Abstract
Mycoplasma hyopneumoniae colonizes the ciliated respiratory epithelium of swine, disrupting mucociliary function and inducing chronic inflammation. P97 and P102 family members are major surface proteins of M. hyopneumoniae and play key roles in colonizing cilia via interactions with glycosaminoglycans and mucin. The p102 paralog, mhp683, and homologs in strains from different geographic origins encode a 135-kDa pre-protein (P135) that is cleaved into three fragments identified here as P45683, P48683, and P50683. A peptide sequence (TTKF↓QE) was identified surrounding both cleavage sites in Mhp683. N-terminal sequences of P48683 and P50683, determined by Edman degradation and mass spectrometry, confirmed cleavage after the phenylalanine residue. A similar proteolytic cleavage site was identified by mass spectrometry in another paralog of the P97/P102 family. Trypsin digestion and surface biotinylation studies showed that P45683, P48683, and P50683 reside on the M. hyopneumoniae cell surface. Binding assays of recombinant proteins F1683–F5683, spanning Mhp683, showed saturable and dose-dependent binding to biotinylated heparin that was inhibited by unlabeled heparin, fucoidan, and mucin. F1683–F5683 also bound porcine epithelial cilia, and antisera to F2683 and F5683 significantly inhibited cilium binding by M. hyopneumoniae cells. These data suggest that P45683, P48683, and P50683 each display cilium- and proteoglycan-binding sites. Mhp683 is the first characterized glycosaminoglycan-binding member of the P102 family.
Keywords: Adhesion, Bacteria, Heparin-binding Protein, Protease, Protein Motifs, Protein Processing, Mycoplasma hyopneumoniae, P102, Cilium-binding Proteins
Introduction
Mycoplasma hyopneumoniae is a global pathogen that inflicts severe economic losses on swine production (1, 2). Current treatment relies on the application of bacterin vaccine formulations that fail to block colonization of respiratory tract cilia and strategic antibiotic therapy. The development of more effective vaccines requires a detailed understanding of the molecular interactions that govern adherence and colonization of the porcine respiratory tract and involve adjuvant formulations that stimulate mucosal immunity (3–8).
M. hyopneumoniae is strictly a pathogen of swine, and alternate hosts or intermediary vectors have not been identified. Electron microscopic studies of infected lung tissues show that M. hyopneumoniae interacts almost exclusively with cilia on the epithelial surfaces that line the trachea, bronchi, and bronchioles in the porcine upper respiratory tract. Specifically, this microorganism is found attached along the entire length of the cilia but rarely to the epithelial cell body (9–11). To survive and proliferate as an infectious agent, M. hyopneumoniae must enter the respiratory tract of its host, traverse mucous layers, resist the mucociliary escalator, adhere and colonize epithelial cilia, secure essential nutrients for growth and replication, evade immune responses, and repeat the cycle of infection by transmission to new hosts via airborne mucosal droplets. Colonization of the upper respiratory tract by M. hyopneumoniae results in the destruction of the mucociliary escalator via ciliostasis, loss of cilia, and eventual epithelial cell death. The underlying mechanisms, however, are poorly understood (9). Once colonized, swine become chronically infected, but the strategies required to maintain a chronic infection state are ill defined. The mechanism utilized by M. hyopneumoniae to colonize respiratory cilia is likely to require multiple adhesins and strategies for avoiding immune detection.
Unlike the human respiratory pathogen Mycoplasma pneumoniae, M. hyopneumoniae does not display a complex terminal organelle and is not known to be motile, yet it is able to circumvent the protective effects of the mucociliary escalator in the porcine respiratory tract. P97, P102, and paralogs of these two molecules play important roles in interactions between M. hyopneumoniae and receptors in the porcine respiratory tract. mRNA transcripts representative of all members of the P97 and P102 paralog families (except Mhp280) are known to be expressed in vivo (12). Previous proteomic studies have determined that cleavage products of Mhp182 (P102), Mhp183 (P97), Mhp493 (P159), and Mhp494 (P216) are prominently featured on the surface of M. hyopneumoniae (13–15), whereas others (Mhp271, Mhp107, and Mhp108) are expressed at lower levels (16–18). Tandem pentapeptide repeats (AAKP(V/E)) in the R1 domain of P97 (19, 20) play an important role in cilium binding. Non-R1-containing members of the P97 family also bind porcine cilia (15, 18) and epithelial cell surfaces (13, 18). The P97 paralog Mhp107 also binds plasminogen, fibronectin, and the glycosaminoglycan heparin (18). Mhp108, a member of the P102 family, binds cilia, fibronectin, and plasminogen (17), and domains within Mhp183, Mhp494, Mhp493, and Mhp271 bind heparin. Heparan sulfate includes regions within proteoglycan side chains that have been identified at the surface of porcine respiratory cilia (21). Heparin, a structural analog of the highly sulfated regions of heparan sulfate proteoglycans, effectively blocks the binding of M. hyopneumoniae to porcine cilia and to porcine kidney epithelium-like cells used previously as a cilial binding model (13, 22, 23). These observations underscore the diverse nature of cilium- and extracellular matrix binding domains presented by the P97 and P102 family of adhesins.
A hallmark feature of these two adhesin families is their tendency to be targets for precise proteolytic cleavage events that generate a complex array of binding domains. In the absence of recognized membrane anchorage motifs, the cleavage fragments adhere to the external membrane surface of M. hyopneumoniae and define discrete domains that bind cilia and a variety of host molecules, including fibronectin, various glycosaminoglycans, and plasminogen. Proteolytic processing is observed in all currently characterized members of the P97/P102 family.
Bioinformatic analysis of the genome sequence of M. hyopneumoniae has identified six P102 paralogs, defined as 30% identity over 70% of the sequence (24). Many of these P102 paralogs comprise two-gene structures with a P97 paralog (24, 25). Compared with P97 and its paralogs, the functions of P102 and related molecules are poorly understood. Cell lysates probed with antisera raised against recombinant P102 identified three proteins with masses of 102 kDa (pre-protein), 72 kDa (P72), and 42 kDa (P42) (12, 14). Immunogold electron microscopy studies of M. hyopneumoniae (14) harvested from broth culture and associated with cilia in infected lung tissue identified P102 and its cleavage fragments on the external surface of Mycoplasma cells and on the surface of porcine cilia (12). These data indicate that P102 is subject to proteolytic cleavage and plays a role in the colonization of the porcine respiratory tract. Many bacterial adhesins display complex cleavage or degradation patterns, which have not been extensively characterized, including several adhesins expressed by the phylogenetically related streptococci (26–29). Recently, we showed that the P102 paralog, Mhp108, is a proteolytically processed, multifunctional adhesin that binds fibronectin and plasminogen and adheres to porcine respiratory cilia (17). In this study, we have extensively characterized Mhp683, a member of the P102 family that forms part of a two-gene operon with P146, as an adhesin-like P97 paralog (24, 25).
EXPERIMENTAL PROCEDURES
M. hyopneumoniae Strains and Culture
The source and conditions used to culture M. hyopneumoniae strains J, 232, and field isolates 00MP1301, 2-2241, and 95MP1509 have been described previously (16, 30). M. hyopneumoniae isolate C1735-2 was isolated from a piggery in Queensland, Australia, and was provided by J. Forbes-Faulkner (Oonoonba Veterinary Laboratory, Queensland, Australia). M. hyopneumoniae cells were centrifuged at 10,000 × g and washed three times in PBS. Final pellets were stored at −20 °C until required.
Proteomics
Separation of Mycoplasma proteins into hydrophobic and hydrophilic fractions using Triton X-114 was performed prior to electrophoresis as described previously (31). Hydrophilic proteins of the aqueous phase were precipitated with cold acetone and resuspended in SSS buffer (8 m urea, 100 mm DTT, 4% (w/v) CHAPS, 0.8% (w/v) 3–10 carrier ampholytes, 40 mm Tris-HCl) for separation by electrophoresis.
