Background: The precise molecular function of Isa1 and Isa2 proteins in Fe/S protein biogenesis is unknown.
Results: The Isa1-Isa2 complex binds iron in vivo and acts late in the synthesis of [4Fe-4S] clusters.
Conclusion: Mitochondria possess specialized factors for maturation of [4Fe-4S] proteins.
Significance: This study defines the role of Isa proteins in mitochondrial Fe/S protein biogenesis.
Keywords: Biosynthesis, Enzymes, Iron, Iron-Sulfur Protein, Mitochondria, Vitamins and Cofactors
Abstract
Most eukaryotes contain iron-sulfur cluster (ISC) assembly proteins related to Saccharomyces cerevisiae Isa1 and Isa2. We show here that Isa1 but not Isa2 can be functionally replaced by the bacterial relatives IscA, SufA, and ErpA. The specific function of these “A-type” ISC proteins within the framework of mitochondrial and bacterial Fe/S protein biogenesis is still unresolved. In a comprehensive in vivo analysis, we show that S. cerevisiae Isa1 and Isa2 form a complex that is required for maturation of mitochondrial [4Fe-4S] proteins, including aconitase and homoaconitase. In contrast, Isa1-Isa2 were dispensable for the generation of mitochondrial [2Fe-2S] proteins and cytosolic [4Fe-4S] proteins. Targeting of bacterial [2Fe-2S] and [4Fe-4S] ferredoxins to yeast mitochondria further supported this specificity. Isa1 and Isa2 proteins are shown to bind iron in vivo, yet the Isa1-Isa2-bound iron was not needed as a donor for de novo assembly of the [2Fe-2S] cluster on the general Fe/S scaffold proteins Isu1-Isu2. Upon depletion of the ISC assembly factor Iba57, which specifically interacts with Isa1 and Isa2, or in the absence of the major mitochondrial [4Fe-4S] protein aconitase, iron accumulated on the Isa proteins. These results suggest that the iron bound to the Isa proteins is required for the de novo synthesis of [4Fe-4S] clusters in mitochondria and for their insertion into apoproteins in a reaction mediated by Iba57. Taken together, these findings define Isa1, Isa2, and Iba57 as a specialized, late-acting ISC assembly subsystem that is specifically dedicated to the maturation of mitochondrial [4Fe-4S] proteins.
Introduction
Iron-sulfur (Fe/S) proteins perform central tasks in electron transport, catalysis, and the regulation of environmental responses (1–3). The complex bacterial biosynthetic systems that assist in the assembly of Fe/S clusters and their transfer into apoproteins fall into three classes: the widely distributed housekeeping ISC2 assembly system; the NIF system, a dedicated assembly apparatus for the nitrogenase of nitrogen-fixing bacteria; and the SUF system, which is present in several bacterial taxa and in plastids (3–7). In eukaryotes, mitochondria are central for Fe/S protein biogenesis (8–12). They harbor the ISC assembly machinery that is closely related to the bacterial ISC system and is essential for maturation of all cellular Fe/S proteins, whether located in the mitochondria, cytosol, or nucleus. Biosynthesis of extramitochondrial Fe/S proteins further depends on the mitochondrial “ISC export machinery” that exports an unknown component required for maturation of cytosolic and nuclear proteins, a step carried out by the cytosolic Fe/S protein assembly (CIA) system (9, 10, 13). The components involved in Fe/S maturation in eukaryotes are highly conserved, and most are essential for cell viability, underscoring the importance of Fe/S protein function for the eukaryotic cell.
Fe/S cluster assembly in mitochondria and bacteria is initiated by a cysteine desulfurase (in yeast termed Nfs1-Isd11) that abstracts sulfur from cysteine and transfers it to the essential U-type ISC assembly proteins that serve as scaffolds for the de novo synthesis of the Fe/S co-factor (5, 6, 14–16). In yeast, de novo Fe/S cluster synthesis on the U-type proteins Isu1 and Isu2 involves an electron transfer chain consisting of the ferredoxin reductase Arh1 and the ferredoxin Yah1. In addition, the desulfurase and the Isu proteins interact with frataxin (yeast Yfh1), which may serve as an iron donor and/or a regulator of desulfurase activity (17, 18). The subsequent cluster transfer to recipient apoproteins is facilitated by the Hsp70 chaperone Ssq1 (bacterial HscA), its cognate J-type co-chaperone Jac1 (bacterial HscB), and the monothiol glutaredoxin Grx5 (7, 19, 20).
The function of two additional members of the yeast mitochondrial ISC assembly machinery, the matrix proteins Isa1 and Isa2, is poorly understood. They are related to the ubiquitous “A-type” ISC assembly factors from bacteria and other eukaryotes (21) and interact with the ISC assembly protein Iba57 (22). In contrast to most other ISC components, Isa1 and Isa2 are not required for cell viability, but low levels of either protein induce virtually identical phenotypes, including respiratory incompetence and glutamate-lysine auxotrophy, demonstrating their involvement in cellular ISC assembly (23–25). Deletion of Iba57 elicits similar phenotypes as the lack of either Isa1 or Isa2 (22). In bacteria, A-type ISC proteins are diverse and widespread (3, 6, 21). Escherichia coli harbors three canonical A-type proteins, IscA, SufA, and ErpA, and the non-canonical NfuA. Deletion of iscA is not associated with a clear phenotype, whereas that of ErpA or a double deletion of iscA and sufA results in strong growth defects (26, 27). These phenotypes frequently aggravate under special growth conditions, such as iron limitation or oxidative stress, and are linked to the loss of function of crucial cellular Fe/S proteins (3, 21, 26–33). Canonical A-type ISC proteins contain three highly conserved cysteine residues in two segments close to the terminus that are essential for function in vivo (21, 23, 25, 30). These residues are part of a flexible structure that was predicted to bind iron or an Fe/S cluster (34, 35). In fact, bacterial and eukaryotic A-type proteins were shown to bind Fe/S clusters upon chemical reconstitution in vitro, and in vivo Fe/S co-factor binding has been demonstrated for SufA (3, 6, 36). The Fe/S cluster assembled on A-type proteins by can be transferred to recipient Fe/S apoproteins in vitro. Collectively, these observations may indicate that bacterial A-type ISC proteins play a scaffold role for de novo synthesis of Fe/S clusters before their transfer to apoproteins (3, 6, 37–39). In addition, E. coli ISCA and SufA and human ISCA have been demonstrated to bind mononuclear iron (27, 30, 32, 40–42). Based on in vitro studies, this iron moiety was proposed to be used for the de novo synthesis of Fe/S co-factors on U-type ISC assembly proteins, suggesting an iron chaperone function for the A-type ISC proteins.
Here, we address several key questions concerning the physiological roles of the Isa proteins in Fe/S protein assembly in yeast. For instance, we asked by yeast complementation experiments whether the two yeast Isa proteins and the functionally diverse bacterial A-type ISC proteins perform orthologous or distinct functions. We then comprehensively investigated numerous cellular Fe/S proteins for their in vivo dependence on Isa1 and Isa2 function to gain insights into the substrate specificity of the Isa proteins. A major goal of our study was the understanding of the functional relation between the Isa proteins and the major Fe/S scaffold proteins Isu1-Isu2. We asked whether de novo Fe/S cluster synthesis on Isu proteins depends on Isa proteins, as proposed for bacterial A-type ISC proteins (3, 6). We also investigated what kind of metal co-factor is bound to the Isa proteins and what role this co-factor might play. Because we found in vivo evidence for iron rather than Fe/S cluster binding to the Isa proteins, we investigated its potential physiological role. In this respect, we were interested in the particular task of Iba57. Together, our findings show that the Isa proteins, in close cooperation with Iba57, form a specialized ISC assembly subcomplex that is dedicated to the maturation of virtually all mitochondrial proteins carrying a [4Fe-4S] cluster, presumably by transferring iron from the Isa proteins in an Iba57-facilitated fashion to recipient apoproteins.
EXPERIMENTAL PROCEDURES
Yeast Strains, Cell Growth, and Plasmid Constructs
Yeast strains used in this study are listed in supplemental Table SI. Cells were cultivated in rich medium (YP) or minimal medium containing a minimal set of supplements (SC) and 2% (w/v) glucose or galactose (43). Iron-depleted minimal media were prepared using yeast nitrogen base lacking FeCl3 (ForMedium). Media for anaerobic cell growth were supplemented with 0.2% Tween 80, 25 μg/ml ergosterol, and 20 μg/ml methionine. Gal-ISA1 and Gal-ISA2 and derivatives of these strains were grown in SD minimal medium for 4 days to deplete the Isa proteins to critical levels. Repression of other Gal strains was performed as described in the corresponding literature (see supplemental Table SI). Plasmids used in this study are listed in supplemental Table SII. Constructs were verified by DNA sequencing and/or functional complementation of corresponding yeast mutant.
