned into pcDNA3, and the resulting plasmid was partially digested with EcoRI and filled in with the Klenow enzyme, followed by ligation and transformation. Clones whose EcoRI site at the 5′ end of the insert was destroyed by filling in were selected and used for the subsequent step that includes an insertion of a 3.4-kb EcoRI-XbaI fragment containing the 3′ portion of the wild type or mutant JAK3 coding sequence and internal ribosome entry site followed by enhanced GFP. These fragments were obtained by complete digestion of JAK3 retroviral constructs with XbaI and partial digestion with EcoRI. JAK3 deletion constructs were created by restriction digestion at appropriate sites and ligation into the pcDNA3 vector or MIGR1 retroviral vector or by PCR amplification of desired fragment using HPC4-JAK3 as a template, followed by insertion into pcDNA3 or MIGR1.
Human ASB2 with a 3×FLAG tag was kindly supplied by Dr. Junya Kohroki (Tokyo University of Science), and the deletion constructs were as described (33). FLAG-DN-Cullin1 and DN-Cullin5 constructs were obtained from Addgene. SKP2-T7/pcDNA3 was a gift from Dr. H. Zhang (Nevada Cancer Institute, Las Vegas, NV). The Myc-tagged NOTCHΔE/pCS2+MT construct was provided generously by Dr. Raphael Kopan (Washington University School of Medicine, St. Louis, MO) (40). YFP-ubiquitin/pK was kindly supplied by Dr. Scott Plafker (Oklahoma Medical Research Foundation, Oklahoma City, OK).
Antibodies and Chemicals
Anti-FLAG monoclonal antibody (M2) was purchased from Sigma Aldrich. Monoclonal anti-HPC4 antibody was kindly provided by Dr. Charles Esmon (Oklahoma Medical Research Foundation) (41). Anti-Myc (9E10) and anti-HA (12CA5) tissue culture supernatants were produced from hybridoma cell lines. Monoclonal anti-GFP and rabbit polyclonal anti-SKP2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polycolonal antibodies against STAT5a and phospho-STAT5 were from Zymed Laboratories, Inc. (Invitrogen). MG-132 was from Calbiochem. IL-7 was purchased from R&D Systems.
Transfection, Immunoprecipitation, and Immunoblotting Assays
NIH3T3 and 293T cells were co-transfected using the calcium phosphate method. NIH3T3 cells were treated with 15% (v/v) glycerol in 1× HBS 3–4 h post transfection to increase transfection efficiency. To constitute the intact JAK3/STAT5 signaling pathway, 2 μg of each plasmids expressing IL-7Rα chain and IL-2Rγ chain were included in transfection assays for measuring JAK3 degradation. Typically, 1 μg of HPC4-JAK3 and 0.5 μg of STAT5a were co-transfected into NIH3T3 cells with or without 2 μg of each FLAG-ASB2 and/or SKP2. Alternatively, 1 μg of NΔE was co-transfected. To rescue ASB2 and/or SKP2 or NΔE induced-degradation of JAK3, 3 μg of FLAG-DN-Cullin1 or DN-Cullin5 were included. Twenty-six to thirty hours after transfection, cells were fed with fresh DMEM containing 10% FBS and 5 ng/ml IL-7 and harvested 1.5 h later. The cells were lysed in RIPA buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS in phosphate-buffered saline), containing a mixture of protease inhibitors (2 ng/ml leupeptin, 2 ng/ml aprotinin, 2 ng/ml pepstatin, 1 mm PMSF), 1 mm DTT, and phosphatase inhibitors (25 mm NaF and 1 mm Na3VO4). Cell lysates were analyzed using 8.5–9% SDS-PAGE followed by immunoblotting with appropriate antibodies. Immunoprecipitation was usually performed with 5 μg of antibodies. Immunoprecipitates were analyzed using 8.5–9% SDS-PAGE followed by immunoblotting with the appropriate antibodies.
In Vivo Ubiquitination Assay
Wild type or mutant full-length JAK3 (5 μg) were co-transfected into 293T cells with 10 μg of HA-tagged or YFP-tagged ubiquitin along with appropriate constructs followed by the treatment with 5 μm MG-132 for 3–5 h before harvest. Treated cells were lysed in RIPA buffer containing protease inhibitors, DTT, and 5 μm MG-132. Immunoprecipitation was performed with anti-HPC4 or anti-GFP antibodies. The precipitate was analyzed using 7.5% SDS-PAGE followed by immunoblotting with anti-HA or anti-HPC4 antibodies.
RESULTS
Induction of JAK3 Turnover by NOTCH and Its Downstream Targets, ASB2 and SKP2
We have shown previously that signaling from NOTCH receptors by co-culturing bone marrow-derived mouse primary pro-B cells with OP9 stromal cells expressing Delta-like-1 significantly reduced the levels of JAK3 (34). However, the underlying mechanism has not been investigated. To do so, we utilized a NOTCH1 mutant, called NΔE, whose extracellular domain is N-terminal truncated and can be activated constitutively in a ligand-independent manner. In addition, NΔE lacks the PEST domain at the C terminus, which mediates the degradation of the intracellular domain, thus resulting in its accumulation in the nucleus (40). In co-transfection experiments, HPC4-tagged JAK3 with or without an increasing amount of NΔE were introduced into NIH3T3 cells. As controls for transfection efficiency, the cells also received a construct expressing STAT5, whose stability was previously shown to be insensitive of NOTCH (34). In addition, constructs expressing IL-7Rα and IL-2Rγ were included to facilitate JAK3 phosphorylation upon IL-7 treatment. Twenty-eight hours post-transfection, cells were harvested following incubation with IL-7 for 1.5 h, and immunoblotting was performed to measure the levels of JAK3 and NΔE (Fig. 1A). The doublets detected with the anti-Myc antibody against the tag on NΔE likely represent the full-length NΔE and NICDΔE produced by spontaneous cleavages of the receptor by γ-secretases. Levels of STAT5 served as controls of transfection efficiency. As shown in Fig. 1A, JAK3 levels dramatically decreased as the levels of NΔE increased. This result could be interpreted to mean that NOTCH signaling accelerates the turnover of JAK3.