Materials and methods used for one- and two-dimensional gel electrophoresis, immunoblotting, trypsin digestion, reduction and alkylation, Zip-Tip® clean-up, and peptide mass-mapping using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF MS) have been described previously (14, 16, 32) with the following adjustments. First dimension immobilized pH-gradient strips (ReadyStripTM IPG Strips, 170 mm in length, nonlinear pH 3–10; Bio-Rad) were used. Strips were rehydrated overnight with 500 μg of Triton X-114 aqueous fraction protein extract in 360 μl of SSS buffer overlaid with paraffin oil. M. hyopneumoniae proteins were reduced prior to one-dimensional gel electrophoresis. Gel slices prepared from one-dimensional SDS-PAGE for tandem mass spectrometry analysis were processed as described previously (16). Protein spots excised from two-dimensional gels were processed as described previously (33, 34). Briefly, spots were excised using a sterile scalpel blade and washed in a destain solution (60:40 solution of 40 mm ammonium bicarbonate (pH 7.8), 100% acetonitrile) for 1 h at room temperature. The solution was removed from the wells, and the gel pieces were vacuum-dried for 1 h. The gel spots were rehydrated in 8 μl of trypsin solution (2 ng μl−1 (sequencing-grade modified trypsin (Promega, Madison WI) in 40 mm ammonium bicarbonate)) at 4 °C for 1 h. Excess trypsin was removed, and the gel pieces were resuspended in 25 μl of 40 mm ammonium bicarbonate and incubated overnight at 37 °C. Peptides were concentrated and desalted using C18 Perfect PureTM tips (Eppendorf, Hamburg, Germany) and eluted in matrix (α-cyano-4-hydroxycinnamic acid (Sigma), 8 mg ml−1 in 70% (v/v) acetonitrile, 1% (v/v) formic acid) directly onto a target plate. Peptide mass maps were generated by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using a Voyager DE-STR (Applied Biosystems, Framingham MA). Mass calibration was performed using trypsin autolysis peaks, m/z 2211.11 and m/z 842.51 as internal standards. Data from peptide mass maps were used to perform searches of the NCBI, Swiss-Prot, and TrEMBL databases, via the program MASCOT. Identification parameters included peptide mass accuracy within 50 ppm, one possible missed tryptic cleavage per peptide, and with the methionine sulfoxide and cysteine acrylamide modifications checked. Identifications were based on MASCOT score and E-values, the observed pI and molecular mass (kDa) of the protein, the number of matching peptide masses, and the total percentage of the amino acid sequence that those peptides covered. Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analysis of one- and two-dimensional electrophoresis gel excisions were performed as described previously (35) except that MASCOT searches were performed with additional variable modifications of deamidation (Asn, Gln) and Gln→pyro-Glu (N-terminal Gln and Glu).
Surface Localization
Graded trypsin digestion of intact M. hyopneumoniae for Western blotting was performed as described previously (14). Trypsin concentrations of 0, 0.1, 0.5, 1, 3, 5, 10, 50, and 300 μg ml−1 were used in this study. Trypsin digestion of intact M. hyopneumoniae for mass spectrometry was performed as described previously (16).
For surface biotinylation experiments, freshly harvested M. hyopneumoniae cells were washed extensively (>3 times) in PBS (4000 × g, 30 min, 4 °C) and pelleted by centrifugation (9000 × g, 10 min, 4 °C). Cells were resuspended in PBS (pH 7.8) and biotinylated with sulfo-NHS-LC biotin (Thermo Scientific) for 30 s on ice. The reaction was then quenched with the addition of a final concentration of 50 mm Tris-HCl (pH 7.4) and incubated for 15 min. Cells were washed in three changes of PBS and pelleted by centrifugation. A 0.1 g pellet of M. hyopneumoniae cells was resuspended in solubilization buffer (7 m urea, 2 m thiourea, 40 mm Tris (pH 8.8), 1% (w/v) C7bZ0) and disrupted with four rounds of sonication at 50% power for 30 s bursts on ice. Proteins were reduced and alkylated with 20 mm acrylamide monomers, 5 mm tributylphosphine for 90 min. Insoluble material was pelleted by centrifugation. Soluble proteins were precipitated in 5 volumes of ice-cold acetone for 30 min, and the pellet was air-dried and then resuspended in 7 m urea, 2 m thiourea, 1% (w/v) C7bZ0. Biotinylated proteins were purified by avidin column affinity chromatography performed as described previously (36). Biotinylated proteins were separated by two-dimensional gel electrophoresis and identified by Western blotting. Spots corresponding to biotinylated proteins were cut from simultaneously run two-dimensional gels and examined using LC-MS/MS.
Bioinformatic Analysis
Bioinformatic analysis of Mhp683 was performed with the use of several on-line resources. Sequence similarity was compared by BLASTP analysis of Mhp683 at the National Center for Biotechnology Information (37, 38) (www.ncbi.nlm.nih.gov). Physical data such as theoretical pI, molecular weight, and extinction coefficients were collected using the ProtParam tool at ExPASy (39). Identification of coiled-coil domains was performed using the COILS2 algorithm at the Swiss node of EMBnet (40) as well as Multicoil and Paircoil2 at the Massachusetts Institute of Technology (41, 42). Theoretical transmembrane domain and signal peptide scores were identified using the TMHMM and SignalP web services, respectively, at the Center for Biological Sequence Analysis, Technical University of Denmark (43–45). Prediction of disordered regions was performed using the Predictor of Naturally Disordered Regions (PONDR), VSL1 algorithm (46, 47).
Expression of Recombinant Proteins and Creation of Polyclonal Antisera
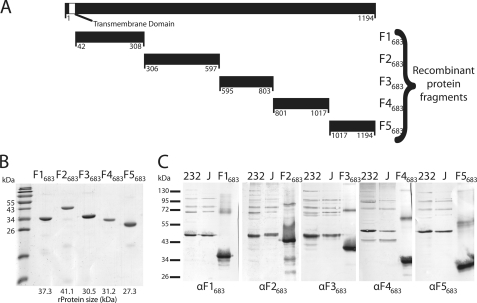
As Mycoplasma species use the UGA codon to translate tryptophan instead of signaling the end of translation, the expression of Mycoplasma proteins in Escherichia coli results in truncations. Cloning of mhp683 was performed in five fragments of varying length with minimal overlap (Fig. 3A) and was based on the M. hyopneumoniae strain 232 homolog (24); fragment size was largely determined by the presence of eight in-frame TGA codons. Fragments were labeled F1683 through F5683 and ranged from amino acids 42–308, 306–597, 595–803, 801–1017, and 1018–1194, respectively. All in-frame TGA codons were mutated to TGG by using mutated primers or site-directed mutagenesis (supplemental Table 1). Fragments were amplified by PCR from chromosomal M. hyopneumoniae strain 232 DNA using Pwo polymerase (Roche Applied Science) and cloned into the pET161/GW/D-TOPO vector (Invitrogen). The reaction mixture was then transformed into TOP10 chemically competent E. coli and incubated overnight at 37 °C in LB agar containing 100 μg ml−1 ampicillin (Sigma). Positive colonies were cultured further in LB media containing 100 μg ml−1 ampicillin (Sigma) and plasmids extracted with the QIAprep Spin Miniprep Kit (Qiagen, Netherlands). Purified plasmids were screened for correct orientation by PCR and sequenced to check for mutations.
FIGURE 3.
Cloning of mhp683 and immunoblot analysis of its protein fragments. A, mhp683 (3582 bp encoding 1194 amino acids) was cloned from M. hyopneumoniae strain 232 as five fragments with sizes ranging from 178 to 292 amino acids. The hydrophobic putative transmembrane domain (41 amino acids) was removed from the N terminus of F1683. All in-frame TGA codons were mutated to TGG by using either mutated flanking primers or site-directed mutagenesis. B, Coomassie-stained SDS-PAGE of the five recombinant protein fragments in ascending order. Recombinant protein F3683 runs at a slightly higher molecular mass than expected on SDS-PAGE, and F1683 runs slightly lower. C, immunoblots of M. hyopneumoniae strain 232 and J whole cell lysates, and recombinant protein fragments (F1683–F5683) were probed with their respective antisera (αF1683–αF5683).
Protein expression was achieved by use of the E. coli BL21StarTM strain (Invitrogen). Plasmids were transformed and inoculated into LB media containing 100 μg ml−1 ampicillin and cultured at 37 °C overnight. A subculture was performed the following day and allowed to grow to mid-log phase (A600 = 0.5–0.8) before induction of expression with 1 mm isopropyl β-d-1-thiogalactopyranoside and incubation for 3–4 h. F1683–F5683 were purified by nickel affinity chromatography and dialyzed in PBS containing 0.1% SDS (Fig. 3B), and their concentrations were estimated as described previously (48).
Polyclonal antisera to recombinant proteins F1683–F5683 were prepared by immunization of New Zealand White Rabbits as described previously (49). All antisera were tested for activity by immunoblots with recombinant protein (Fig. 3C).
Heparin Binding Assays
Heparin binding, inhibition, and competitive immunoassays were performed in 96-well, flat-bottomed microtiter plates (Linbro/Titertek; ICN Biomedicals Inc., Aurora, OH) with binding steps all performed in a volume of 100 μl. For heparin binding assays, proteins (F1683–F5683) were diluted to 10 μg ml−1 in carbonate coating buffer (18 mm NaHCO3, 27 mm Na2CO3 (pH 9.5)) and bound to plates by shaking on a Titramax 1000 microtiter plate shaker (Heidolph, Schwabach, Germany) at room temperature for 2 h; unbound and excess protein were removed by immersing wells five times in wash buffer (0.05% Tween 20 in PBS). Pre-diluted biotinylated heparin (Calbiochem) was added to wells in serial 2-fold dilutions starting from 100 μg ml−1, and plates were incubated at room temperature for 1 h with shaking. The plates were again immersed five times in wash buffer followed by addition of streptavidin/peroxidase (Roche Applied Science) at a dilution of 1:3000 in PBS and incubated with shaking for 1 h. After a final wash step, the plates were developed with 1 mm 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (Sigma) in citrate buffer (100 mm citric acid, 200 mm sodium hydrogen phosphate di-basic (pH 4.2)) treated with hydrogen peroxide. Plates were developed with shaking, and the absorbance at 414 nm was measured at 7-, 15-, 25-, and 45-min intervals.