Miscellaneous Methods
In vivo radiolabeling of yeast cells with 55FeCl3 (ICN) and measurement of 55Fe incorporation into Fe/S proteins by immunoprecipitation and scintillation counting were carried out as described previously (44, 45). Antibodies were raised in rabbits against recombinant proteins expressed in E. coli. Labeling of Fe/S proteins in mitochondrial extracts with 35S sulfur was performed using the protocol described in Ref. 46, except that 10 μCi of [35S]cysteine and 0.3 mm FeCl2 were used. The following published methods were used: manipulation of DNA and PCR (47); transformation of yeast cells (48); PCR-mediated gene replacement in yeast (49); preparation of yeast mitochondria (50); immunological techniques (51); and enzyme activities of succinate dehydrogenase, malate dehydrogenase, and cytochrome c oxidase (44, 45), aconitase (52), and homoaconitase (53). Error bars represent the standard error of the mean (n > 4).
RESULTS
Yeast Isa1 but Not Isa2 Is the Functional Orthologue of Bacterial A-type ISC Proteins
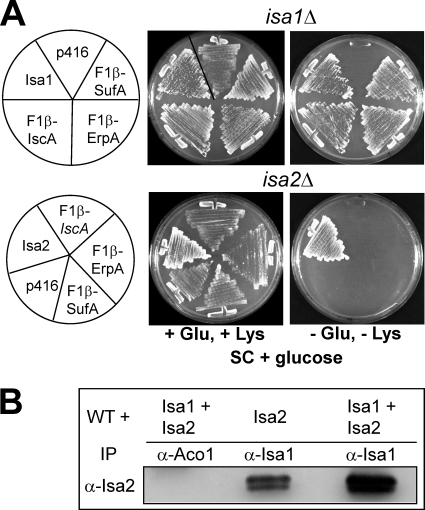
Most of the recent work on A-type ISC proteins has been carried out with bacterial members (3, 6). Whether the bacterial and eukaryotic A-type proteins are functional orthologues has not been established. We therefore tested if the bacterial homologues can complement the glutamate and lysine auxotrophies of Saccharomyces cerevisiae isa1Δ and isa2Δ cells (23–25). When fused to a mitochondrial targeting sequence, all three A-type members from E. coli, IscA, SufA, and ErpA, rescued the lysine and glutamate auxotrophies of isa1Δ cells (Fig. 1A). This identifies Isa1 as a functional orthologue of bacterial A-type ISC proteins. In contrast, neither bacterial protein was able to rescue isa2Δ cells. Apparently, S. cerevisiae Isa2, which carries a unique sequence insertion that is not found in other A-type ISC proteins, cannot be functionally replaced by the bacterial proteins, indicating that it performs a eukaryote-specific function.
FIGURE 1.
Bacterial A-type ISC proteins are functional orthologues of S. cerevisiae Isa1 but not of Isa2. A, S. cerevisiae strains isa1Δ and isa2Δ harboring p416-MET25 plasmids for the expression of E. coli IscA, SufA, or ErpA, each fused to the mitochondrial targeting sequence of the F1-ATPase subunit β (F1β), were grown on SC minimal medium supplemented with glucose (SD) in the presence or absence of Glu and Lys. Cells harboring either the empty vector p416-MET25 (p416) or vectors containing yeast ISA1 or ISA2 genes were cultivated in parallel as a control. B, WT cells overproducing Isa1 and/or Isa2, as indicated, were grown in liquid SD medium. Cell extracts were prepared and subjected to immunoprecipitation (IP) with immobilized antibodies against Isa1 or Aco1. The immunoprecipitated material was analyzed by immunostaining with antibodies against Isa2.
Isa1 and Isa2 Form a Complex in Yeast
Bacterial A-type proteins form oligomers. In order to test the complex formation between yeast Isa1 and Isa2, we immunoprecipitated Isa1 from cell lysates with specific antibodies. Indeed, upon overproduction, Isa2 co-purified with anti-Isa1 immunobeads (Fig. 1B). This association was specific, because it was not detected with antibodies that do not recognize Isa1 (Fig. 1B). When Isa1 was immunoprecipitated from cells overproducing both Isa proteins, the amount of co-immunoprecipitated Isa2 was largely increased, indicating a concentration-dependent complex formation. The inverse analysis was not possible, because Isa1 co-migrates with the small IgG subunits (data not shown). We have shown previously that Isa1 and Isa2 interact with Iba57 in S. cerevisiae (22). Deletions of either ISA1, ISA2, or IBA57 elicit virtually identical phenotypes (22–25). Therefore, it is likely that the hetero-oligomeric complex of Isa1, Isa2, and Iba57 is the functional unit in S. cerevisiae. The fact that this complex includes a component, Isa2, that cannot be functionally replaced by bacterial A-type ISC proteins, raises the interesting question of whether the physiological roles of bacterial A-type proteins are conserved in eukaryotes.
Fe/S Cluster Formation on Isu1 and Yah1 Does Not Require Isa1 or Isa2 in Vivo
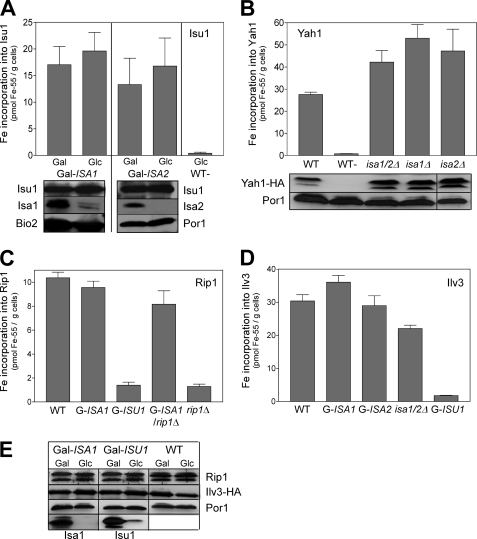
Recent work on bacterial A-type proteins suggests two different models for the function of this protein class. Bacterial IscA and SufA were shown to bind a transient Fe/S co-factor that can be transferred to recipient Fe/S apoferredoxins. Hence, it was suggested that the A-type ISC proteins may perform a role as an Fe/S scaffold or Fe/S transfer protein that operates in parallel or downstream, respectively, of U-type ISC proteins (3, 5, 6, 36). Alternatively, the proteins were demonstrated to bind mononuclear iron, and thus they were suggested to serve as iron chaperones promoting Fe/S cluster assembly on the U-type ISC scaffold proteins (30, 32, 40–42). In order to test the relevance of these observations for the yeast Isa proteins, we analyzed the de novo synthesis of the Fe/S cluster of Isu1 by 55Fe radiolabeling of Gal-ISA1 and Gal-ISA2 cells overproducing Isu1. In these Gal strains, the expression of ISA1 and ISA2 can be repressed upon cultivation in glucose (see supplemental Table SII). Following the radiolabeling, Isu1 was immunoprecipitated from cell lysates with specific antibodies. 55Fe bound to the immunobeads reflects the amount of de novo Fe/S cluster assembly on Isu1 and was quantified by scintillation counting (19). Remarkably, the amount of 55Fe co-immunoprecipitated with Isu1 from these cells slightly increased in both Gal-ISA1 and Gal-ISA2 under depleting conditions, indicating that the maturation of Isu1 can occur independently of the Isa proteins (Fig. 2A). Because Isu1 binds a [2Fe-2S] cluster, we compared these findings for 55Fe binding to the mitochondrial [2Fe-2S] ferredoxin Yah1 by in vivo radiolabeling of wild-type, isa1Δ, isa2Δ, and isa1/2Δ cells upon overexpression of Yah1. The amounts of 55Fe associated with Yah1 were 1.5-fold higher in cells lacking the Isa proteins compared with wild-type cells (Fig. 2B). As judged by immunostaining, levels of Yah1 and Isu1 remained unaltered in all strains analyzed. Similar results were obtained with Gal-ISA1 cells (not shown). Collectively, these findings demonstrate that the Isa proteins are not involved in the formation of the [2Fe-2S] clusters on Isu1 and Yah1 in vivo and thus do not serve as iron chaperones for mitochondrial Fe/S protein biogenesis. Moreover, these findings are compatible with the non-essential character of the ISA genes in yeast, because Yah1 and Isu1 are the only known essential mitochondrial Fe/S proteins.
FIGURE 2.