FIGURE 1.
NOTCH induces the degradation of JAK3 by promoting the polyubiquitination of JAK3. A, NIH3T3 cells were co-transfected with JAK3 plus or minus increasing amounts of a ligand-independent form of NOTCH1, NΔE, along with constructs expressing IL-7 receptor α and IL-2 receptor common γ chain. Twenty eight hours post transfection, cells were treated with 5 ng/ml IL-7 for 1.5 h and then harvested. Cell lysates were subjected to immunoblot analysis with antibodies against the HPC4 tag on JAK3, and the Myc tag on NOTCH1. As a control for transfection efficiency, STAT5 was also co-transfected and immunoblotted (IB). NICD means Notch intracellular domain. B, 293T cells were co-transfected with YFP-tagged ubiquitin plus empty vector (V), JAK3, or JAK3 and NΔE. Twenty four hours post transfection, cells were treated with 5 μm MG-132 for 3 h, and then cell lysates were subjected to immunoprecipitation with anti-GFP antibodies. The precipitates were blotted with the anti-HPC4 antibody for JAK3. Input was analyzed for indicated proteins.
To determine whether NOTCH stimulates the poly-ubiquitination of JAK3, in vivo ubiquitination assays were performed in 293T cells. HPC4-tagged JAK3 was co-transfected with or without NΔE along with YFP-tagged ubiquitin. Total ubiquitinated proteins were then immunoprecipitated from whole cell lysates with anti-GFP antibodies. The precipitates were analyzed by immunoblotting with the anti-HPC4 antibody to detect ubiquitinated JAK3. Expression of NΔE significantly increased the amount of polyubiquitinated JAK3 compared with cells transfected with the empty vector (Fig. 1B). As a negative control, no signal was detected in the sample without HPC4-JAK3 expression. These results revealed that NOTCH significantly enhanced the polyubiquitination of JAK3, suggesting that enhanced ubiquitination of JAK3 stimulated by NOTCH mediates its degradation.
NOTCH signaling has been shown to stimulate the transcription of genes encoding ASB2 and SKP2, which are thought to bridge the formation of a dimeric E3 ligase complex (33, 35, 36). To test whether ASB2 and SKP2 are involved in JAK3 degradation, similar co-transfection assays were carried out as described for Fig. 1A. Co-transfection of JAK3 with either ASB2 or SKP2 led to an obvious reduction of JAK3 levels, suggesting that both proteins stimulate the turnover of JAK3 (Fig. 2A). Furthermore, when both proteins were co-expressed, a greater effect was observed (Fig. 2B). More importantly, incubation with a proteasome inhibitor, MG-132, for 1.5 h prior to harvest significantly recovered JAK3 (Fig. 2B, compare lanes 3 and 4). Admittedly, the level of JAK3 did not return to that of cells without ASB2 and SKP2 expression (lane 2). This may be due to a nonspecific toxic effect of MG-132 on cells as the levels of JAK3 in cells transfected with or without ASB2 but both treated with MG-132 (Fig. 2B, compare lanes 4 and 5) were comparable after normalization with the internal control. This result thus suggests that ASB2 and SKP2-induced JAK3 degradation is proteasome-dependent.
FIGURE 2.
ASB2 and SKP2 lead to the degradation of JAK3 in a proteasome-dependent manner. A, co-transfection assays were performed as described in the legend to Fig. 1A except that 2 μg of ASB2 or SKP2 replaced NΔE. B, NIH3T3 cells were co-transfected with or without JAK3 plus or minus both ASB2 and SKP2 along with IL-7 receptors and internal controls. Twenty eight hours later, cells were treated with 5 ng/ml IL-7 along with DMSO vehicle (−) or 50 μm MG-132 (+) for 1.5 h. Cells were then harvested, and lysates were subjected to immunoblot analysis with antibodies for indicated proteins.