Heparin binding specificity and competitive binding assays were performed as described previously (48). All heparin binding assays were performed in triplicate with previously stated controls (48) and graphed by GraphPad Prism Version 4.02 for Microsoft Windows (Graphpad Software, CA) by nonlinear regression.
Heparin binding dot-blots were performed using the Bio-Dot® microfiltration apparatus (Bio-Rad). Hybond Super-C nitrocellulose membrane (Amersham Biosciences) was equilibrated in PBS and fastened into the apparatus. Proteins were added to the wells in serial 2-fold dilutions starting from 1 μg. Proteins were allowed to bind for 1 h or until the solution had drained through the membrane. Unbound protein was removed by washing with 100 μl of PBS. The membrane was removed from the manifold and blocked with 5% skim milk blocking buffer (5% skim milk, 10 mm Tris, 150 mm sodium chloride (pH 7.4)) for 1 h with shaking. The membrane was then immersed in 0.1% skim milk wash buffer (0.1% skim milk, 10 mm Tris, 150 mm sodium chloride (pH 7.4)) containing 30 μg ml−1 biotinylated heparin with shaking for 90 min. After three vigorous washes, the membrane was immersed in a solution of 1 part streptavidin and 3000 parts 0.1% skim milk wash buffer for 1 h with shaking. Following another wash step (as before), the membrane was equilibrated in 100 mm Tris (pH 7.6) solution and developed with 0.05% diaminobenzidine dissolved in 100 mm Tris (pH 7.6) and treated with hydrogen peroxide.
Cilium Binding and Inhibition Assays
The binding of Mhp683 to porcine cilia was examined as described previously using a microtiter plate adherence assay developed for the identification of the cilium-binding protein P97 (15). Inhibition of M. hyopneumoniae adherence to porcine cilia by Mhp683 antisera was examined using the microtiter plate adherence assay with the following adjustments. Plates were coated in cilia as described previously (15) and blocked for 1 h with 1% gelatin (Sigma) in PBS. Freshly cultured M. hyopneumoniae cells were washed twice, resuspended in PBS at 1:200 of the original culture volume, and then incubated with a 1:50 dilution of antisera for 1 h. Following antisera treatment, cells were added directly to cilia-coated wells, incubated for 1.5 h, and washed three times with PBS. Mycoplasmas were detected by subsequent addition of mouse monoclonal antibody F1B6 diluted to 1:250 and alkaline phosphatase-conjugated anti-mouse antibodies (1:1000) absorbed against rabbit antibodies.
RESULTS
Mhp683 Is Proteolytically Cleaved
In strain 232, mhp683 encodes a P102 paralog with a theoretical molecular mass of 135 kDa and a theoretical pI of 7.2. Homologs of mhp683 exist within other M. hyopneumoniae genome sequences and are referred to as mhj_0662 in strain J and mhp7448_0662 in strain 7448. These homologs encode proteins that share 94% amino acid sequence identity that are referred to collectively here as Mhp683. The TMHMM algorithm identified a transmembrane domain (p = 0.973) from residues 17 to 39 in both M. hyopneumoniae strains J and 232, indicating the presence of a putative signal peptide; however, analysis of Mhp683 with SignalP resulted in a low signal peptide probability (p = 0.409, signal peptide cutoff of p > 0.5).
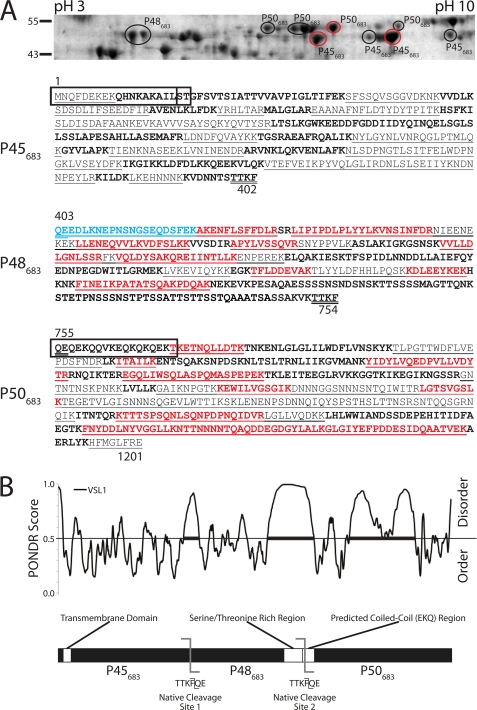
To determine whether Mhp683 is expressed during growth in broth culture, we adopted an approach combining one-dimensional SDS-PAGE and LC-MS/MS. Global analysis of strain J proteins that migrated in an SDS-polyacrylamide gel with masses from ∼45 to 55 kDa identified a panel of tryptic peptides that mapped across the Mhp683 sequence (Fig. 1). These data suggested that Mhp683 was a target of post-translational cleavage events that cleave the molecule in three fragments each with a mass of 45–55 kDa. MALDI-TOF-MS and LC-MS/MS of protein spots separated by two-dimensional gel electrophoresis identified several distinct groups of protein spots that mapped to three nonoverlapping regions of Mhp683. An N-terminal cleavage product (P45683; Fig. 2A) was matched from a linear pattern of four spots at ∼45 kDa with pI values ranging from 8 to 10. Proteins that mapped to the central region of Mhp683 (P48683; Fig. 2A) were identified in two spots with pI between 5 and 6 and molecular mass of ∼48 kDa. A linear array of five protein spots resolving at pI 7–9 and with a mass of ∼50 kDa mapped to the C terminus of Mhp683 (P50683; Fig. 2A). A peptide (1MNQFDEKEK9) was identified that spanned the first nine amino acids from the putative N-terminal methionine residue indicating that P45683 includes the N terminus of Mhp683. This was confirmed by Edman sequencing of proteins from two spots representing P45683 (Fig. 2A, circled in red) that generated the sequence 1MNQFDEKEKQHNKAKAIL18. Edman sequencing of protein spots that generated tryptic peptides spanning amino acids 773–1201 (P50683) produced the sequence 755QEQEKQQVKEQKQKQEKT772 indicating that the cleavage site that creates P50683 resides between Phe-754 and Gln-755.
FIGURE 1.
Expected molecular weight of MHJ_0622 (the Mhp683 homolog in strain J) is not reflected in peptide analysis. Gel image showing cell lysate (100 μg) of M. hyopneumoniae strain J separated by SDS-PAGE and stained with FlamingoTM fluorescent stain. The gel lane was sectioned into pieces representing 14 molecular weight ranges, and each piece was digested with trypsin. Peptides from the gel piece representing a 40–55 kDa mass range were identified by LC-MS/MS (underlined in bold) and matched to regions of MHJ_0662. No peptides matching to MHJ_0662 were identified in proteins resolving at ∼135 kDa, the predicted mass of the MHJ_0662.
FIGURE 2.
A, two-dimensional gel electrophoresis, mass spectrometry, and N-terminal sequencing confirms MHJ_0662 is cleaved into three fragments of approximately equal size. Coomassie-stained two-dimensional gel of a cell lysate of M. hyopneumoniae strain J depicts proteins with masses between 43 and 55 kDa. Protein spots were excised from the gel, digested with trypsin, and identified by peptide-mass fingerprinting with MALDI-TOF-MS, where trypsin-digested peptides are identified by matching peaks to the calculated masses of predicted fragments. Protein spots were also identified by LC-MS/MS, where trypsin-digested peptides are directly sequenced using tandem mass spectrometry. Peptides identified by MALDI-TOF-MS are underlined; peptides identified by LC-MS/MS are shown in bold, red, and underlined; sequences not identified by either technique are in bold. Peptides from MHJ_0662 (Mhp683 homolog in strain J) were identified in spots localizing to three distinct regions (circled) of the gel with masses of 45–55 kDa and varying isoelectric points. Cleavage fragments spanning the amino, central, and carboxyl portions of MHJ_0662 are labeled P45683, P48683, and P50683, respectively. N-terminal sequences were obtained by Edman degradation and LC-MS/MS. Sequences obtained by Edman degradation are shown in boxes and were taken from gel spots circled in red. Two Edman sequences from P45683 show an intact N terminus with no signal peptide cleavage. The N terminus of P48683 was identified using LC-MS/MS (shown in blue). Mass spectrum of this fragment shows an N-terminal pyroglutamate, derived from the original glutamine, which blocks the free N terminus and explains the failure of Edman degradation for this fragment (supplemental Fig. 1). B, molecular analysis of mhp683. For PONDR VSL1 analysis (top), regions above the line at 0.5 denote disordered regions within Mhp683. Thick bars denote disordered regions spanning 40 or more amino acids (55). Mhp683 is predicted to contain three regions of significant disorder, two corresponding with the cleavage sites of Mhp683. The TMHMM algorithm predicts that Mhp683 contains a transmembrane domain (p = 0.973). Multiple coiled-coil prediction algorithms identified an EKQ repeat region as a putative coiled coil. A serine/threonine-rich region has also been identified at the C terminus of cleavage fragment P48683. Experimentally determined cleavage site positions are also shown.