The Isa proteins are not required for the maturation of mitochondrial [2Fe-2S] proteins. A, Gal-ISA1 and Gal-ISA2 cells overproducing Isu1 were cultivated in iron-poor SC minimal medium containing Gal or Glc. Cells were labeled with 10 μCi of 55Fe for 2 h, and Isu1 was immunoprecipitated from crude cell extracts with specific antibodies. The amount of radioactivity co-precipitated with the immunobeads was quantified by liquid scintillation counting. Wild-type cells containing the empty plasmid (WT−) were analyzed in parallel. B, WT, isa1Δ, isa2Δ, and isa1/2Δ cells harboring plasmid p426-YAH1-HA were labeled with 55Fe, and radioactivity bound to Yah1 was determined by immunoprecipitation with α-HA antibodies as described above. The bottom panels of A and B are immunostains of the indicated proteins. C and D, 55Fe binding to the Rieske Fe/S protein (Rip1) (C) or Ilv3 (D) was determined in WT, the indicated Gal-regulatable mutant strains (Gal-ISA1, Gal-ISA2, and Gal-ISU1/Δisu2), and isa1/2Δ cells overproducing either a soluble, matrix-targeted Rieske Fe/S protein (F1β-ΔN95-Rip1) or Ilv3-HA, respectively. rip1Δ cells were analyzed as a control. E, levels of the indicated proteins in extracts from the respective cells were assessed by immunostaining. Gal strains were depleted to critical levels by cultivation in SD minimal medium for 3 days prior to analysis. Error bars, S.E. (n > 4). Por1, porin.
Fe/S Cluster Assembly on the Rieske Fe/S Protein and Ilv3 Does Not Require the Isa Proteins
The data above suggest that the Isa proteins are specialized ISC assembly factors that are required only for a subset of Fe/S proteins. In order to comprehensively define this subset, we next analyzed the de novo maturation of the Rieske Fe/S protein (Rip1) and of dihydroacid dehydratase Ilv3, the two other known mitochondrial [2Fe-2S] proteins in S. cerevisiae (see supplemental Fig. S1 for data indicating that Ilv3 is a [2Fe-2S] protein). In order to distinguish the insertion of 55Fe into the Fe/S cluster of Rip1 from that into cytochromes of complex III, we constructed vector p426-ΔN-RIP1, which allows the synthesis of a soluble Rieske protein in the mitochondrial matrix (54).
Wild-type levels of 55Fe were immunoprecipitated with anti-Rip1 antibodies from extracts of depleted Gal-ISA1 cells, whereas only background levels of 55Fe were found after depletion of Isu1 in Gal-ISU1/isu2Δ cells (Fig. 2C). As a control, we constructed a Gal-ISA1/rip1Δ strain that completely lacks an endogenous Rieske protein. In this strain, 55Fe binding to the matrix-targeted Rieske Fe/S protein remained at more than 80% of wild-type levels upon depletion of Isa1, indicating that the majority of 55Fe precipitated with our antibodies was indeed inserted into this form of the Rieske Fe/S protein in an Isa1-independent fashion. Iron binding to Ilv3 was determined in cells overproducing the protein with a C-terminal HA tag and gave a qualitatively similar picture. Although the depletion of Isu1 caused a strong reduction in 55Fe binding to Ilv3, the depletion of Isa1 or Isa2 had virtually no effect, and only a slight decrease was observed in isa1/2Δ cells (Fig. 2D). In all of these experiments, the levels of the investigated Fe/S proteins did not significantly change (Fig. 2E). Taken together, our analyses show that the Isa proteins are dispensable for the maturation of all yeast mitochondrial [2Fe-2S] proteins tested. Apparently, this type of Fe/S cluster is assembled on the Isu scaffold proteins and can be inserted into mitochondrial apoproteins without further assistance from the Isa proteins in vivo.
The de Novo Assembly of Aconitase-type Fe/S Proteins Requires the Isa Proteins
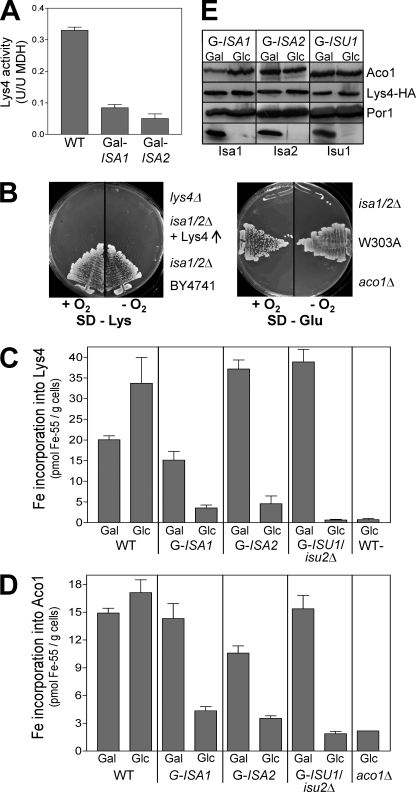
S. cerevisiae deleted for ISA1, ISA2, or IBA57 display auxotrophies for lysine and glutamate (Fig. 1A) (22). Glutamate biosynthesis requires the Fe/S protein aconitase (Aco1), and the biosynthesis of lysine involves both aconitase and homoaconitase (Lys4). Both proteins contain a [4Fe-4S] cluster and are predominantly located in mitochondria. The activity of aconitase is strongly diminished in cells depleted for Isa1 or Isa2 (24, 25). We therefore analyzed whether the Isa proteins are also required for Fe/S cluster formation on homoaconitase. Wild-type mitochondria displayed robust homoaconitase activities upon overexpression of a C-terminal HA-tagged version of Lys4 (Fig. 3A). In contrast, mitochondria from Isa-depleted Gal-ISA1 or Gal-ISA2 cells showed more than 4-fold lower enzyme activities, although Lys4-HA protein levels did not change upon Isa depletion (Fig. 3E). Oxidative stress could be excluded as a reason for the loss of activity of aconitase-type Fe/S proteins, because the auxotrophies for lysine and glutamate of isa1/2Δ cells that lack both ISA genes were not cured under anaerobic conditions, even upon overproduction of Lys4 (Fig. 3B). Depletion of Isa1 in Gal-ISA1 caused a 4.5-fold decrease in 55Fe incorporation into overproduced Lys4-HA in vivo (Fig. 3C). In Isa2-depleted Gal-ISA2 cells in vivo, 55Fe binding was 8-fold diminished, and only background levels remained in depleted Gal-ISU1/isu2Δ cells. Similar results were obtained with aconitase for 55Fe incorporation and protein levels (Fig. 3, D and E). Taken together, these data establish an essential and specific function of the Isa proteins in the de novo maturation of mitochondrial Fe/S proteins of the aconitase family. The auxotrophies of cells deleted for ISA1 or ISA2 indicate a strict requirement for A-type ISC assembly proteins for the de novo synthesis of aconitase-type Fe/S proteins in eukaryotes. The same loss of function of aconitase-type proteins was seen in cells with low levels of Iba57, consistent with the observation that Iba57 interacts with Isa1 and Isa2 (22).
FIGURE 3.
The Isa proteins are essential for the maturation of aconitase-type Fe/S proteins. A, homoaconitase (Lys4) enzyme activities were determined in mitochondria isolated from WT and from depleted Gal-ISA1 or Gal-ISA2 cells overproducing Lys4-HA from vector p426-TDH3. Activities were normalized to those of malate dehydrogenase (MDH). B, isa1/2Δ cells with or without a vector overproducing Lys4p-HA were cultivated in parallel with wild-type cells (W303-1A or BY4741), lys4Δ, and aco1Δ cells for 3 days under aerobic (+O2) or anaerobic conditions (−O2) on SD minimal medium lacking either Lys or Glu. C, 55Fe binding to overproduced Lys4-HA was determined in WT, Gal-ISA1, Gal-ISA2, and Gal-ISU1/Δisu2 cells cultivated under inducing (Gal) or repressive conditions (Glc), as described in the legend to Fig. 2. Wild-type cells containing the empty plasmid (WT−) were analyzed in parallel. D, 55Fe binding to aconitase was determined in WT and the indicated Gal strains cultivated under inducing (Gal) or repressive conditions (Glc). E, the total amounts of aconitase (Aco1), Lys4-HA, porin (Por1), Isa1, Isa2, and Isu1 in extracts from the indicated cells were assessed by immunostaining. Gal-ISA strains were repressed to critical levels by cultivation in SD minimal medium for 3 days prior to analysis. Gal-ISU1/Δisu2 cells were depleted for 16 h. aco1Δ cells served as a control. Error bars, S.E. (n > 4).