Interaction between JAK3 and ASB2
Because ASB2 is able to induce the degradation of JAK3, the next logical questions are whether JAK3 physically interacts with ASB2 and which regions in JAK3 mediate the interaction. Therefore, co-immunoprecipitation assays were performed in 293T cells, where JAK3 and ASB2 were overexpressed. To map the regions in JAK3 that mediate the interaction with ASB2, truncation mutants were generated as diagramed in Fig. 3A. These mutants contain one of the known functional domains, the FERM domain responsible for receptor binding, the pseudo-kinase domain known to regulate JAK function or the kinase domain with catalytic activities (42, 43). In a series of co-IP assays (Fig. 3, B–D), HPC4-tagged full-length or mutant Jak3 were co-transfected with or without FLAG-tagged Asb2. Immunoprecipitation was performed with the anti-FLAG antibody or control IgG in RIPA buffer, which is a stringent condition for protein-protein interaction. The precipitates were immunoblotted with the anti-HPC4 antibody. The results showed that not only the full-length JAK3, but also all three mutants were readily co-precipitated by the anti-FLAG antibody from extracts of ASB2-expressing cells. In contrast, control IgG did not bring down full-length JAK3, demonstrating the specificity of the assays. In addition, we have also created JAK3 mutants lacking one of the functional domains and found that these mutants were capable of association of ASB2, consistent with the observation that multiple ASB2-interacting domains exist in JAK3 (data not shown). We have also replaced the HPC4 tag with the HA tag and obtained the same results, indicating that the interaction was not mediated by the epitope tag (data not shown).
FIGURE 3.
Mapping the ASB2-interacting domains in JAK3. A, schematic diagram of mouse JAK3 and its mutant constructs. Functional domains of JAK3 are marked as gray boxes. Numbers indicate the positions in amino acid sequence of JAK3. All constructs contain a HPC4 tag at the N terminus. B–D, interaction between JAK3 and ASB2. Full-length JAK3 or indicated mutants were co-transfected into 293T cells with or without FLAG-tagged ASB2 as indicated. Immunoprecipitation was performed with anti-FLAG or control antibodies in RIPA buffer. The precipitates and inputs were probed with anti-HPC4 and anti-FLAG antibodies. Expected positions of the wild type and mutant proteins are labeled along the side of the gels. The asterisk marks the signal resulted from reactions with Ig heavy chain. Slow-migrating bands were sometimes observed in precipitates with the FERM domain mutant for unknown reasons.
To further investigate the interaction between ASB2 and JAK3, we searched for the sequences in ASB2 that are required for interaction with full-length JAK3. Five ASB2 deletion mutants containing either the ankyrin repeats or SOCS box or both were analyzed in co-IP assays by pulling down FLAG-tagged ASB2 and immunoblotting for JAK3 (Fig. 4, A and B). All of these ASB2 mutants were able to co-precipitate with JAK3, suggesting that several regions in ASB2 can mediate the interaction with JAK3.
FIGURE 4.
Defining the regions of ASB2 mediating interaction with different functional domains of JAK3. A, schematic diagram of human ASB2 and its mutant constructs. N-terminal ankyrin repeats are highlighted by a gray bar, and the black box represents the C-terminal SOCS box. Numbers denote the positions in amino acid sequence of ASB2. All constructs are fused to a 3×FLAG tag at the C terminus. Regions known to mediate the interaction with SKP2 and ElonginB/C/Cullin5 are labeled on top of the full-length construct. B, interaction between wild type JAK3 and ASB2 mutants. 293T cells were co-transfected with or without full-length JAK3 plus or minus full-length (FL) or mutant ASB2 as indicated. Immunoprecipitation was performed with anti-FLAG or control (Con) antibodies in RIPA buffer. The precipitates and inputs were probed with anti-HPC4 and anti-FLAG antibodies. Association of full-length JAK3 (FL) and deletion mutants with ASB2-ΔN (C) and ASB2-(1–328) (D) were examined as described for A. Eln, elongin.
Next, we attempted to define the regions of the ASB2-mediating interaction with different functional domains of JAK3 by using two ASB2 mutants, 1–328 and ΔN, which contain the N-terminal ankyrin repeats and C-terminal SOCS box, respectively. Both constructs lack the region previously shown to be involved in SKP2 interaction (Fig. 4A). They were co-transfected with full-length JAK3 or mutants possessing only the FERM, pseudo-kinase, or kinase domain (Fig. 4, C and D). In comparison with the behavior of full-length JAK3, it appeared that the FERM domain interacted with both 1–328 and ΔN, whereas the pseudo-kinase domain did not associate with ΔN. More interestingly, the kinase domain interacted very poorly with 1–328 and did not bind ΔN. As both constructs lack the region necessary for SKP2 interaction, and the kinase domain is capable of interacting with full-length ASB2 (Fig. 3D), it raises a possibility that the association of the kinase domain with ASB2 depends on SKP2.
Establishment of JAK3 Mutant Resistant to NOTCH or ASB2/SKP2-induced Degradation
To aid future in vivo studies of the biological significance of NOTCH-induced JAK3 degradation and to further understand the biochemical mechanism underlying JAK3 ubiquitination, we sought to create JAK3 point mutants that are resistant to NOTCH-induced degradation while retaining cytokine-regulated kinase activities. To design point mutations in JAK3, we consulted with knowledge accumulated from structural studies of JAK proteins in the literature (44). Although no information is available for JAK3, several structures have been solved in the context of JAK2 or glycoprotein 130 bound to SOCS proteins (44–46). It is known that the activation loop in the kinase domain of JAK2 is critical not only for its kinase activity but also for SOCS-mediated ubiquitination (31, 47). Because JAK3 shares extensive homology with JAK2 and ASB2 belongs to the extended SOCS family, we decided first to introduce point mutations into the kinase domain. Several residues of the activation loop in the kinase domain of JAK2 were predicted to interact with the SH2 domain of SOCS1 (44). The best known residue in this loop is Tyr-1007, whose phosphorylation is essential for degradation. However, mutation of this residue also destroys the enzyme and thus become unsuitable for future biological studies. Another key residue in this region is Lys-1011, which is equivalent to Arg-980 in the kinase domain of JAK3. We therefore generated the R980W mutant by substituting Arg-980 with tryptophan, which has drastically different properties. To test whether this mutation dampens cytokine-regulated kinase activity, wild type JAK3 or R980W plus IL-7Rα and IL-2Rγ chains were transfected into 293T cells along with their downstream target, STAT5a. The cells were then treated with or without IL-7 to activate JAK3. In the presence but not absence of IL-7, both wild type or mutant JAK3 phosphorylated STAT5 (Fig. 5A), indicating that R980W was enzymatically active and able to respond to cytokine stimulation.