We were unsuccessful in attempts to determine the N-terminal sequence of P48683 by Edman degradation. Peptide mass fingerprinting analyses of protein spots containing P48683 indicated that the P45683/P48683 cleavage site is situated in the region spanning two lysine residues at positions 392 and 423 in Mhp683. To identify the N terminus of P48683, tryptic digests of spots representing P48683 were subjected to LC-MS/MS. This analysis identified a 21-residue semi-tryptic peptide 403Q(−17)EEDLKNEPNSN(+1)GSEQDSFEK423 in spot variants of J and 232 lysates representing P48683 (Fig. 2A; supplemental Fig. 1) and defined the N terminus of P48683. The alteration of the N-terminal glutamine to pyroglutamate (−17 Da) is consistent with our inability to sequence the N terminus of P48683 by Edman degradation (50). Our data indicate that P50683 and P48683 are released from the Mhp683 pre-protein by a protease that recognizes the motif TTKF↓QE, with cleavage occurring immediately after the phenylalanine residue. Based on this hypothesis, P45683 spans amino acids 1–402 generating a cleavage fragment with a predicted mass of 46 kDa (pI = 8.3); P48683 spans amino acids 403–754 with a predicted mass of 39 kDa (pI = 5.9), and P50683 spans amino acids 755–1201 with a predicted mass of 50 kDa (pI = 7.9). Compared with P45683 and P50683, P48683 is rich in glutamic acid residues, and this is likely to contribute to P48683 resolving at a pI of 5.9 and showing abnormal migration during SDS-PAGE (Figs. 1–3) (51).
Molecular Analysis of Mhp683
Apart from the previously reported similarity with other members of the P102 family (24), BLASTP identified LppT (expect = 2e-04), the operon partner of the LppS adhesin from Mycoplasma conjunctivae (52, 53) as the molecule with the greatest degree of similarity to Mhp683. Sequence similarity was confined to ∼350 residues of the N terminus. Examination of the remaining ∼850 residues of the Mhp683 sequence, which encompasses both P48683 and P50683, showed no significant similarity to other proteins. These data show that endoproteolysis generates protein fragments with completely novel sequence on the surface of M. hyopneumoniae.
Analysis of Mhp683 with several coiled-coil prediction algorithms identified a 30-residue putative coiled-coil region between residues 742 and 776 in M. hyopneumoniae strain 232 and 756–785 in strain J (Fig. 2B). These regions correspond with an EKQ repeat domain with the motif ((QK))(EQ)(QK)(X) identified previously (54), which is in close proximity to the N terminus of P50683. The COILS2 algorithm (p > 0.9) and the Paircoil2 algorithm (p = 0.0243, coiled-coil cutoff of p < 0.025) both indicated that the EKQ region forms a coiled coil.
The PONDR VSL1 algorithm was used to predict regions of structural disorder present within Mhp683 (Fig. 2B). Four areas of significant structural disorder spanning more than 40 amino acids were predicted to occur at residues 381–428, 636–777, 875–987, and 990–1084 in the strain 232 homolog (55). Similar regions were identified in the strain J homolog MHJ_0662. The predicted cleavage motif (TTKF↓QE) at positions 402–403 and 749–750 resided within disordered regions spanning residues 381–428 and 636–777. An S/T-rich repeat region in the C terminus of P48683 also lies within a disordered region spanning residues 636–777 (Fig. 2B).
Mhp683 Is Processed in Strains with Different Geographic Origins
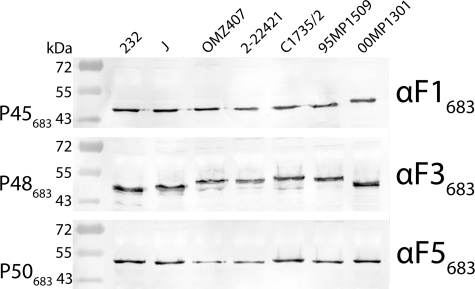
To determine whether Mhp683 is subject to proteolytic processing in field strains of M. hyopneumoniae from geographically diverse locations, immunoblots (Fig. 4) of whole cell lysates of strains sourced from Australia and the United States were probed with antisera raised against recombinant fragments F1683–F5683. Using αF1683, αF3683, and αF5683 sera, fragments P45683, P48683, and P50683 were detected in all strains examined.
FIGURE 4.
Processing of Mhp683 homologs is conserved across geographically diverse strains of M. hyopneumoniae. Immunoblots containing whole cell lysates of different M. hyopneumoniae field isolates were separately probed with αF1683, αF3683, and αF5683 sera to assess the consistency of protein expression and processing. Cleavage fragments equivalent to P45683, P48683, and P50683 are conserved across all isolates.
P45683, P48683, and P50683 Reside on the Surface of M. hyopneumoniae
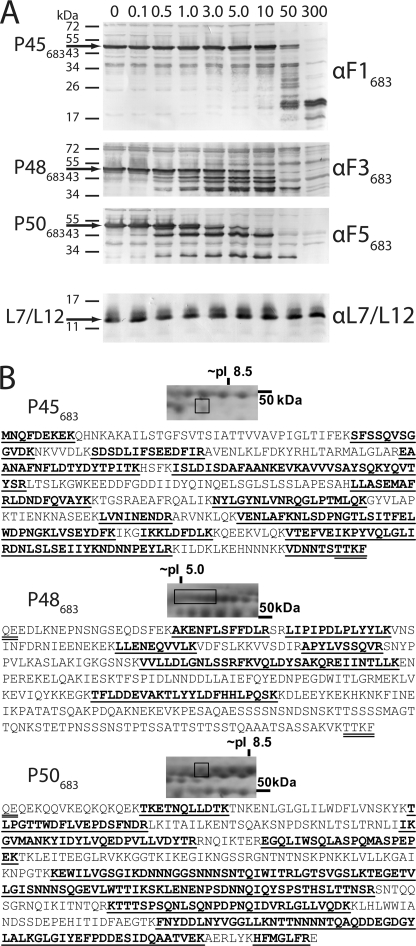
To determine whether P45683, P48683, and P50683 reside on the surface of M. hyopneumoniae, immunoblots containing cell lysates of freshly cultured cells that have been exposed to different concentrations of trypsin for 15 min (37 °C) were separately probed with αF1683, αF3683, and αF5683 sera detecting cleavage fragments P45683, P48683, and P50683, respectively (Fig. 5A). These protein bands were almost completely digested at a trypsin concentration of 50 μg ml−1. Identical lysates exposed to antisera raised against the ribosomal protein L7/L12 showed that this protein was detected at a trypsin concentration of 300 μg ml−1 suggesting that the integrity of cell membrane remained unaffected in these experiments. In addition to immunoblotting, we combined trypsin digestion of whole cell M. hyopneumoniae with mass spectrometry. After digestion, multiple peptides unique to cleavage fragments P45683, P48683, and P50683 were identified by LC-MS/MS (supplemental Fig. 2).
FIGURE 5.
MHJ_0662 (Mhp683 homolog) is present on the surface of M. hyopneumoniae. A, intact and washed (strain J) cells were digested for 15 min with increasing concentrations of trypsin to determine whether P45683, P48683, and P50683 fragments were present on the cell surface. Immunoblots were probed with αF1683, αF3683, and αF5683 sera and identified P45683, P48683, and P50683 fragments of MHJ_0662, respectively. Trypsin concentrations (above blots) are quoted in μg ml−1. Identical lysates were subsequently probed with antiserum to an intracellular control protein (ribosomal protein L7/L12) to confirm cellular integrity. B, surface biotinylation of M. hyopneumoniae. Intact and washed M. hyopneumoniae (strain J) cells were biotinylated and lysed, and biotin conjugates were purified by affinity chromatography. Following separation by two-dimensional electrophoresis, peptides unique to P45683, P48683, and P50683 were identified from purified biotinylated proteins by LC-MS/MS. Underlined sequences indicate peptides identified by LC-MS/MS; M. hyopneumoniae cleavage site motifs are double underlined.
To further confirm surface localization, we biotinylated intact M. hyopneumoniae cells, purified biotin-conjugated proteins, and examined them using two-dimensional electrophoresis and LC-MS/MS. Peptides unique to P45683, P48683, and P50683 were each identified at masses similar to those determined in previous electrophoresis experiments (Fig. 5B). Biotinylation of each protein spot was confirmed by matching to a streptavidin blot prepared from a simultaneously run two-dimensional gel. We identified a 10-residue semi-tryptic peptide with sequence 393VDNNTSTTKF402 that defines the C terminus of P45683 and provides further evidence of cleavage at the TTFK↓QE motif (supplemental Fig. 3).