Mitochondrial Targeting of Two Bacterial Ferredoxins Supports the Specific Role of Isa Proteins in [4Fe-4S] Protein Maturation
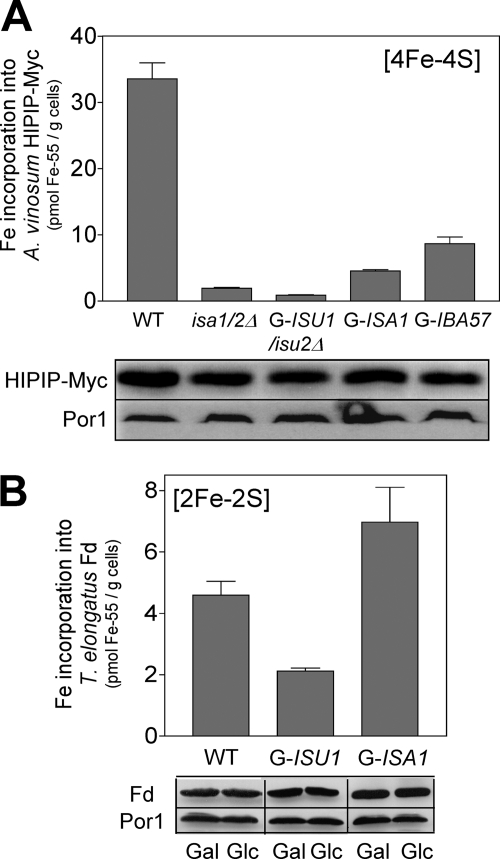
The observations above suggest a simple model for the function of the Isa proteins in eukaryotes. [2Fe-2S] proteins are formed in an Isa-independent manner, whereas the maturation of [4Fe-4S] cluster-containing proteins requires a joint function of the Isa and the Isu proteins. To support this idea, we analyzed the 55Fe binding to two low molecular mass bacterial ferredoxins, a [4Fe-4S] HiPIP from Allochromatium vinosum and a plant-type [2Fe-2S] ferredoxin from the cyanobacterium Thermosynechococcus elongatus. Genes for both proteins were fused to the coding information of a mitochondrial targeting sequence, the fusion genes were over expressed in S. cerevisiae, and the mitochondrial localization of the recombinant ferredoxins was verified by cell fractionation (not shown). In the case of HiPIP, in vivo 55Fe binding strongly declined upon depletion of either Isu1 or Isa1 in Gal-ISU1/isu2Δ and Gal-ISA1 cells, respectively, and in cells lacking both Isa1 and Isa2 (Fig. 4A). Thus, the formation of the [4Fe-4S] cluster of this foreign protein was dependent on both A-type and U-type ISC proteins, similar to the findings for yeast Aco1 and Lys4 (cf. Fig. 3). Remarkably, depletion of the Isa-interacting protein Iba57 caused a similar reduction in 55Fe binding to HiPIP (Fig. 4A). Similar to the Isa proteins, Iba57 is essential for the maturation of aconitase-type proteins in yeast but is not required in the maturation of the [2Fe-2S] protein Yah1 (22). In contrast, 55Fe binding to the plant-type [2Fe-2S] ferredoxin increased almost 2-fold in yeast cells depleted for Isa1 yet declined 2.5-fold upon depletion of Isu1 (Fig. 4B). This behavior is reminiscent of that of the other mitochondrial [2Fe-2S] proteins (Fig. 2) (19), verifying that the maturation of these proteins takes place in an Isa-independent manner in S. cerevisiae.
FIGURE 4.
The Isa proteins are required for incorporation of [4Fe-4S] but not [2Fe-2S] clusters into mitochondrial matrix-targeted, bacterial ferredoxins. The indicated strains overproducing either a Myc-tagged HiPIP [4Fe-4S] protein from A. vinosum (A) or a plant-type [2Fe-2S] ferredoxin (Fd) from the cyanobacterium T. elongatus (B) were cultivated in iron-poor minimal SD medium, and 55Fe binding was determined by radiolabeling and immunoprecipitation with anti-Myc or anti-ferredoxin antibodies. Protein levels in extracts from the respective cells were assessed by immunostaining (bottom panels). Gal-regulated strains were repressed to critical levels prior to the analyses. Error bars, S.E. (n > 4).
What about complex Fe/S proteins with multiple Fe/S clusters? We have shown previously that yeast isa1/2Δ and iba57Δ cells display a biotin auxotrophy and a lipoic acid deficiency that could be assigned to the inactivation of the radical AdoMet Fe/S enzymes Bio2 and Lip5 (22, 55). In addition, respiratory complex II, which contains [2Fe-2S], [3Fe-4S], and [4Fe-4S] clusters was inactive in cells depleted for Isa1 (supplemental Table SIII). This protein complex remained active in respiration-deficient strains, such as aco1Δ or cyt2Δ, indicating an Isa1-specific effect. These data strongly suggest that the function of composite [4Fe-4S]-containing proteins is severely affected in yeast cells with low levels of Isa proteins. Table 1 summarizes the effect of low levels of the Isa proteins on the different types of Fe/S proteins. This overview strongly suggests that the Isa proteins are specific ISC assembly factors dedicated to formation of mitochondrial [4Fe-4S] proteins.
TABLE 1.
Effects of Isa protein depletion on various mitochondrial Fe/S proteins
| [2Fe-2S] proteins (maturation normal) | [4Fe-4S] proteins (no maturation) | Complex Fe/S proteins (enzymes inactive) |
|---|---|---|
| Isu1a | Aconitase (Aco1) | Biotin synthase (Bio2) ([2Fe-2S], [4Fe-4S], AdoMet) |
| Rieske Fe/S | Homoaconitase (Lys4) | Lipoic acid synthase (Lip5) ([4Fe-4S], [4Fe-4S], AdoMet) |
| Ferredoxin (Yah1) | Succinate dehydrogenase (Sdh2) [2Fe-2S], [3Fe-4S], [4Fe-4S] | |
| Ilv3b | ||
| [2Fe-2S] ferredoxinc | HiPIPc |
a See Ref. 63.
b See Ref. 65 and supplemental Fig. S1.
c Mitochondrial matrix-targeted bacterial Fe/S proteins.
The Isa and Iba57 Proteins Are Dispensable for the Maturation of Extramitochondrial Fe/S Proteins
We have shown previously that depletion of Isa1 and Isa2 results in diminished activities of the cytosolic aconitase-type protein Leu1 due to decreased incorporation of its Fe/S co-factor, indicating that the Isa proteins are involved in the maturation of cytosolic Fe/S proteins (24, 25). However, the Isa proteins are not essential for this process, because cells deleted for ISA1 and ISA2 displayed no auxotrophies for leucine or methionine, amino acids that require cytosolic [4Fe-4S] proteins for their biogenesis (supplemental Fig. S2A) and show normal Leu1 activity (supplemental Fig. S2B). Consistent with this non-essential function, cells depleted for Isa1, Isa2, or the Isa-interacting protein Iba57 showed no deregulated cellular iron homeostasis and hardly any mitochondrial iron accumulation, two characteristic features of cells with low levels of components of the mitochondrial ISC assembly and export systems (16, 22). In these cells, the deregulation of cellular iron homeostasis is caused by low cytosolic levels of the (unknown) substrate of the ABC transporter Atm1 and is always associated with a reduced maturation of extramitochondrial Fe/S proteins.
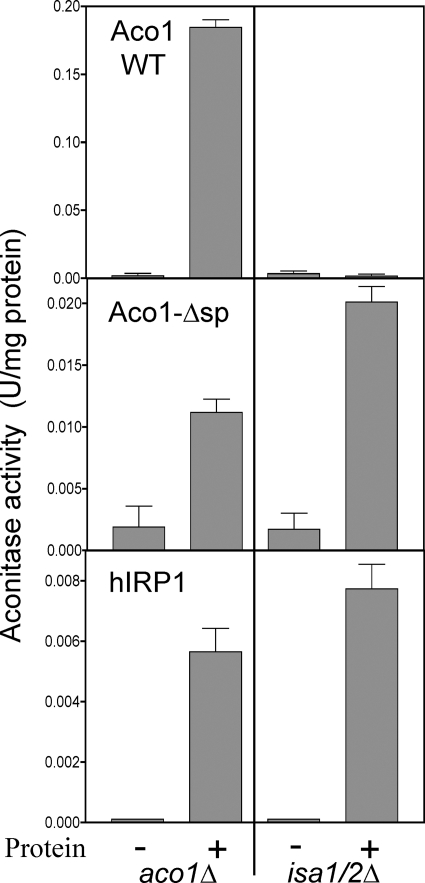
Collectively, these observations suggest that the Isa proteins play qualitatively different roles in the maturation of aconitase-type Fe/S proteins in the cytosol and in mitochondria. In order to explore this further, we analyzed the enzyme activities upon expression of native (Aco1 WT) and cytosolic versions of yeast aconitase (Aco1-Δsp) (56) and of human cytosolic aconitase (hIRP1) in isa1/2Δ cells. Aco1-Δsp lacking the mitochondrial targeting sequence showed detectable aconitase activities in cell extracts of isa1/2Δ cells and, similarly, in aco1Δ cells (Fig. 5). However, these activities were 10–15-fold lower than those for expression of native Aco1 in aco1Δ cells, indicating a comparatively poor maturation of Aco1 in the cytosol (56). Consistent with this idea, Aco1-Δsp did not rescue the glutamate auxotrophy of aco1Δ cells (supplemental Fig. S2D). Expression of wild-type aconitase in isa1/2Δ cells did not restore their glutamate auxotrophy and did not produce detectable aconitase activities, suggesting that wild-type aconitase is transported efficiently into mitochondria, where it cannot be matured due to the absence of the Isa proteins (Fig. 5 and supplemental Fig. S2D). In a similar way as seen for Aco1-Δsp, the expression of the hIRP1 in the cytosol resulted in detectable but low aconitase activity in extracts of both isa1/2Δ and aco1Δ cells (Fig. 5). These data clearly show that maturation of aconitase-type Fe/S proteins in the cytosol is possible without the Isa proteins. However, the low enzyme activities found in Aco1-Δsp- or hIRP1-complemented yeast cells indicated an inefficient maturation of aconitases in the yeast cytosol. Taken together, these data demonstrate that the essential role of the Isa proteins in the assembly of aconitase-type Fe/S proteins is restricted to mitochondria-localized proteins.