FIGURE 5.
Arg-980 residue of JAK3 is required for NOTCH or ASB2/SKP2-mediated degradation. A, to measure cytokine-induced kinase activity, wild type JAK3 and R980W mutant were co-transfected into 293T cells with STAT5a. Transfected cells were treated with or without IL-7 for 1 h before harvest. Phosphorylation of STAT5 was probed with anti-phospho-STAT5 antibodies. Total amounts of STAT5 and JAK3 expressed were examined as controls. Co-transfection assays of wild type and R980W mutant of JAK3 were carried out with NΔE (B), ASB2 plus SKP2 (C), ASB2 alone (D), or SKP2 alone (F) as described in the legend to Fig. 1A. E, in vivo ubiquitination assay. Wild type or R980W JAK3 plus HA-tagged ubiquitin were co-transfected into 293T cells with or without ASB2. Twenty four hours posttransfection, cells were treated with 5 μm MG-132 for 3 h, and HPC4-JAK3 was immunoprecipitated with the anti-HPC4 antibody. The precipitates were immunoblotted with the anti-HA antibody for ubiquitinated JAK3, and the amounts of JAK3 recovered were determined by immunoblotting with the anti-HPC4 antibody. Inputs were analyzed by immunoblotting for the tags on JAK3 and ASB2. Vec, vector; NICD, Notch Intracellular Domain.
To evaluate the sensitivity of R980W to NOTCH-induced degradation, we introduced wild type or mutant JAK3 into NIH3T3 cells with or without NΔE and analyzed whole cell lysates by immunoblotting. In the presence of NΔE, the level of wild type JAK3 protein was dramatically decreased. In contrast, levels of the R980W mutant were similar in the presence or absence of NΔE (Fig. 5B).
We then tested the stability of JAK3 mutants when both ASB2 and SKP2 were overexpressed. Asb2 and Skp2 genes are the downstream target of NOTCH signaling, and encoded proteins are critical substrate recognizing subunits of non-canonical heterodimeric E3 ligase complexes (33). Therefore, overexpression of both proteins is expected to have a powerful effect on JAK3 ubiquitination and degradation. Indeed, the level of wild type JAK3 was reduced significantly in the presence of ASB2 and SKP2. In contrast, the R980W mutant remained unchanged (Fig. 5C). Likewise, R980W was also insensitive to degradation induced by ASB2 alone (Fig. 5D).
Next, the ability of ASB2 to stimulate the ubiquitination of R980W was examined. Wild type and mutant JAK3 plus or minus ASB2 were co-transfected into 293T cells along with a construct expressing HA-tagged ubiquitin. JAK3 proteins were immunoprecipitated with the anti-HPC4 antibody, and the precipitates were analyzed by immunoblotting with the anti-HA antibody to reveal ubiquitinated JAK3. The amounts of JAK3 brought down in each lane were determined by immunoblotting with the anti-HPC4 antibody. Input controls were also analyzed by immunoblotting. As shown in Fig. 5E, in the presence of ASB2, the amount of ubiquitinated wild type JAK3 was increased dramatically. However, the amount of ubiquitinated R980W mutant was unchanged, suggesting that the mutation renders the protein resistant to ASB2-induced degradation by impairing its ubiquitination. Similar to the response of R980W to ASB2, it is also resistant to SKP2-induced degradation (Fig. 5F).
Finally, to understand why the ubiquitination of R980W was diminished, we examined its interaction with components of the E3 ligases. Because the kinase domain of JAK proteins are thought to mediate interaction with SOCS1 and ASB2 also belong to the SOCS superfamily, we first tested the interaction between ASB2 and R980W. Wild type or mutant JAK3 along with or without ASB2 was overexpressed in 293T cells, and co-immunoprecipitation was performed. Surprisingly, R980W displayed similar affinities to ASB2 as wild type JAK3 (Fig. 6A). We then evaluated the physical interaction between wild type or mutant JAK3 proteins with SKP2 by performing similar co-IP assays. Comparing to wild type JAK3, R980W associated poorly with SKP2 (Fig. 6B). These results suggest that mutation at Arg-980 impairs the interaction of JAK3 with SKP2 and thus resulted in its increased stability. Furthermore, this result corroborates with the notion described in Fig. 4 that the kinase domain of JAK3 associates with ASB2 primarily through its direct or indirect interaction with SKP2 and verify the idea that NOTCH-induced JAK3 degradation involves ubiquitination mediated by dimeric E3 ligase complexes containing both SKP2 and ASB2.
FIGURE 6.