Predicting Cleavage Sites in Other Members of the P97 Family of Paralogs
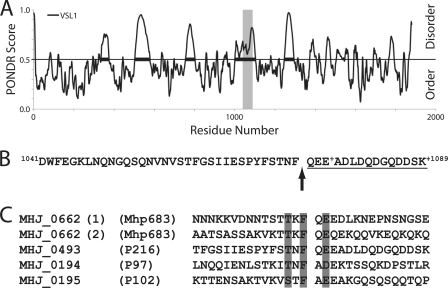
Previously, we showed that the cilium adhesin Mhp493 and its homolog in strain J (MHJ_0493) was subject to a major cleavage event that removes 85 kDa from the C terminus of the 216-kDa pre-protein. Attempts to delineate the N terminus of P85 by Edman degradation were unsuccessful (15). Peptide mass fingerprinting indicated that the N terminus of P85 resided between amino acids 1041 and 1089 in Mhp493 (15). Based on the reiterated cleavage site TTKF↓QE identified in this study, we hypothesized that the sequence STNF↓QE, which also resides in a strongly disordered region of Mhp493, may be recognized by the same putative protease that cleaves Mhp683. A semi-tryptic fragment with the sequence 1075QEEADLDQDGQDDSR1089 was identified by LC-MS/MS and defines the N terminus of P85 (Fig. 6; supplemental Fig. 4).
FIGURE 6.
Peptide motif TTKFQE found in Mhp683 is predictive of other cleavage sites within the P97/P102 family; identification of the Mhp493 cleavage site. A, PONDR analysis of Mhp493 (P216). Cleavage of Mhp493 into P120 and P85 was previously identified, but the precise cleavage site was unable to be accurately determined (15). Gray bar denotes region spanning amino acids 1041–1089 previously shown to house the putative cleavage site. B, amino acids 1041–1089 of the strain J homolog (MHJ_0493) of Mhp493. The N-terminal peptide identified by LC-MS/MS (supplemental Fig. 4) is underlined. Arrow denotes the site of cleavage that separates 85 kDa (P85) from the C terminus of MHJ_0493. + identifies a site of amino acid variability. The comparable sequence in Mhp493 from strain 232 is identical except an aspartic acid residue replaces a glutamic acid residue at position 1077 and an asparagine residue replaces a lysine residue at position 1089. C, comparison of identified P97/P102 family cleavage sites (14). Shaded regions denote highly similar or identical residues.
Mhp683 Binds Heparin
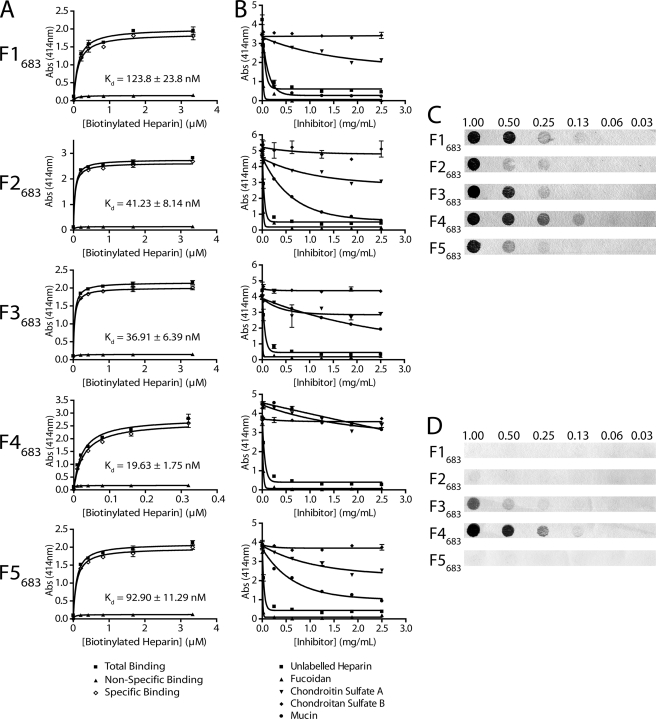
In previous studies we have identified members of the P97 family as heparin-binding proteins (15, 16, 18, 48). To determine whether Mhp683 binds glycosaminoglycans, we performed microtiter plate assays with biotin-labeled heparin. Recombinant fragments F1683–F5683 spanning Mhp683 bound to biotinylated heparin in a dose-dependent and saturable manner (Fig. 7A). Binding was almost completely inhibited by the presence of a large excess of unlabeled heparin indicating that the binding interaction was specific. Recombinant protein F4683 bound heparin with the highest affinity (Kd = 19.63 ± 1.75 nm) and F1683 bound with lowest affinity (Kd = 123.8 ± 23.8 nm). Unlabeled heparin and fucoidan were found to effectively inhibit the binding of biotinylated heparin to all recombinant fragments, whereas chondroitin sulfates A and B did not (Fig. 7B). The ability of porcine mucin II to inhibit heparin binding varied for each recombinant fragment. Mucin dramatically inhibited the ability of F1683 to bind heparin. Mucin also inhibited the binding of heparin to F2683 and F5683, but it only had a moderate effect on the ability of F3683 to bind heparin. Mucin did not significantly inhibit F4683 from binding heparin. The heparin binding observed in microtiter plate assays were largely consistent when examined using dot-blot ligand binding assays with the recombinants bound to nitrocellulose in a native conformation (Fig. 7C). F4683 was again found to have the highest affinity for biotinylated heparin, whereas F2683 displayed the least affinity. The affinity of F1683 for heparin was higher compared with the microtiter plate assays. F1683, F2683, and F5683 completely lost affinity for biotinylated heparin when applied to the membrane after denaturation, and the affinity to F3683 was significantly weakened (Fig. 7D). F4683 retained its ability to bind biotinylated heparin regardless of its conformational state.
FIGURE 7.
Recombinant proteins F1683–F5683 bind heparin. A, binding of recombinant protein to biotinylated heparin. Total binding (■) was determined by coating recombinant protein F1683–F5683 (10 μg ml−1) to 96-well microtiter plates followed by binding with increasing concentrations of biotinylated heparin (x axis). Nonspecific binding (▴) was determined by binding with unlabeled heparin in a 50-fold excess to biotinylated heparin. Specific binding (◊) was determined by subtracting the nonspecific from the total binding. Kd values for all five recombinant proteins were determined from the specific binding data. B, inhibition of heparin binding by glycosaminoglycans. Recombinant protein (10 μg ml−1) was coated to 96-well microtiter plates and bound with biotinylated heparin in the presence of an increasing excess of inhibitor (x axis). Inhibitors included unlabeled heparin (■), fucoidan (▴), chondroitin sulfate A (▾), chondroitin sulfate B (♦), and porcine type II mucin (●). C, nondenatured recombinant protein dot blots. Recombinant proteins were spotted onto nitrocellulose membrane, blocked, and incubated with 30 μg ml−1 biotinylated heparin. Bound recombinant protein amounts (above blots) are listed in micrograms. D, denatured recombinant protein dot blots. Recombinant proteins were denatured by boiling in an SDS-containing reducing solution before spotting onto nitrocellulose membrane and incubation with heparin as with C. Bound recombinant protein amounts (above blots) are listed in micrograms.
Mhp683 Binds Porcine Cilia
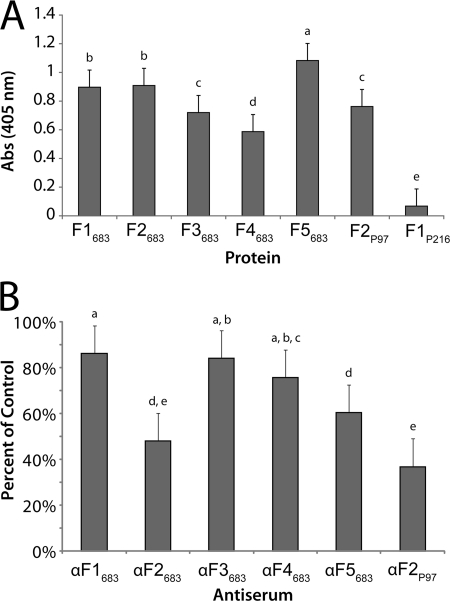
A microtiter plate assay used previously to identify cilium-binding proteins (15) showed that Mhp683 recombinant proteins F1683–F5683 reproducibly bind cilia. The recombinant protein F1216, previously reported to display low cilium binding properties (15), did not bind porcine cilia. F2P97, a recombinant protein that carries the R1 cilium binding domain of cilium adhesin P97, was used as a positive control and bound to porcine cilia as expected (Fig. 8A).
FIGURE 8.