FIGURE 5.
The Isa proteins are not required for the maturation of cytosolic aconitase-type proteins. isa1/2Δ and aco1Δ cells were transformed with empty vector (−) or vector overexpressing (+) yeast aconitase with (Aco1 WT) or without its mitochondrial targeting sequence (Aco1-Δsp) or the human iron regulatory protein 1 (hIRP1). Growth was in SD minimal medium, and cell extracts were prepared and analyzed for aconitase activity. Error bars, S.E. (n > 4).
The Isa Proteins Bind Iron That Accumulates upon Depletion of Iba57
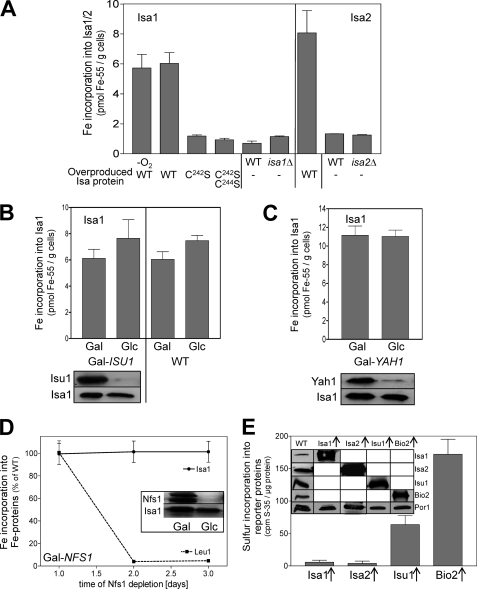
Several studies have demonstrated that A-type ISC proteins from both bacteria and eukaryotes bind iron or Fe/S clusters (3, 5, 6). We used the 55Fe radiolabeling-immunoprecipitation technique to analyze the metal co-factor binding properties of the yeast A-type proteins in vivo. 55Fe binding to both Isa1 and Isa2 was detectable in wild-type cells overproducing Isa1 or Isa2 and showed no sensitivity to chelators, detergents, or ambient oxygen (Fig. 6A). Only background levels of 55Fe were co-immunoprecipitated with the Isa1 mutant proteins C242S and C242S/C244S in which these conserved C-terminal cysteine residues were replaced by serine. The essential character of this conserved cysteine pair for in vivo function of Isa1 and Isa2 suggests that the iron binding capacity of these residues is of physiological importance (6, 23, 25, 30).
FIGURE 6.
Iron binding to Isa1 and Isa2 is independent of the sulfur supply from the mitochondrial ISC assembly machinery. A, 55Fe binding to Isa1 or Isa2 was determined by in vivo radiolabeling of wild-type cells overproducing either Isa1 (WT), the site-directed Isa1 mutant proteins C242S and C242S/C244S, or Isa2 (WT; right panel). Cell disruption and immunoprecipitation were performed under anaerobic (−O2) or aerobic (all other samples) conditions. isa1Δ, isa2Δ and WT cells containing the empty plasmid (−) served as controls. B and C, 55Fe binding to Isa1 was determined in Isa1-overproducing Gal-ISU1/Δisu2 or WT (B) and Gal-YAH1 cells (C) cultivated under inducing (Gal) or repressive conditions (Glc). Bottom panels of B and C show an immunostaining of the indicated proteins in cell extracts. D, Gal-NFS1 cells overproducing Isa1 were cultivated under repressive conditions in SD medium. At the indicated time points, 55Fe binding to Isa1 and Leu1 was determined in parallel by immunoprecipitation and scintillation counting. Inset, levels of the indicated proteins in cell extracts were assessed by immunostaining. E, detergent extracts of isolated mitochondria from wild-type cells overproducing (↑) Isa1, Isa2, Isu1, or Bio2 were radiolabeled with 10 μCi of [35S]cysteine for 2.5 h under anaerobic conditions. [35S]Sulfide binding was determined by immunoprecipitation and scintillation counting. Inset, protein levels in mitochondria from the indicated cells were assessed by immunostaining. Error bars, S.E. (n > 4).
In order to discriminate between the binding of iron or an Fe/S cluster to the Isa proteins, we studied the dependence of the 55Fe association on core members of the mitochondrial ISC assembly system. Surprisingly, 55Fe binding to overproduced Isa1 remained at wild-type levels upon depletion of Isu1 or Yah1 (Fig. 6, B and C). Likewise, depletion of the cysteine desulfurase Nfs1, which provides the sulfur moiety for all cellular Fe/S clusters (57, 58), did not significantly alter 55Fe binding to Isa1, in contrast to the 55Fe incorporation into the Fe/S protein Leu1, which declined more than 50-fold (Fig. 6D). These data suggest that Isa1 binds iron under conditions of low levels of the core components of the mitochondrial ISC assembly machinery. To further explore the character of the iron binding center of the Isa proteins, we attempted to directly measure the incorporation of [35S]sulfide into these proteins. Because [35S]Cys becomes inserted into newly synthesized proteins in in vivo approaches, we developed an in vitro 35S-radiolabeling procedure that was based on a method to detect the incorporation of 55Fe into Fe/S proteins in mitochondrial lysates (46). Isolated mitochondria from iron-starved wild-type yeast cells overproducing Bio2, Isu1, Isa1, or Isa2 were lysed by detergent and incubated with 10 μCi of [35S]cysteine and 0.3 mm FeCl2 under anaerobic conditions. Subsequently, the incorporation of 35S into these proteins was determined by immunoprecipitation of the four proteins and scintillation counting. In each case, a background signal obtained with wild-type mitochondria (no overproduction) was subtracted. Significant amounts of 35S were incorporated into the Fe/S cluster-containing Bio2 and Isu1 by this in vitro technique (Fig. 6E). In striking contrast, only background levels of 35S were associated with both Isa1 and Isa2, although the proteins were highly expressed. Taken together, these findings support the notion that the yeast Isa proteins bind iron rather than an Fe/S cluster.
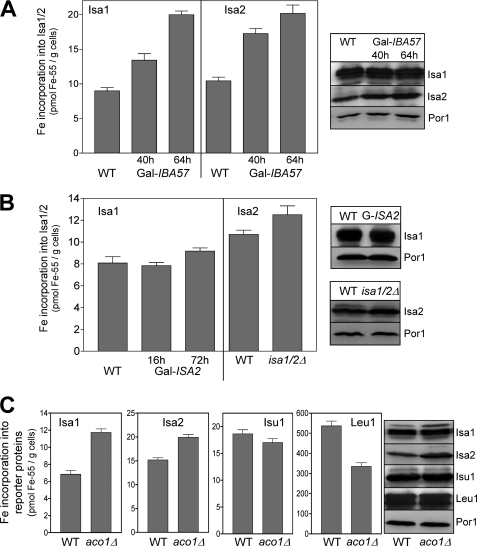
Next, we tested whether iron binding to the Isa proteins is influenced in cells depleted for the Isa-interacting protein Iba57 (22). In the simplest scenario, Iba57 could be involved in either iron loading or dissociation of the Isa proteins. 55Fe binding on Isa1 increased 1.5-fold in Gal-IBA57 cells after 40 h of depletion of Iba57 (Fig. 7A) (i.e. at a time when the Fe/S protein assembly defects developed in these cells) (22). After 64 h of depletion, when the cells showed their full phenotype, a 2.2-fold increase in 55Fe binding to Isa1 was observed. A similar Iba57-dependent iron accumulation was seen for Isa2 (Fig. 7A). These data are reminiscent of the Fe/S cluster accumulation on Isu1 in the absence of the dedicated chaperones Ssq1 and Jac1 or the monothiol glutaredoxin Grx5 (19) and hence suggested that iron bound to the Isa proteins may be transferred to recipient Fe/S proteins in a step involving Iba57. We then tested whether iron binding on Isa1 depends on Isa2 and vice versa (Fig. 7B). Depletion of Isa2 had little effect on iron binding to Isa1, and that on Isa2 was marginally (1.2-fold) increased in cells deleted for ISA2. Apparently, the Isa proteins can bind iron independently of each other.