Interaction of R980W with ASB2 and SKP2. A, wild type and mutant JAK3 were co-transfected into 293 cells with or without FLAG-ASB2 (A) or SKP2 (B). Whole cell lysates were used in immunoprecipitation with anti-FLAG (A) or anti-SKP2 (B) and control IgG antibodies. The precipitates and inputs were analyzed with the anti-HPC4 antibody for wild type and R980W JAK3. Anti-FLAG and anti-SKP2 antibodies were used to detect ASB2 and SKP2 proteins.
Both Cullin1 and Cullin5 Are Involved in NOTCH or ASB2/SKP2-stimulated JAK3 Degradation
The fact that both ASB2 and SKP2 were shown to promote JAK3 degradation suggests a possibility that these two proteins could facilitate the formation of heterodimeric E3 ligase complexes involving both Cullin1 and Cullin5 and enhance JAK3 ubiquitination as they do for JAK2 (33). If so, one would expect that expression of dominant-negative mutants of either Cullin1 or Cullin5 would prevent JAK3 degradation as Cullin1 is known to specifically interact with SKP2, whereas Cullin5 binds to ASB2 (48). On the other hand, if ASB2 and SKP2 act separately, DN-Cullin1 would specifically rescue SKP2-induced JAK3 degradation, whereas DN-Cullin5 would only block ASB2-mediated JAK3 turnover. To test these hypotheses, we made use of two dominant-negative mutants, DN-Cullin1 and DN-Cullin5, which lack the regions responsible for E2 enzyme recruitment and thus are catalytically inactive (49). In co-transfection assays, expression of ASB2 or SKP2 alone or together diminished JAK3 levels (Fig. 7, A and B, compare lanes 3, 7, and 11 with lanes 4, 8, and 12, respectively). However, co-expression of either DN-Cullin1 or DN-Cullin5 was capable of restoring JAK3 protein levels (Fig. 7, A and B, compare lane 4 with lanes 5 and 6; lane 8 with lanes 9 and 10; lane 12 with lanes 13 and 14). These results suggest that JAK3 degradation promoted by ASB2 and SKP2 depends on both Cullin1 and Cullin5, which likely form a dimeric complex together with ASB2 and SKP2 plus other associated molecules as we reported previously (33). As negative controls, the dominant-negative mutants had no obvious effects on JAK3 levels in the absence of ASB2 or SKP2 (Fig. 7, A and B, lanes 1, 2, 15, and 16).
FIGURE 7.
Cullin1 and Cullin5-based E3 ligases are involved in NOTCH or ASB2/SKP2-stimulated JAK3 degradation. Co-transfection experiments were performed using NIH3T3 cells as described in the legend to Fig. 1A except that DN-Cul1 or DN-Cul5 was included in indicated samples to examine their effects on JAK3 degradation induced by SKP2 or ASB2 (A), SKP2 plus ASB2 (B), and NΔE (C).
To determine whether NOTCH signaling utilizes a similar machinery to promote JAK3 degradation, JAK3 plus or minus NΔE were co-transfected into NIH3T3 cells with or without these dominant-negative mutants. Levels of JAK3 and the control were measured by immunoblotting. Expression of either DN-Cullin1 or DN-Cullin5 recovered JAK3 protein levels (Fig. 7C, compare lane 4 with lane 6 and lane 10 with lane 8, respectively). This result is in favor of our hypothesis that NOTCH-induced JAK3 ubiquitination employs a Cullin1 and Cullin5 containing the E3 ligase complex by stimulating the transcription of Asb2 and Skp2 genes.
DISCUSSION
JAK3 is an important player in mediating JAK-STAT signaling in lymphoid cells (19, 50). By binding the IL-2 receptor common γ chain (IL-2Rγ), it acts downstream of all cytokine receptors that utilize IL-2Rγ, for example, the IL-7 receptor (51). Null mutations of the Jak3 gene in mouse and human result in severe combined immunodeficiency (25, 26, 52, 53). Likewise, NOTCH signaling controls several crucial checkpoints in B and T cell development. Data presented here and elsewhere suggest an important cross-talk between the two signaling pathways, which is likely to be critical for regulating various lineage decisions and differentiation programs, as well as leukemogenesis (33, 34). For example, NOTCH is known to suppress B lymphopoiesis in the thymus and down-regulation of JAK3 could dampen IL-7 signaling necessary for B cell development (10, 22). Likewise, NOTCH plays a role in Th2 differentiation, which relies heavily on the fine-tuning of cytokine signaling (54). Therefore, understanding the biochemical mechanisms by which NOTCH signaling promotes the degradation of JAK3 will greatly aid the in vivo studies of the biological significance of the crosstalk.
In this report, we focused on the mechanism underlying NOTCH-induced JAK3 degradation. JAK3 belongs to a subfamily of non-receptor tyrosine kinases, consisting of JAK1, JAK2, JAK3, and TYK2. JAK2 has served as a prototype for this family of kinases and thus much of the available biochemical data have been about JAK2. For example, JAK2 is known to be bound to SOCS proteins, and structural studies predict that the SH2 domain of SOCS proteins bind to the activation loop of JAK2 (44). Interaction between JAK2 and SOCS proteins not only leads to inhibition of the kinase activity but also results in the ubiquitination and degradation of JAK2 by employing the ElonginB/C/Cullin5 (ECS)-containing E3 ubiquitin ligases (31, 55). Recently, we have shown that NOTCH-induced JAK2 degradation involves its downstream targets, the products of the Asb2 and Skp2 genes, which are transcriptionally activated by NOTCH signaling. By displacing SOCS1, ASB2 binds to elongins and Cullin5 and also recruits SKP2, which in turn brings SKP1 and Cullin1. Together, these molecules, along with the RING finger proteins and possibly other ancillary proteins, form a dimeric non-canonical E3 ligase complex for JAK2 ubiquitination (33).