Mhp683 recombinant proteins bind porcine cilia, and antisera blocks cilium binding by M. hyopneumoniae. A, cilium binding assay. Microtiter plate binding assay showing Mhp683 recombinant fragments binding to porcine cilia. Recombinant fragment derived from P97 encoding the cilium binding domain R1 and R2 (F2P97) acted as a positive control for cilium binding in this experiment (48). The recombinant fragment that has previously been reported to have low cilium binding (F1P216) acted as a negative control for cilium binding in this experiment (15). Data represent means plus least significant difference at p < 0.05. Means with any superscript (a–e) in common are not different; those with no superscript in common are different at p < 0.05. B, M. hyopneumoniae adherence to porcine cilia is blocked by Mhp683 antisera. Microtiter plate assay showing inhibition of M. hyopneumoniae binding to porcine cilia after pretreatment with αF1683–αF5683 sera diluted at 1:50. Data are presented as a percentage of binding control, where cells were incubated in rabbit sera with no specificity to M. hyopneumoniae proteins. Antisera (αF2P97) derived from the recombinant fragment F2P97 was used as a blocking control. Data represent means plus least significant difference at p < 0.05. Means with any superscript (a–e) in common are not different; those with no superscript in common are different at p < 0.05. Binding control (100%) was in statistical group a.
Mhp683 Antibodies Inhibit Adherence of M. hyopneumoniae to Cilia
Antibodies that bind the R1 region of the cilium adhesin P97 have been previously shown to block the adherence of M. hyopneumoniae to porcine cilia (20). To determine whether this was the case with Mhp683 antibodies, we incubated M. hyopneumoniae with αF1683–αF5683 sera and examined binding to porcine cilia in a microtiter-based assay. M. hyopneumoniae binding to porcine cilia was significantly and reproducibly inhibited by both αF2683 and αF5683 sera (Fig. 8B). Binding of M. hyopneumoniae coated in αF2683 and αF5683 sera was reduced by 52.0 ± 6.1% and 39.5 ± 7.5%, respectively. Binding of M. hyopneumoniae to cilia was not significantly inhibited by treatment with αF1683, αF3683, or αF4683 sera. The adherence blocking control, αF2P97, an antiserum recognizing the R1 cilium binding domain of cilium adhesin P97, was also used and inhibited M. hyopneumoniae binding to porcine cilia by 63.1 ± 3.3% consistent with previously reported observations (20).
DISCUSSION
mhp683, the second gene in a putative two-gene operon with mhp684, encodes a protein with a predicted mass of 135 kDa, but a protein with this mass and a tryptic cleavage pattern matching to Mhp683 was not found when high mass proteins resolved by SDS-PAGE were characterized by MALDI-TOF MS (15). Data from SDS-PAGE and LC-MS/MS and two-dimensional immunoblotting using sera raised against recombinant fragments spanning Mhp683 provide strong evidence that Mhp683 and homologs in strains of M. hyopneumoniae from geographically diverse regions are subject to two post-translational cleavage events that produce proteins P45683, P48683, and P50683. Trypsin digestion and surface biotinylation experiments show that P45683, P48683 and P50683 reside on the surface of M. hyopneumoniae, suggesting that in the absence of membrane spanning domains, the fragments must bind to other surface-localized components of either Mycoplasma or host origin. Recombinant proteins (F1683–F5683) spanning Mhp683 bind heparin and porcine cilia suggesting that P45683, P48683, and P50683 each play an important role in facilitating colonization of the respiratory tract of swine.
During broth culture, the P97 and P102 paralogs are among the most highly expressed proteins,3 and mRNAs derived from genes that encode these proteins have been detected in M. hyopneumoniae recovered from the lungs of infected swine (12). We have previously established that proteolytic processing of members of the P97 and P102 paralog families generates a complex array of cleavage fragments that are displayed on the surface of M. hyopneumoniae and perform key roles in adhesion to structurally diverse host molecules (13–18, 48). However, the rationale for the extensive proteolytic processing of these adhesins by M. hyopneumoniae remains elusive. The respiratory pathogen Bordetella pertussis also proteolytically cleaves its FHA adhesin, and although mutants lacking the responsible protease (SphB1) retained respiratory cell adhesion, they had a decreased ability to colonize lung tissue (56). This suggested that processing of FHA facilitated the detachment of individual bacteria from microcolonies, allowing infection to efficiently spread within the host (56). Adhesin processing in M. hyopneumoniae may have a similar role; unfortunately, due to the present absence of a targeted gene knock-out technique for this bacterium, this hypothesis cannot be tested using this approach.
Although considerable progress has been made in determining the precise cleavage sites for a number of proteolytic cleavage events (13, 14), attempts to define the N-terminal sequences of a number of cleavage products have failed (13, 15) presumably due to chemical modifications to N-terminal residues that block Edman degradation. Here, we identify a reiterated cleavage motif with sequence TTKF↓QE in Mhp683. Cleavage after the phenylalanine residue was confirmed by a combination of Edman sequencing of P50683 and LC-MS/MS of both the P48683 N terminus and P45683 C terminus. The semi-tryptic peptide 403Q(−17)EEDLKNEPNSN(+1)GSEQDSFEK423, where −17 denotes the presence of pyroglutamate and +1 the conversion of asparagine to aspartic acid through deamidation (57), was identified on numerous occasions by mass spectrometry and defined the N terminus of P48683. Previous attempts to perform Edman degradation on P48683 were unsuccessful presumably due to the presence of pyroglutamate, which is known to block Edman sequencing (50). Further evidence for cleavage at this position was provided by the identification of the semi-tryptic peptide, 393VDNNTSTTKF402 representing the C terminus of P45683.
Although unsuccessful in previous attempts to accurately define cleavage sites within the P97 paralog Mhp493 (15) and in the P102-related molecule Mhp494 (13) by Edman sequencing, we were able to accurately localize the cleavage site to within short stretches of sequence using peptide mapping strategies. We hypothesized that the identification of the reiterated cleavage sequence TTKF↓QE in Mhp683 may facilitate accurate prediction of cleavage sites within other P97 and P102 paralogs. To test this hypothesis, we selected the P97 paralog Mhp493 (P216). Mhp493 undergoes a cleavage event generating fragments P120 and P85 (15), which display cilium binding properties on the surface of M. hyopneumoniae. Peptide mapping analyses delineated the N terminus to reside within a stretch of 49 amino acids spanning positions 1041 to 1089. A sequence 1071STNF↓QE1076 identified in this region of Mhp493 closely resembled the TTKF↓QE cleavage motif in Mhp683. A semi-tryptic peptide with the sequence 1075QEEADLDQDGQDDSR1089 was identified by LC-MS/MS, which defined the N terminus of P85. Interestingly, the two TTKF↓QE cleavage sites and the STNF↓QE cleavage site each reside within intrinsically disordered regions (Figs. 2 and 6) in Mhp683 and Mhp493, respectively. Regions within proteins displaying intrinsic disorder are often targets for proteolytic attack and other post-translational modifications (55, 58). These data suggest that a protease in M. hyopneumoniae responsible for processing the P97 and P102 family of cilium adhesins recognizes a peptide motif with a sequence similar to TTKF↓QE.
We have previously identified several cleavage sites within P97 and P102 by Edman sequencing (14, 20). Analysis of the sequences surrounding these cleavage sites reveals two motifs that closely resemble those we have identified in this study (Fig. 6C). The sequence 192ITNF↓AD197 in Mhp183 spans the cleavage site responsible for the maturation of the cilium adhesin P97. This cleavage event removes 195 amino acids from the N terminus of the 125-kDa pre-protein (Mhp183), generating the mature P97 adhesin and the N-terminal cleavage product P22 (14, 20, 59). The sequence 553VSTF↓AE558 spans the cleavage site in Mhp182 (P102) that generates the N-terminal 72-kDa fragment (P72) and the C-terminal 42-kDa fragment (P42) (14). Comparison of these cleavage motifs with those of this study identifies three key features as follows: (i) the amino side residue of the cleavage site (−1 position) is phenylalanine; (ii) the −3 position (amino cleavage fragment) is an alcohol-containing residue (S/T); (iii) the +2 position (carboxyl cleavage fragment) is negatively charged (D/E) (Fig. 6C). Our data suggest that a putative protease(s) responsible for processing in M. hyopneumoniae recognizes some or all of these key residues. The characterization of further N-terminal sequences either by Edman degradation or mass spectrometry would facilitate the definition of a robust protease recognition motif, which would be valuable in the global prediction of proteolytic cleavage sites from M. hyopneumoniae genome sequences.