FIGURE 7.
Depletion of Iba57 results in iron accumulation on the Isa proteins. A, 55Fe binding to Isa1 or Isa2 was determined in WT and Gal-IBA57 cells overproducing Isa1 or Isa2 as in Fig. 2. Gal-IBA57 cells were cultivated for 40 or 64 h in SD minimal medium prior to analysis. B, 55Fe binding to the Isa proteins was determined for Gal-ISA2 cells overproducing Isa1 and isa1/2Δ cells overproducing Isa2. Gal-ISA2 cells were cultivated for either 16 or 72 h in SD medium prior to analysis. Protein levels were assessed by immunostaining of whole cell extracts. C, 55Fe binding to the indicated proteins was determined in WT and aco1Δ cells overproducing Isa1, Isa2, or Isu1. Protein levels were assessed by immunostaining. Error bars, S.E. (n > 4).
Finally, we investigated whether a similar iron accumulation on the Isa proteins as that seen in the absence of Iba57 was observable when the abundant mitochondrial [4Fe-4S] protein aconitase was missing. In such a situation iron may accumulate on the Isa proteins, because they are underemployed. Consistent with this idea, 55Fe binding on Isa1 was 1.7-fold higher in aco1Δ than in wild-type cells, and a smaller (1.3-fold) increase was observed for Isa2 (Fig. 7C). This increase was significant, because the strong phenotype of aco1Δ cells generally results in a decline of iron incorporation into Fe/S proteins, such as Leu1 (Fig. 7C). Collectively, the results suggest that the iron bound to the Isa proteins accumulates when either Iba57 or major target Fe/S proteins such as aconitase are depleted.
DISCUSSION
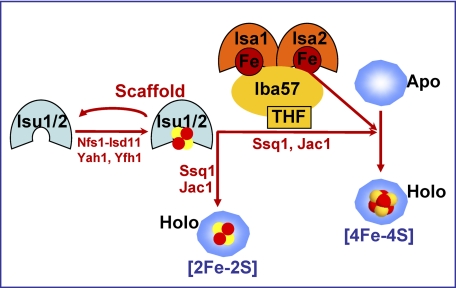
A-type ISC assembly proteins related to Isa1 and Isa2 of S. cerevisiae are found in most organisms (21). Despite their ubiquitous nature and high degree of structural conservation, their role in Fe/S protein assembly, in particularly in eukaryotes, is not resolved (15, 16). Here, we have carried out a comprehensive analysis of the requirement of Isa1 and Isa2 for the maturation of mitochondrial Fe/S proteins in S. cerevisiae. Our results can be summarized in a simple working model for the function of the Isa proteins in mitochondrial Fe/S protein biogenesis (Fig. 8 and Table 1). The maturation of [2Fe-2S] proteins was unaffected in the absence of the Isa proteins, whereas that of mitochondrial proteins containing [4Fe-4S] clusters was dependent on their function. The functional requirement of the Isa proteins included complex proteins with both types of Fe/S clusters. In the latter case, the reason for their inactivity may also be the defective assembly of the [4Fe-4S] cluster, although no clear de novo synthesis defects were found for the radical AdoMet enzymes Bio2 and Lip5 (22, 55). Probably, the assembly of the [2Fe-2S] co-factor remains normal, and/or an inactive co-factor is inserted into the [4Fe-4S] site in the absence of the Isa proteins.
FIGURE 8.
Working model for the assembly of mitochondrial [4Fe-4S] proteins. The maturation of all mitochondrial Fe/S proteins requires the de novo synthesis of a [2Fe-2S] cluster on the central scaffold proteins Isu1-Isu2 (cf. Ref. 10). This co-factor is released from Isu1-Isu2 with the help of the Hsp70 chaperone Ssq1 and its co-chaperone Jac1 and is inserted into [2Fe-2S] proteins. The maturation of [4Fe-4S] proteins additionally involves the specialized complex of Isa1, Isa2, and the tetrahydrofolate (THF)-binding protein Iba57. The Isa proteins bind iron that may be used for the formation of mitochondrial [4Fe-4S] proteins in a step involving Iba57. The biochemical mechanism of the generation of the [4Fe-4S] cluster, including the precise fate of Isa-bound iron, is unknown. (See “Discussion” for further details).
The specificity of Isa protein function for mitochondrial [4Fe-4S] proteins explains most of the key phenotypes displayed by isa1Δ or isa2Δ cells. First, the [2Fe-2S] proteins Yah1 and Isu1 are the only known essential mitochondrial Fe/S proteins in S. cerevisiae. Their Isa-independent maturation fits to the non-essential character of the ISA genes in yeast. Second, the respiratory deficiency and the auxotrophies for glutamate and lysine of isa1Δ or isa2Δ cells are caused by loss of function of aconitase-type proteins (23–25). Third, the biotin auxotrophy and lipoic acid deficiency are caused by impairment of the radical AdoMet, [4Fe-4S] cluster-containing enzymes Bio2 and Lip5 (22, 55). Finally, the specific defects in the maturation of [4Fe-4S] proteins may explain the loss of mtDNA in isa1Δ or isa2Δ cells, because, for example, the deletion of LIP5 induces the loss of mitochondrial DNA in yeast (59). Furthermore, aconitase is a component of the mitochondrial nucleoid and thus important for mtDNA maintenance (60). The complex phenotype of isa1Δ or isa2Δ cells is virtually indistinguishable from that of cells lacking IBA57 (22). Aconitase-type Fe/S proteins and radical AdoMet proteins are matured in an Iba57-dependent manner, whereas the maturation of the [2Fe-2S] protein Yah1 remains normal in the absence of Iba57. Because Iba57, Isa1, and Isa2 physically interact, these three ISC proteins form a functional ISC assembly subcomplex that is dedicated to the maturation of [4Fe-4S] proteins. Removal of any of its constituents completely inactivates this complex (22–25). The fact that the auxotrophies of isa1/2Δ and iba57Δ cells for metabolites involving [4Fe-4S] proteins are not cured upon cultivation under anaerobic conditions demonstrates that oxidative stress does not significantly contribute to the inactivation of mitochondrial [4Fe-4S] proteins in these cells. Rather, a bona fide Fe/S protein maturation defect appears to explain this characteristic phenotype. Taken together, our results demonstrate an essential requirement of Isa1, Isa2, and Iba57 for the maturation of mitochondrial [4Fe-4S] co-factors in yeast.
The maturation of mitochondrial and cytosolic aconitase-like [4Fe-4S] proteins appears to differ markedly with respect to its Isa protein dependence. We report here that yeast cells lacking the Isa proteins (isa1/2Δ) show normal Leu1 enzyme activity and normal maturation of cytosolic Fe/S proteins (supplemental Fig. S2). Further, isa1/2Δ cells do not elicit signs of a defective core ISC assembly machinery, such as the induction of the yeast iron regulon (22). Maturation of the two tested aconitases, yeast Aco1 and human IRP1, in the yeast cytosol did not involve the Isa proteins and was rather inefficient compared with the process in mitochondria, although it is of physiological importance for the glyoxylate shunt (56). Recent work on the Isa proteins from the protist T. brucei supports the conclusion that the function of the Isa proteins is restricted to the maturation of Fe/S proteins within mitochondria (61). Previously, we have shown a dependence of the maturation of Leu1 on the Isa proteins upon depletion of these proteins in Gal-ISA1 and Gal-ISA2 cells in lactate minimal medium (24, 25). The results presented here are based on isa1/2Δ deletion strains cultivated in glucose-containing medium. Hence the effects on Leu1 observed previously with our Gal strains are probably indirect, secondary consequences of the specific depletion conditions used in our previous studies. The situation seems to be comparable with recent results obtained for the siRNA-mediated depletion of human Isa proteins in HeLa cells. Fe/S cluster assembly of cytosolic aconitase (IRP1) was significantly affected (40% residual activity), whereas the enzyme activity and/or levels of two other cytosolic Fe/S proteins (GPAT and DPYD) remained unchanged (62),3 although they are sensitive markers for cytosolic Fe/S protein assembly. Together, it seems likely to us that, under certain depletion conditions, the deficiency of mitochondrial Isa proteins can cause moderate assembly defects and/or a damage of cytosolic Fe/S proteins, in particular of aconitase-like proteins, such as yeast Leu1 and IRP1, which can readily lose their forth iron (e.g. under oxidative stress conditions). Based on the results presented here, these effects seem to be indirect consequences of the physiological changes during Isa protein depletion rather than mechanistically linked to Isa protein function.