Results from our analyses of JAK3 ubiquitination and degradation suggest that a similar E3 ligase complex also is utilized for JAK3 ubiquitination because both ASB2 and SKP2 associate with JAK3 and promote its degradation (Fig. 8). More importantly, functional inhibition of either Cullin1 or Cullin5, which binds SKP2 and ASB2, respectively, was able to prevent JAK3 degradation induced by NOTCH or ASB2 plus SKP2. By examining the interaction between ASB2 and JAK3, we provided further insights into the relationship between JAK3 and its E3 ligases. We show that multiple interfaces exist in both JAK3 and ASB2. The FERM domain of JAK3 is capable of interaction with ASB2 via either the N or C terminus. In contrast, the pseudo-kinase domain associates with the N terminus avidly but has no affinity for the C terminus. Interestingly, the kinase domain interacts poorly with either the N or C termini of ASB2 and mostly associates with ASB2 through SKP2. Although the interacting domains between ASB2 and JAK2 have not been thoroughly mapped, one distinction between JAK2 and JAK3 is that JAK2 does not appear to bind to the N-terminal region of ASB2 (33). Therefore, these results raise a question if the multiple interfaces of JAK3 with ASB2 strengthen the interaction of JAK3 with its E3 ligases, thus increasing the efficiency of its ubiquitination. To answer this question, a parallel comparison between the susceptibility of JAK2 and JAK3 to ASB2/SKP2-induced ubiquitination is necessary.
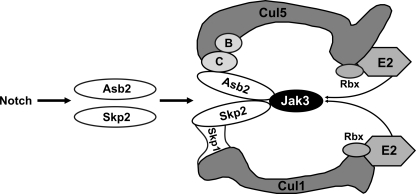
FIGURE 8.
Molecular interaction of JAK3 with its E3 ligase components. The model outlines NOTCH-induced JAK3 ubiquitination by transcriptional activation of the genes encoding ASB2 and SKP2, which mediate the assembly of a non-canonical dimeric E3 ligase complex. Subunits of the complex are as labeled. B and C represent for ElonginB and ElonginC, respectively. Rbx, ring box protein.
One of the motivations of this study is to develop NOTCH-resistant mutants for investigating the biological significance of NOTCH-mediated JAK3 degradation. We designed a mutation, R980W, based on structural information obtained in examining the interaction between SOCS1 and JAK2. The Arg-980 residue is equivalent to Lys-1011 in JAK2, which interacts with the SH2 domain of SOCS1 (44). Data from the analyses of this mutant turned out to be extremely interesting for several reasons. First, from a mechanistic perspective, the fact that the R980W mutant is resistant to ASB2-induced ubiquitination/degradation although defective in interaction with SKP2 but not ASB2 suggests that ASB2 and SKP2 indeed cooperate in JAK3 ubiquitination, further supporting our dimeric E3 ligase model. Second, the fact that the R980W mutant retains cytokine-regulated kinase activity but maintains resistance to degradation suggests that the ability of JAK3 to efficiently interact with its E3 ligases rather than its enzymatically active configuration is crucial for its ubiquitination (47, 56). It has been shown that a JAK2 mutant, Y1007F, which lacks cytokine-induced kinase activity and is thus unable to auto-phosphorylate at Tyr-1007, is also resistant to degradation induced by either SOCS1 or NOTCH (31).3 Because both phospho-Tyr-1007 and Lys-1011 (equivalent to Arg-980 in JAK3) are predicted to make direct contacts with the SH2 domain of SOCS1 (44), these residues are essential for JAK protein ubiquitination. However, the questions that remain are whether the Arg-980 residue mediates the interaction with SKP2 through a SH2 domain and if so, whether such a domain is present in SKP2 or in unknown intermediates.
Finally, the properties of the R980W mutant, which include the resistance to NOTCH or ASB2/SKP2-induced degradation while maintaining cytokine-regulated kinase activities, make it an excellent tool for investigations into the biological significance of NOTCH-induced degradation of JAK3. For example, we are particularly interested in learning whether R980W could act in concert with a NOTCH-resistant E2A mutant to reverse the inhibitory effects of NOTCH on B cell development and the stimulatory effect of NOTCH on marginal zone B cell formation. It would also be interesting to determine whether this mutant has any leukemogenic potential (57, 58).
Acknowledgments
We thank Drs. Raphael Kopan, Scott Plafker, Junya Kohroki, and Hui Zhang for reagents. We are grateful to Dr. Lei Nie for advice and Ying Zhao for help in creating Jak3 cDNA.
This work was supported, in whole or in part, by National Institutes of Health Grant AI56129 (to X.-H. S.).
L. Nie and X.-H. Sun, unpublished data.
- IC
- intracellular domain
- SOCS
- suppressors of cytokine signaling
- JAK3
- Janus kinase 3
- SOCS
- suppressors of cytokine signaling
- RIPA
- radioimmune precipitation assay buffer
- SCF
- SKP1/Cullin1/F-box protein FERM, 4.1, ezrin, radixin, moesin
- DN
- dominant negative.