Primary bacterial pathogens are capable of overcoming the protective effects of the mucosal barrier and colonize epithelial sites in the respiratory tract. These processes expose previously inaccessible colonization sites that provide opportunities for secondary bacterial pathogens to exploit (60). The identification and characterization of the molecules involved in the adherence to both cilia and extracellular matrix components are important first steps in understanding the pathogenic armory of M. hyopneumoniae. Epithelial surfaces are awash with mucins, antimicrobial peptides, and mucopolysaccharides that act as decoys for microbial adhesins (60). Glycosaminoglycans are present in the extracellular matrix and include regions of proteoglycans that are exposed on the surface of almost all eukaryotic cells (61). Heparan sulfate, an important component of proteoglycans, has been identified on the cilial surface in the swine upper respiratory tract (21). In other bacterial pathogens, heparan sulfate has been identified as a key target for adhesive proteins that either bind directly or via recruitment of other glycosaminoglycan-binding host molecules (62–64). In M. hyopneumoniae, we have previously identified a number of glycosaminoglycan-binding proteins from the P97 family (13, 15, 16, 18, 48). Here, we show that recombinant fragments F1683–F5683 of Mhp683 bind heparin at physiologically significant levels in a saturable and dose-dependent manner. Heparin is structurally analogous to the sulfated regions of heparan sulfate found on the surfaces of epithelial cells and is a reliable substrate for the identification of glycosaminoglycan-binding proteins (65). Because of the high negative charge of heparin, interactions with bacterial adhesins are largely mediated via lysine and arginine residues, although polar amino acids such as asparagine and glutamine also contribute (66). The loss of binding due to denaturation of F1683, F2683, F3683, and F5683 indicates that adherence to heparin was largely dependent on conformational epitopes. F4683, however, retained an ability to bind heparin after boiling in Laemmli buffer indicating that a linear heparin-binding motif may be present in this sequence. Analysis of the isoelectric points of the F1683–F5683 indicates that F4683 is the most alkaline with a theoretical pI of 9.70. Arginine and lysine make up 15% of the total amino acid complement of F4683 and are likely to play an important role in binding heparin.
Competitive binding assays showed that the ability of F1683–F5683 to bind heparin was effectively inhibited by fucoidan but not significantly by chondroitin sulfate A or B, indicating that the presence and placement of sulfate functional groups are important (48, 65). Porcine mucin also inhibited the ability of F1683, F2683, and F5683 and to a lesser extent F3683 to bind heparin, but this effect was not observed with heparin binding to F4683. These data suggest heparin binding domains are scattered throughout Mhp683 and that all three cleavage fragments P45683, P48683, and P50683 bind proteoglycans and play important roles in colonizing mucosal epithelial cilia.
P97 uses variable tandem repeat regions to facilitate adherence to both cilia and extracellular matrix components. The R1 and R2 regions of P97 and Mhp271 have both been identified as key sequences involved in the binding of cilia and heparin (16, 48, 67). Mhp683 possesses a variable tandem repeat region rich in EKQ residues (54). Our results suggest that it is not critical in facilitating binding of Mhp683 to either heparin or cilia. The EKQ region includes a KEKE-like motif that has been implicated in protein-protein interactions (68–70). Analysis of other P97/P102 paralogs demonstrates that this motif is present in Mhp684 (P146 adhesin-like protein), Mhp493 (P216), and Mhp494 (P159 adhesin). KEKE motifs are often associated with a coiled-coil protein structure (70), and the EKQ region in Mhp683 is predicted to display a coiled-coil conformation. Coiled-coil regions are associated with adhesins such as the Vaa adhesin of Mycoplasma hominis (71). The function of putative coiled-coil regions in M. hyopneumoniae surface proteins remains unknown.
Mhp683 is one of a growing number (Mhp493, Mhp107, and Mhp108) of cilial adhesins of M. hyopneumoniae that do not display an R1 cilium binding domain. In this study, recombinant fragments F1683–F5683 were observed to bind porcine cilia in a microtiter-based assay previously used to identify the P97 adhesin. Critically, we have also demonstrated that antisera to Mhp683 recombinant proteins significantly and reproducibly inhibited the adherence of M. hyopneumoniae to porcine cilia underlining the biological importance of this protein to the bacterium. Both αF2683 and αF5683 sera significantly blocked this interaction, and inhibition by αF2683 was observed at levels similar to antiserum made from a recombinant protein containing the R1 binding domain of the cilium adhesin P97. Inhibition of M. hyopneumoniae adherence to porcine cilia was not absolute as ∼50% of Mycoplasma cells were able to bind cilia after coating with Mhp683 antisera, a result consistent with the redundancy previously observed in the P97 and P102 families (15–18, 20). Although αF2683 and αF5683 sera significantly inhibited binding of M. hyopneumoniae to porcine cilia; antisera to recombinant fragments F1683, F3683, and F4683 did not, despite evidence that they directly bound porcine cilia, indicating that F2683 and F5683 contain critical and exposed binding sites that play significant roles in M. hyopneumoniae pathogenesis.
Our analysis of members of the P97 and P102 families show that they are highly expressed, subject to proteolytic cleavage and other post-translational modification events, and often display unusual sequence motifs (13–18). Although we do not understand how adhesin cleavage fragments remain attached to the cell surface, the identification of a proteolytic cleavage motif represents a significant development that should facilitate the prediction of other processed surface proteins and their cleavage sites and assist in the identification of the putative protease(s). The growing number of surface proteins dedicated to binding epithelial cilia attests to the importance colonization of this niche plays in the survival, proliferation, and spread of this ubiquitous and economically important pathogen. It is clear that proteolytic cleavage fragments derived from Mhp683 and other members of the P97 and P102 families are critical components of the surface architecture of M. hyopneumoniae.
Acknowledgments
We thank Ben Crossett for the preliminary matrix-assisted laser desorption/ionization-time-of-flight MS work done to locate Mhp683 and David C. Oneal for original mutagenesis work on mhp683.
Footnotes
This work was supported by ARC-Linkage Grant LP776711 and a grant from the McGarvie Smith Trust (to S. P. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1–4, and Data 1–5.
M. P. Padula and S. P. Djordjevic, unpublished data.
REFERENCES
- 1. Clark L., Armstrong C., Scheidt A., VanAlstine W. G. (1993) J. Swine Health Prod. 1, 10–14 [Google Scholar]
- 2. Haesebrouck F., Pasmans F., Chiers K., Maes D., Ducatelle R., Decostere A. (2004) Vet. Microbiol. 100, 255–268 [DOI] [PubMed] [Google Scholar]
- 3. Chin J., San Gil F., Novak M., Eamens G., Djordjevic S., Simecka J., Duncan J., Mullbacher A. (1996) J. Biotechnol. 44, 13–19 [DOI] [PubMed] [Google Scholar]
- 4. Djordjevic S. P., Eamens G. J., Romalis L. F., Nicholls P. J., Taylor V., Chin J. (1997) Aust. Vet. J. 75, 504–511 [DOI] [PubMed] [Google Scholar]
- 5. Fagan P. K., Djordjevic S. P., Chin J., Eamens G. J., Walker M. J. (1997) Infect. Immun. 65, 2502–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fagan P. K., Djordjevic S. P., Eamens G. J., Chin J., Walker M. J. (1996) Infect. Immun. 64, 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fagan P. K., Walker M. J., Chin J., Eamens G. J., Djordjevic S. P. (2001) Microb. Pathog. 30, 101–110 [DOI] [PubMed] [Google Scholar]
- 8. Matic J. N., Terry T. D., Van Bockel D., Maddocks T., Tinworth D., Jennings M. P., Djordjevic S. P., Walker M. J. (2009) Infect. Immun. 77, 1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeBey M. C., Ross R. F. (1994) Infect. Immun. 62, 5312–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mebus C. A., Underdahl N. R. (1977) Am. J. Vet. Res. 38, 1249–1254 [PubMed] [Google Scholar]
- 11. Tajima M., Yagihashi T. (1982) Infect. Immun. 37, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams C., Pitzer J., Minion F. C. (2005) Infect. Immun. 73, 7784–7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Burnett T. A., Dinkla K., Rohde M., Chhatwal G. S., Uphoff C., Srivastava M., Cordwell S. J., Geary S., Liao X., Minion F. C., Walker M. J., Djordjevic S. P. (2006) Mol. Microbiol. 60, 669–686 [DOI] [PubMed] [Google Scholar]