Our current investigation clarifies the role of several candidate Fe/S scaffold proteins in the mitochondrial matrix. As strongly supported by previous and current work, the U-type ISC assembly proteins Isu1 and Isu2 serve as the central physiological Fe/S scaffold proteins for the de novo synthesis of virtually all Fe/S proteins in S. cerevisiae (10). In contrast, the suggested role of the A-type ISC assembly proteins as alternative Fe/S scaffold proteins that work in parallel to the Isu proteins seems unlikely in yeast. Rather, the Isa proteins and Iba57 form a specialized ISC subcomplex that operates downstream of the central components of the mitochondrial ISC assembly system and is specifically required for maturation of mitochondrial [4Fe-4S] proteins. Nevertheless, the Isa proteins cooperate with the Isu scaffold proteins in the generation of [4Fe-4S] clusters. Recent work on bacterial Fe/S protein biogenesis systems suggests that A-type assembly proteins function as Fe/S transfer proteins (A-type carriers) that accept ready-made Fe/S co-factors from a scaffold protein, such as IscU, and transfer them to specific target proteins (3, 6, 36). The existence of four different A-type assembly proteins in E. coli is consistent with the idea that A-type proteins represent specialized assembly factors that are each optimized to assist maturation of a specific subset of target Fe/S proteins (21). In the context of the eukaryotic cell, these A-type ISC assembly proteins may have been optimized for the maturation of proteins with [4Fe-4S] co-factors by the invention of a novel A-type protein, Isa2, that has no obvious functional orthologue in bacteria. Apparently, this target specificity for [4Fe-4S] proteins resembles that of bacteria, because a deficiency of the A-type ISC proteins SufA and IscA in E. coli results in the loss of activity of [4Fe-4S] proteins but not of [2Fe-2S] proteins under aerobic growth conditions (27, 32).
We provide in vivo evidence that the yeast Isa1 and Isa2 bind iron, in analogy to some of their bacterial orthologues (3, 5, 6, 32). Iron binding to the Isa proteins appears to be intimately linked to their physiological function, because mutation of the conserved C-terminal cysteine pair resulted in both loss of function of Isa1 and loss of iron binding (25). Cysteine-coordinated iron binding has been shown to be characteristic for bacterial rubredoxins. Our data provide evidence that the iron bound to the Isa proteins is probably transferred to recipient [4Fe-4S] proteins in vivo. First, only small amounts of iron were detected on simple [4Fe-4S] apoproteins, when any of the three dedicated assembly factors, Isa1, Isa2, or Iba57, was missing. Second, iron accumulated on the Isa proteins in mitochondria with low amounts of the abundant [4Fe-4S] protein aconitase. Third, iron gradually accumulated on the Isa proteins upon depletion of Iba57. This iron accumulation on Isa proteins under Iba57 deficiency is reminiscent of the Fe/S cluster accumulation on Isu1 in the absence of the dedicated chaperones Ssq1 and Jac1 or Grx5 (19), which had allowed us to dissect the pathway of Fe/S protein biogenesis into two main reactions (10). Similar to the chaperones and Grx5, the ISC protein Iba57 may facilitate the transfer of the Isa-bound iron to recipient apoproteins in vivo (Fig. 8).
The transfer of iron from bacterial A-type proteins to target apoproteins has been frequently observed by Fe/S co-factor reconstitution experiments in vitro; however, there is a discrepancy concerning the chemical nature of the transferred iron (3, 6). First, based on the observation that bacterial and eukaryotic A-type proteins can bind [2Fe-2S] co-factors, the A-type proteins have been proposed to function as Fe/S transfer proteins that accept ready-made Fe/S clusters from the core components of the ISC assembly systems for a subsequent insertion into apotargets (3, 6, 36–39). The chemical combination of [2Fe-2S] clusters transiently bound to both U-type and A-type ISC proteins might yield a [4Fe-4S] cluster and hence provide a simple explanation for the specific A-type function in [4Fe-4S] cluster formation. On the other hand, bacterial A-type proteins were shown to bind mononuclear iron that can serve as an iron source during the maturation or repair of Fe/S apoproteins in vitro (27, 32, 40–42). Taken at face value, our data on Isa1 and Isa2 from S. cerevisiae favor a role as an iron donor, because iron binding to Isa proteins does not require the sulfur donor Nfs1 or the central ISC assembly proteins Isu1 and Yah1, indicating that the iron is probably bound at a mononuclear site in vivo (Fig. 8). In strong support of this hypothesis, we were unable to detect any sulfide bound to the Isa proteins in an in vitro Fe/S cluster reconstitution system that was able to generate Fe/S clusters on Bio2 and Isu1 (Fig. 6E). Unfortunately, an in vivo confirmation of this result is currently impossible, because under physiological conditions, the sensitivity of 35S incorporation into Isa proteins (and Fe/S proteins in general) is too low to address this question. Furthermore, the Isa proteins are too low in abundance in wild-type mitochondria to allow the isolation of sufficient amounts of Isa protein complex and the spectroscopic characterization of the type of bound iron.
Without a final in vivo verification, any definitive conclusion concerning the nature of the iron bound to the Isa proteins under undisturbed conditions has to be made with a note of caution. It is conceivable that the Isa proteins bind an Fe/S co-factor under normal circumstances in vivo and may only switch to an unphysiological iron-only binding mode in cells with a compromised mitochondrial ISC assembly system or upon Isa overproduction (i.e. the conditions that allowed us to unambiguously show iron-only binding). The well documented findings of Fe/S cluster and/or iron-only binding to the variety of different A-type ISC proteins may even indicate that this protein class is flexible, accommodating either Fe/S clusters or iron at the same binding site (6).
Taken together, our work demonstrates that the complex of the three ISC assembly proteins Isa1, Isa2, and Iba57 forms a specialized ISC assembly subcomplex that acts late in mitochondrial Fe/S protein biogenesis and is essential for and specifically dedicated to the maturation of mitochondrial [4Fe-4S] proteins. The complex operates downstream of the central components of the mitochondrial ISC assembly system, such as Nfs1-Isd11, Yah1, and Yfh1 (Fig. 8). The Isa proteins bind iron, and Iba57 may facilitate its dislocation and utilization for the synthesis of the [4Fe-4S] cluster on target apoproteins. The Isa1/2-Iba57 subcomplex cooperates with the central U-type Fe/S scaffold proteins Isu1 and Isu2 that provide the core of the Fe/S cluster, probably in form of a [2Fe-2S] cluster (63). Furthermore, two chaperones, the Hsp70 Ssq1 and, in the case of aconitase, also the chaperonin Hsp60 (related to bacterial GroEL), are involved (64). The biochemical mechanism converting the Isu-generated [2Fe-2S] clusters into functional [4Fe-4S] clusters and the putative role of iron delivered by Iba57 from the Isa proteins are currently unknown and will require future biochemical studies.
Acknowledgments
We thank Drs. W. Buckel and M. Brock for a sample of homoaconitate, T. Brüser for a DNA probe from A. vinosum, and J.-M. Moulis for plasmid YepL-IRP1.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 593, Gottfried-Wilhelm Leibniz program, and GRK 1216), von Behring-Röntgen Stiftung, the LOEWE program of state Hessen, Max-Planck Gesellschaft, the German-Israeli Foundation, and Fonds der Chemischen Industrie.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables SI–SIII and Figs. S1 and S2.
A. Sheftel and R. Lill, unpublished results.
- ISC
- iron-sulfur cluster
- hIRP1
- human IRP1
- HiPIP
- high potential iron-sulfur protein.