REFERENCES
- 1. Artavanis-Tsakonas S., Rand M. D., Lake R. J. (1999) Science 284, 770–776 [DOI] [PubMed] [Google Scholar]
- 2. Simpson P. (1998) Semin. Cell Dev. Biol. 9, 581–582 [DOI] [PubMed] [Google Scholar]
- 3. Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., Furukawa T., Honjo T. (1995) Curr. Biol. 5, 1416–1423 [DOI] [PubMed] [Google Scholar]
- 4. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 5. Callahan R., Raafat A. (2001) J. Mammary. Gland. Biol. Neoplasia 6, 23–36 [DOI] [PubMed] [Google Scholar]
- 6. Allman D., Punt J. A., Izon D. J., Aster J. C., Pear W. S. (2002) Cell 109, S1–11 [DOI] [PubMed] [Google Scholar]
- 7. Maillard I., Fang T., Pear W. S. (2005) Annu. Rev. Immunol. 23, 945–974 [DOI] [PubMed] [Google Scholar]
- 8. Tanigaki K., Honjo T. (2007) Nat. Immunol. 8, 451–456 [DOI] [PubMed] [Google Scholar]
- 9. Radtke F., Fasnacht N., Macdonald H. R. (2010) Immunity 32, 14–27 [DOI] [PubMed] [Google Scholar]
- 10. Wilson A., MacDonald H. R., Radtke F. (2001) J. Exp. Med. 194, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H. R., Aguet M. (1999) Immunity 10, 547–558 [DOI] [PubMed] [Google Scholar]
- 12. Tanigaki K., Han H., Yamamoto N., Tashiro K., Ikegawa M., Kuroda K., Suzuki A., Nakano T., Honjo T. (2002) Nat. Immunol. 3, 443–450 [DOI] [PubMed] [Google Scholar]
- 13. Saito T., Chiba S., Ichikawa M., Kunisato A., Asai T., Shimizu K., Yamaguchi T., Yamamoto G., Seo S., Kumano K., Nakagami-Yamaguchi E., Hamada Y., Aizawa S., Hirai H. (2003) Immunity 18, 675–685 [DOI] [PubMed] [Google Scholar]
- 14. Maillard I., Weng A. P., Carpenter A. C., Rodriguez C. G., Sai H., Xu L., Allman D., Aster J. C., Pear W. S. (2004) Blood 104, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 15. Leonard W. J. (2001) Int. J. Hematol. 73, 271–277 [DOI] [PubMed] [Google Scholar]
- 16. Murray P. J. (2007) J. Immunol. 178, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 17. Gurniak C. B., Berg L. J. (1996) Blood 87, 3151–3160 [PubMed] [Google Scholar]
- 18. Thomis D. C., Berg L. J. (1997) Curr. Opin. Immunol. 9, 541–547 [DOI] [PubMed] [Google Scholar]
- 19. O'Shea J. J., Husa M., Li D., Hofmann S. R., Watford W., Roberts J. L., Buckley R. H., Changelian P., Candotti F. (2004) Mol. Immunol. 41, 727–737 [DOI] [PubMed] [Google Scholar]
- 20. von Freeden-Jeffry U., Vieira P., Lucian L. A., McNeil T., Burdach S. E., Murray R. (1995) J. Exp. Med. 181, 1519–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moore T. A., von Freeden-Jeffry U., Murray R., Zlotnik A. (1996) J. Immunol. 157, 2366–2373 [PubMed] [Google Scholar]
- 22. Milne C. D., Paige C. J. (2006) Semin. Immunol. 18, 20–30 [DOI] [PubMed] [Google Scholar]
- 23. He Y. W., Nakajima H., Leonard W. J., Adkins B., Malek T. R. (1997) J. Immunol. 158, 2592–2599 [PubMed] [Google Scholar]
- 24. Noguchi M., Nakamura Y., Russell S. M., Ziegler S. F., Tsang M., Cao X., Leonard W. J. (1993) Science 262, 1877–1880 [DOI] [PubMed] [Google Scholar]
- 25. Thomis D. C., Gurniak C. B., Tivol E., Sharpe A. H., Berg L. J. (1995) Science 270, 794–797 [DOI] [PubMed] [Google Scholar]
- 26. Park S. Y., Saijo K., Takahashi T., Osawa M., Arase H., Hirayama N., Miyake K., Nakauchi H., Shirasawa T., Saito T. (1995) Immunity 3, 771–782 [DOI] [PubMed] [Google Scholar]
- 27. Cornejo M. G., Kharas M. G., Werneck M. B., Le Bras S., Moore S. A., Ball B., Beylot-Barry M., Rodig S. J., Aster J. C., Lee B. H., Cantor H., Merlio J. P., Gilliland D. G., Mercher T. (2009) Blood 113, 2746–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alexander W. S., Hilton D. J. (2004) Annu. Rev. Immunol. 22, 503–529 [DOI] [PubMed] [Google Scholar]
- 29. O'shea J. J. (2004) Ann. Rheum. Dis. 63, ii67–ii71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wormald S., Hilton D. J. (2004) J. Biol. Chem. 279, 821–824 [DOI] [PubMed] [Google Scholar]