- 14. Djordjevic S. P., Cordwell S. J., Djordjevic M. A., Wilton J., Minion F. C. (2004) Infect. Immun. 72, 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilton J., Jenkins C., Cordwell S. J., Falconer L., Minion F. C., Oneal D. C., Djordjevic M. A., Connolly A., Barchia I., Walker M. J., Djordjevic S. P. (2009) Mol. Microbiol. 71, 566–582 [DOI] [PubMed] [Google Scholar]
- 16. Deutscher A. T., Jenkins C., Minion F. C., Seymour L. M., Padula M. P., Dixon N. E., Walker M. J., Djordjevic S. P. (2010) Mol. Microbiol. 78, 444–458 [DOI] [PubMed] [Google Scholar]
- 17. Seymour L. M., Deutscher A. T., Jenkins C., Kuit T. A., Falconer L., Minion F. C., Crossett B., Padula M., Dixon N. E., Djordjevic S. P., Walker M. J. (2010) J. Biol. Chem. 285, 33971–33978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seymour L. M., Falconer L., Deutscher A. T., Minion F. C., Padula M. P., Dixon N. E., Djordjevic S. P., Walker M. J. (2011) J. Biol. Chem. 286, 10097–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Minion F. C., Adams C., Hsu T. (2000) Infect. Immun. 68, 3056–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q., Young T. F., Ross R. F. (1995) Infect. Immun. 63, 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erlinger R. (1995) Cell Tissue Res. 281, 473–483 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Q., Young T. F., Ross R. F. (1994) Infect. Immun. 62, 1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zielinski G. C., Young T., Ross R. F., Rosenbusch R. F. (1990) Am. J. Vet. Res. 51, 339–343 [PubMed] [Google Scholar]
- 24. Minion F. C., Lefkowitz E. J., Madsen M. L., Cleary B. J., Swartzell S. M., Mahairas G. G. (2004) J. Bacteriol. 186, 7123–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasconcelos A. T., Ferreira H. B., Bizarro C. V., Bonatto S. L., Carvalho M. O., Pinto P. M., Almeida D. F., Almeida L. G., Almeida R., Alves-Filho L., Assunção E. N., Azevedo V. A., Bogo M. R., Brigido M. M., Brocchi M., Burity H. A., Camargo A. A., Camargo S. S., Carepo M. S., Carraro D. M., de Mattos Cascardo J. C., Castro L. A., Cavalcanti G., Chemale G., Collevatti R. G., Cunha C. W., Dallagiovanna B., Dambrós B. P., Dellagostin O. A., Falcão C., Fantinatti-Garboggini F., Felipe M. S., Fiorentin L., Franco G. R., Freitas N. S., Frías D., Grangeiro T. B., Grisard E. C., Guimarães C. T., Hungria M., Jardim S. N., Krieger M. A., Laurino J. P., Lima L. F., Lopes M. I., Loreto E. L., Madeira H. M., Manfio G. P., Maranhão A. Q., Martinkovics C. T., Medeiros S. R., Moreira M. A., Neiva M., Ramalho-Neto C. E., Nicolás M. F., Oliveira S. C., Paixão R. F., Pedrosa F. O., Pena S. D., Pereira M., Pereira-Ferrari L., Piffer I., Pinto L. S., Potrich D. P., Salim A. C., Santos F. R., Schmitt R., Schneider M. P., Schrank A., Schrank I. S., Schuck A. F., Seuanez H. N., Silva D. W., Silva R., Silva S. C., Soares C. M., Souza K. R., Souza R. C., Staats C. C., Steffens M. B., Teixeira S. M., Urmenyi T. P., Vainstein M. H., Zuccherato L. W., Simpson A. J., Zaha A. (2005) J. Bacteriol. 187, 5568–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Courtney H. S., Hasty D. L., Li Y., Chiang H. C., Thacker J. L., Dale J. B. (1999) Mol. Microbiol. 32, 89–98 [DOI] [PubMed] [Google Scholar]
- 27. Jung C. J., Zheng Q. H., Shieh Y. H., Lin C. S., Chia J. S. (2009) Mol. Microbiol. 74, 888–902 [DOI] [PubMed] [Google Scholar]
- 28. Fischetti V. A., Jones K. F., Scott J. R. (1985) J. Exp. Med. 161, 1384–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelly C., Evans P., Bergmeier L., Lee S. F., Progulske-Fox A., Harris A. C., Aitken A., Bleiweis A. S., Lehner T. (1989) FEBS Lett. 258, 127–132 [DOI] [PubMed] [Google Scholar]
- 30. Scarman A. L., Chin J. C., Eamens G. J., Delaney S. F., Djordjevic S. P. (1997) Microbiology 143, 663–673 [DOI] [PubMed] [Google Scholar]
- 31. Bordier C. (1981) J. Biol. Chem. 256, 1604–1607 [PubMed] [Google Scholar]
- 32. Scott N. E., Marzook N. B., Deutscher A., Falconer L., Crossett B., Djordjevic S. P., Cordwell S. J. (2010) Proteomics 10, 277–288 [DOI] [PubMed] [Google Scholar]
- 33. Cordwell S. J., Len A. C., Touma R. G., Scott N. E., Falconer L., Jones D., Connolly A., Crossett B., Djordjevic S. P. (2008) Proteomics 8, 122–139 [DOI] [PubMed] [Google Scholar]
- 34. Zhang K., McKinlay C., Hocart C. H., Djordjevic M. A. (2006) J. Proteome Res. 5, 3355–3367 [DOI] [PubMed] [Google Scholar]
- 35. Szczepanek S. M., Frasca S., Jr., Schumacher V. L., Liao X., Padula M., Djordjevic S. P., Geary S. J. (2010) Infect. Immun. 78, 3475–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nunomura K., Nagano K., Itagaki C., Taoka M., Okamura N., Yamauchi Y., Sugano S., Takahashi N., Izumi T., Isobe T. (2005) Mol. Cell. Proteomics 4, 1968–1976 [DOI] [PubMed] [Google Scholar]
- 37. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Altschul S. F., Wootton J. C., Gertz E. M., Agarwala R., Morgulis A., Schäffer A. A., Yu Y. K. (2005) FEBS J. 272, 5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J. M. ed) pp. 571–607, Humana Press Inc., Totowa, NJ [Google Scholar]
- 40. Lupas A., Van Dyke M., Stock J. (1991) Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 41. McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Bioinformatics 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 42. Wolf E., Kim P. S., Berger B. (1997) Protein Sci. 6, 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 44. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 45. Sonnhammer E. L., von Heijne G., Krogh A. (1998) Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 46. Peng K., Radivojac P., Vucetic S., Dunker A. K., Obradovic Z. (2006) BMC Bioinformatics 7, 208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A. K. (2005) Proteins 61, Suppl. 7, 176–182 [DOI] [PubMed] [Google Scholar]
- 48. Jenkins C., Wilton J. L., Minion F. C., Falconer L., Walker M. J., Djordjevic S. P. (2006) Infect. Immun. 74, 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott N. E., Bogema D. R., Connolly A. M., Falconer L., Djordjevic S. P., Cordwell S. J. (2009) J. Proteome Res. 8, 4654–4664 [DOI] [PubMed] [Google Scholar]
- 50. Hellström J. L., Vehniäinen M., Mustonen M., Lövgren T., Lamminmäki U., Hellman J. (2006) Biochim. Biophys. Acta 1764, 1735–1740 [DOI] [PubMed] [Google Scholar]
- 51. Proft T., Hilbert H., Plagens H., Herrmann R. (1996) Gene 171, 79–82 [DOI] [PubMed] [Google Scholar]
- 52. Belloy L., Vilei E. M., Giacometti M., Frey J. (2003) Microbiology 149, 185–193 [DOI] [PubMed] [Google Scholar]
- 53. Zimmermann L., Peterhans E., Frey J. (2010) J. Bacteriol. 192, 3773–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Castro L. A., Rodrigues Pedroso T., Kuchiishi S. S., Ramenzoni M., Kich J. D., Zaha A., Henning Vainstein M., Bunselmeyer Ferreira H. (2006) Vet. Microbiol. 116, 258–269 [DOI] [PubMed] [Google Scholar]
- 55. Uversky V. N., Dunker A. K. (2010) Biochim. Biophys. Acta 1804, 1231–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Coutte L., Alonso S., Reveneau N., Willery E., Quatannens B., Locht C., Jacob-Dubuisson F. (2003) J. Exp. Med. 197, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wright H. T. (1991) Protein Eng. 4, 283–294 [DOI] [PubMed] [Google Scholar]
- 58. Dunker A. K., Obradovic Z. (2001) Nat. Biotechnol. 19, 805–806 [DOI] [PubMed] [Google Scholar]
- 59. Hsu T., Artiushin S., Minion F. C. (1997) J. Bacteriol. 179, 1317–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Linden S. K., Sutton P., Karlsson N. G., Korolik V., McGuckin M. A. (2008) Mucosal. Immunol. 1, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Menozzi F. D., Pethe K., Bifani P., Soncin F., Brennan M. J., Locht C. (2002) Mol. Microbiol. 43, 1379–1386 [DOI] [PubMed] [Google Scholar]
- 62. Moelleken K., Hegemann J. H. (2008) Mol. Microbiol. 67, 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Agarwal V., Asmat T. M., Luo S., Jensch I., Zipfel P. F., Hammerschmidt S. (2010) J. Biol. Chem. 285, 23486–23495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baron M. J., Filman D. J., Prophete G. A., Hogle J. M., Madoff L. C. (2007) J. Biol. Chem. 282, 10526–10536 [DOI] [PubMed] [Google Scholar]
- 65. Rabenstein D. L. (2002) Natural Product Reports 19, 312–331 [DOI] [PubMed] [Google Scholar]
- 66. Sobel M., Soler D. F., Kermode J. C., Harris R. B. (1992) J. Biol. Chem. 267, 8857–8862 [PubMed] [Google Scholar]
- 67. Hsu T., Minion F. C. (1998) Infect. Immun. 66, 4762–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gonciarz-Swiatek M., Rechsteiner M. (2006) Mol. Immunol. 43, 1993–2001 [DOI] [PubMed] [Google Scholar]
- 69. Realini C., Rechsteiner M. (1995) J. Biol. Chem. 270, 29664–29667 [DOI] [PubMed] [Google Scholar]
- 70. Realini C., Rogers S. W., Rechsteiner M. (1994) FEBS Lett. 348, 109–113 [DOI] [PubMed] [Google Scholar]
- 71. Boesen T., Fedosova N. U., Kjeldgaard M., Birkelund S., Christiansen G. (2001) Protein Sci. 10, 2577–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]