REFERENCES
- 1. Beinert H. (2000) J. Biol. Inorg. Chem. 5, 2–15 [DOI] [PubMed] [Google Scholar]
- 2. Fontecave M. (2006) Nat. Chem. Biol. 2, 171–174 [DOI] [PubMed] [Google Scholar]
- 3. Py B., Barras F. (2010) Nat. Rev. Microbiol. 8, 436–446 [DOI] [PubMed] [Google Scholar]
- 4. Fontecave M., Choudens S. O., Py B., Barras F. (2005) J. Biol. Inorg Chem. 10, 713–721 [DOI] [PubMed] [Google Scholar]
- 5. Johnson D. C., Dean D. R., Smith A. D., Johnson M. K. (2005) Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 6. Fontecave M., Ollagnier-de-Choudens S. (2008) Arch. Biochem. Biophys. 474, 226–237 [DOI] [PubMed] [Google Scholar]
- 7. Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008) Biochem. Soc. Trans. 36, 1112–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rouault T. A., Tong W. H. (2008) Trends Genet. 24, 398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheftel A. D., Lill R. (2009) Ann. Med. 41, 82–99 [DOI] [PubMed] [Google Scholar]
- 10. Lill R. (2009) Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 11. Balk J., Pilon M. (2011) Trends Plant Sci. 16, 218–226 [DOI] [PubMed] [Google Scholar]
- 12. Ye H., Rouault T. A. (2010) Biochemistry 49, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma A. K., Pallesen L. J., Spang R. J., Walden W. E. (2010) J. Biol. Chem. 285, 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng L., Dean D. R. (1994) J. Biol. Chem. 269, 18723–18726 [PubMed] [Google Scholar]
- 15. Lill R., Mühlenhoff U. (2006) Annu. Rev. Cell Dev. Biol. 22, 457–486 [DOI] [PubMed] [Google Scholar]
- 16. Lill R., Mühlenhoff U. (2008) Annu. Rev. Biochem. 77, 669–700 [DOI] [PubMed] [Google Scholar]
- 17. Stemmler T. L., Lesuisse E., Pain D., Dancis A. (2010) J. Biol. Chem. 285, 26737–26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsai C. L., Barondeau D. P. (2010) Biochemistry 49, 9132–9139 [DOI] [PubMed] [Google Scholar]
- 19. Mühlenhoff U., Gerber J., Richhardt N., Lill R. (2003) EMBO J. 22, 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vickery L. E., Cupp-Vickery J. R. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 95–111 [DOI] [PubMed] [Google Scholar]
- 21. Vinella D., Brochier-Armanet C., Loiseau L., Talla E., Barras F. (2009) PLoS Genet. 5, e1000497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gelling C., Dawes I. W., Richhardt N., Lill R., Mühlenhoff U. (2008) Mol. Cell Biol. 28, 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen L. T., Culotta V. C. (2000) Mol. Cell Biol. 20, 3918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pelzer W., Mühlenhoff U., Diekert K., Siegmund K., Kispal G., Lill R. (2000) FEBS Lett. 476, 134–139 [DOI] [PubMed] [Google Scholar]
- 25. Kaut A., Lange H., Diekert K., Kispal G., Lill R. (2000) J. Biol. Chem. 275, 15955–15961 [DOI] [PubMed] [Google Scholar]
- 26. Loiseau L., Gerez C., Bekker M., Ollagnier-de Choudens S., Py B., Sanakis Y., Teixeira de Mattos J., Fontecave M., Barras F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13626–13631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan G., Lu J., Bitoun J. P., Huang H., Ding H. (2009) Biochem. J. 420, 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson D. C., Unciuleac M. C., Dean D. R. (2006) J. Bacteriol. 188, 7551–7561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balasubramanian R., Shen G., Bryant D. A., Golbeck J. H. (2006) J. Bacteriol. 188, 3182–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lu J., Yang J., Tan G., Ding H. (2008) Biochem. J. 409, 535–543 [DOI] [PubMed] [Google Scholar]
- 31. Angelini S., Gerez C., Ollagnier-de Choudens S., Sanakis Y., Fontecave M., Barras F., Py B. (2008) J. Biol. Chem. 283, 14084–14091 [DOI] [PubMed] [Google Scholar]
- 32. Wang W., Huang H., Tan G., Si F., Liu M., Landry A. P., Lu J., Ding H. (2010) Biochem. J. 432, 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Py B., Moreau P. L., Barras F. (2011) Curr. Opin. Microbiol 14, 218–223 [DOI] [PubMed] [Google Scholar]
- 34. Cupp-Vickery J. R., Silberg J. J., Ta D. T., Vickery L. E. (2004) J. Mol. Biol. 338, 127–137 [DOI] [PubMed] [Google Scholar]
- 35. Morimoto K., Yamashita E., Kondou Y., Lee S. J., Arisaka F., Tsukihara T., Nakai M. (2006) J. Mol. Biol. 360, 117–132 [DOI] [PubMed] [Google Scholar]
- 36. Gupta V., Sendra M., Naik S. G., Chahal H. K., Huynh B. H., Outten F. W., Fontecave M., Ollagnier de Choudens S. (2009) J. Am. Chem. Soc. 131, 6149–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu G., Mansy S. S., Hemann C., Hille R., Surerus K. K., Cowan J. A. (2002) J. Biol. Inorg. Chem. 7, 526–532 [DOI] [PubMed] [Google Scholar]
- 38. Ollagnier-de Choudens S., Nachin L., Sanakis Y., Loiseau L., Barras F., Fontecave M. (2003) J. Biol. Chem. 278, 17993–18001 [DOI] [PubMed] [Google Scholar]
- 39. Ollagnier-de-Choudens S., Sanakis Y., Fontecave M. (2004) J. Biol. Inorg. Chem. 9, 828–838 [DOI] [PubMed] [Google Scholar]
- 40. Yang J., Bitoun J. P., Ding H. (2006) J. Biol. Chem. 281, 27956–27963 [DOI] [PubMed] [Google Scholar]
- 41. Ding H., Yang J., Coleman L. C., Yeung S. (2007) J. Biol. Chem. 282, 7997–8004 [DOI] [PubMed] [Google Scholar]
- 42. Lu J., Bitoun J. P., Tan G., Wang W., Min W., Ding H. (2010) Biochem. J. 428, 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sherman F. (2002) Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 44. Stehling O., Smith P. M., Biederbick A., Balk J., Lill R., Mühlenhoff U. (2007) Methods Mol. Biol. 372, 325–342 [DOI] [PubMed] [Google Scholar]
- 45. Molik S., Lill R., Mühlenhoff U. (2007) Methods Cell Biol. 80, 261–280 [DOI] [PubMed] [Google Scholar]
- 46. Mühlenhoff U., Richhardt N., Gerber J., Lill R. (2002) J. Biol. Chem. 277, 29810–29816 [DOI] [PubMed] [Google Scholar]
- 47. Sambrook J., Russel D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 48. Gietz R. D., Woods R. A. (2002) Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 49. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. (2002) Nucleic Acids Res. 30, e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Diekert K., de Kroon A. I., Kispal G., Lill R. (2001) Methods Cell Biol. 65, 37–51 [DOI] [PubMed] [Google Scholar]
- 51. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 52. Kennedy M. C., Emptage M. H., Dreyer J. L., Beinert H. (1983) J. Biol. Chem. 258, 11098–11105 [PubMed] [Google Scholar]
- 53. Wallace M. A., Liou L. L., Martins J., Clement M. H., Bailey S., Longo V. D., Valentine J. S., Gralla E. B. (2004) J. Biol. Chem. 279, 32055–32062 [DOI] [PubMed] [Google Scholar]
- 54. Link T. A., Saynovits M., Assmann C., Iwata S., Ohnishi T., von Jagow G. (1996) Eur. J. Biochem. 237, 71–75 [DOI] [PubMed] [Google Scholar]
- 55. Mühlenhoff U., Gerl M. J., Flauger B., Pirner H. M., Balser S., Richhardt N., Lill R., Stolz J. (2007) Eukaryot. Cell 6, 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Regev-Rudzki N., Karniely S., Ben-Haim N. N., Pines O. (2005) Mol. Biol. Cell 16, 4163–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kispal G., Csere P., Prohl C., Lill R. (1999) EMBO J. 18, 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., Lill R. (2004) J. Biol. Chem. 279, 36906–36915 [DOI] [PubMed] [Google Scholar]
- 59. Sulo P., Martin N. C. (1993) J. Biol. Chem. 268, 17634–17639 [PubMed] [Google Scholar]
- 60. Chen X. J., Wang X., Butow R. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Long S., Changmai P., Tsaousis A. D., Skalický T., Verner Z., Wen Y. Z., Roger A. J., Lukeš J. (2011) Mol. Microbiol. 81, 1403–1418 [DOI] [PubMed] [Google Scholar]
- 62. Song D., Tu Z., Lee F. S. (2009) J. Biol. Chem. 284, 35297–35307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raulfs E. C., O'Carroll I. P., Dos Santos P. C., Unciuleac M. C., Dean D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8591–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chaudhuri T. K., Farr G. W., Fenton W. A., Rospert S., Horwich A. L. (2001) Cell 107, 235–246 [DOI] [PubMed] [Google Scholar]
- 65. Flint D. H., Emptage M. H. (1988) J. Biol. Chem. 263, 3558–3564 [PubMed] [Google Scholar]