- 31. Ungureanu D., Saharinen P., Junttila I., Hilton D. J., Silvennoinen O. (2002) Mol. Cell Biol. 22, 3316–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. (2004) Genes Dev. 18, 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nie L., Zhao Y., Wu W., Yang Y. Z., Wang H. C., Sun X. H. (2011) Cell Res. 21, 754–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nie L., Perry S. S., Zhao Y., Huang J., Kincade P. W., Farrar M. A., Sun X. H. (2008) Mol. Cell Biol. 28, 2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nie L., Xu M., Vladimirova A., Sun X. H. (2003) EMBO J. 22, 5780–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sarmento L. M., Huang H., Limon A., Gordon W., Fernandes J., Tavares M. J., Miele L., Cardoso A. A., Classon M., Carlesso N. (2005) J. Exp. Med. 202, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jackson P. K., Eldridge A. G. (2002) Mol. Cell 9, 923–925 [DOI] [PubMed] [Google Scholar]
- 38. Hilton D. J., Richardson R. T., Alexander W. S., Viney E. M., Willson T. A., Sprigg N. S., Starr R., Nicholson S. E., Metcalf D., Nicola N. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pui J. C., Allman D., Xu L., DeRocco S., Karnell F. G., Bakkour S., Lee J. Y., Kadesch T., Hardy R. R., Aster J. C., Pear W. S. (1999) Immunity 11, 299–308 [DOI] [PubMed] [Google Scholar]
- 40. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 41. Rezaie A. R., Fiore M. M., Neuenschwander P. F., Esmon C. T., Morrissey J. H. (1992) Protein Expr. Purif. 3, 453–460 [DOI] [PubMed] [Google Scholar]
- 42. Saharinen P., Silvennoinen O. (2002) J. Biol. Chem. 277, 47954–47963 [DOI] [PubMed] [Google Scholar]
- 43. Yamaoka K., Saharinen P., Pesu M., Holt V. E., 3rd, Silvennoinen O., O'Shea J. J. (2004) Genome Biol. 5, 253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giordanetto F., Kroemer R. T. (2003) Protein Eng. 16, 115–124 [DOI] [PubMed] [Google Scholar]
- 45. Bergamin E., Wu J., Hubbard S. R. (2006) Structure 14, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 46. Bullock A. N., Rodriguez M. C., Debreczeni J. E., Songyang Z., Knapp S. (2007) Structure 15, 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Feng J., Witthuhn B. A., Matsuda T., Kohlhuber F., Kerr I. M., Ihle J. N. (1997) Mol. Cell Biol. 17, 2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yan Q., Kamura T., Cai Y., Jin J., Ivan M., Mushegian A., Conaway R. C., Conaway J. W. (2004) J. Biol. Chem. 279, 43019–43026 [DOI] [PubMed] [Google Scholar]
- 49. Jin J., Ang X. L., Shirogane T., Wade Harper J. (2005) Methods Enzymol. 399, 287–309 [DOI] [PubMed] [Google Scholar]
- 50. Candotti F., Oakes S. A., Johnston J. A., Giliani S., Schumacher R. F., Mella P., Fiorini M., Ugazio A. G., Badolato R., Notarangelo L. D., Bozzi F., Macchi P., Strina D., Vezzoni P., Blaese R. M., O'Shea J. J., Villa A. (1997) Blood 90, 3996–4003 [PubMed] [Google Scholar]
- 51. Pesu M., Laurence A., Kishore N., Zwillich S. H., Chan G., O'Shea J. J. (2008) Immunol. Rev. 223, 132–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Macchi P., Villa A., Giliani S., Sacco M. G., Frattini A., Porta F., Ugazio A. G., Johnston J. A., Candotti F., O'Shea J. J. (1995) Nature 377, 65–68 [DOI] [PubMed] [Google Scholar]
- 53. Russell S. M., Tayebi N., Nakajima H., Riedy M. C., Roberts J. L., Aman M. J., Migone T. S., Noguchi M., Markert M. L., Buckley R. H., O'Shea J. J., Leonard W. J. (1995) Science 270, 797–800 [DOI] [PubMed] [Google Scholar]
- 54. Tu L., Fang T. C., Artis D., Shestova O., Pross S. E., Maillard I., Pear W. S. (2005) J. Exp. Med. 202, 1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Waiboci L. W., Ahmed C. M., Mujtaba M. G., Flowers L. O., Martin J. P., Haider M. I., Johnson H. M. (2007) J. Immunol. 178, 5058–5068 [DOI] [PubMed] [Google Scholar]
- 56. Chatti K., Farrar W. L., Duhé R. J. (2004) Biochemistry 43, 4272–4283 [DOI] [PubMed] [Google Scholar]
- 57. Walters D. K., Mercher T., Gu T. L., O'Hare T., Tyner J. W., Loriaux M., Goss V. L., Lee K. A., Eide C. A., Wong M. J., Stoffregen E. P., McGreevey L., Nardone J., Moore S. A., Crispino J., Boggon T. J., Heinrich M. C., Deininger M. W., Polakiewicz R. D., Gilliland D. G., Druker B. J. (2006) Cancer Cell 10, 65–75 [DOI] [PubMed] [Google Scholar]
- 58. Vainchenker W., Dusa A., Constantinescu S. N. (2008) Semin. Cell Dev. Biol. 19, 385–393 [DOI] [PubMed] [Google Scholar]