Background: NADH and NADPH are critically important but labile coenzymes.
Results: We identified an enzymatic repair system for hydrated NAD(P)H consisting of an ATP- or ADP-dependent dehydratase and an epimerase.
Conclusion: The extreme conservation of this repair system suggests its importance for many species.
Significance: This work indicates that searches for other enzymes involved in metabolite and coenzyme repair might be fruitful.
Keywords: Enzyme Kinetics, Enzyme Purification, Enzymes, Metabolism, NADH
Abstract
The reduced forms of NAD and NADP, two major nucleotides playing a central role in metabolism, are continuously damaged by enzymatic or heat-dependent hydration. We report the molecular identification of the eukaryotic dehydratase that repairs these nucleotides and show that this enzyme (Carkd in mammals, YKL151C in yeast) catalyzes the dehydration of the S form of NADHX and NADPHX, at the expense of ATP, which is converted to ADP. Surprisingly, the Escherichia coli homolog, YjeF, a bidomain protein, catalyzes a similar reaction, but using ADP instead of ATP. The latter reaction is ascribable to the C-terminal domain of YjeF. This represents an unprecedented example of orthologous enzymes using either ADP or ATP as phosphoryl donor. We also show that eukaryotic proteins homologous to the N-terminal domain of YjeF (apolipoprotein A-1-binding protein (AIBP) in mammals, YNL200C in yeast) catalyze the epimerization of the S and R forms of NAD(P)HX, thereby allowing, in conjunction with the energy-dependent dehydratase, the repair of both epimers of NAD(P)HX. Both enzymes are very widespread in eukaryotes, prokaryotes, and archaea, which together with the ADP dependence of the dehydratase in some species indicates the ancient origin of this repair system.
Introduction
Recent work from our and other laboratories indicates the importance of an emerging group of enzymes serving to eliminate abnormal metabolites that result from side activities of enzymes of the intermediary metabolism (1, 2). This variety of metabolite repair enzymes, which we propose to call “metabolite-proofreading enzymes,” plays a similar role in intermediary metabolism as the proofreading activities of DNA polymerases and aminoacyl-tRNA synthases in replication and translation, namely to increase the fidelity of the respective biosynthetic processes by correcting for the lack of specificity of the biosynthetic enzymes involved. l-2-Hydroxyglutarate dehydrogenase, a mitochondrial FAD-dependent enzyme, serves to repair a side activity of l-malate dehydrogenase consisting of the reduction of α-ketoglutarate to l-2-hydroxyglutarate (1). l-2-Hydroxyglutarate dehydrogenase deficiency causes l-2-hydroxyglutarate accumulation in tissues and body fluids, leading to a severe neurological disorder (1). Other examples of metabolite-proofreading enzymes are GDP-glucose phosphorylase (2) and ethylmalonyl-CoA decarboxylase (21) which correct side activities of GDP-mannose pyrophosphorylase and acetyl-CoA carboxylase, respectively.
The earliest known proofreading enzyme in intermediary metabolism is probably the one that dehydrates a hydrated form of NAD(P)H known as NAD(P)HX. Studies in the 1950s showed that glyceraldehyde 3-phosphate dehydrogenase slowly catalyzes the formation of NADHX4 from NADH (3–5). The equilibrium of this hydration reaction is in favor (100/1) of the hydrated form (6), making this reaction virtually irreversible. NADHX and NADPHX are inhibitors of several dehydrogenases (7, 8), and it is therefore important to eliminate them. Reconversion of NADHX to NADH is catalyzed by an ATP-dependent dehydratase, which is present in yeast extracts (6, 9) but has never been molecularly identified, making it difficult to know whether this repair mechanism is widespread or restricted to yeast and what consequences its absence may have.
In view of the importance of nicotinamide nucleotides in numerous cellular processes, the purpose of the present work was to identify the enzyme that catalyzes the ATP-dependent repair of NAD(P)HX. This identification led us to discover a highly conserved enzymatic system, consisting not only of a dehydratase, which is specific for the S form of NADHX or NADPHX and, depending on the species, uses ATP or ADP as energy source, but also of an epimerase that converts the R form of NADHX or NADPHX to the form used by the dehydratase.
EXPERIMENTAL PROCEDURES
Preparation of NADHX and NADPHX
NADHX and NADPHX were produced by incubating NADH or NADPH (∼28 mm) in the presence of 0.5 m NaPi, pH 6.0, for 30 min at 35 °C. The pH was then brought to pH 8 by the addition of NaOH to inhibit further degradation of NAD(P)H. Under these conditions, mixtures containing ∼3.7 mm (S)-NADHX, 2.8 mm (R)-NADHX, and 2.8 mm cyclic NADHX were obtained. Similar amounts of NADPH derivatives were measured after incubation of NADPH. These mixtures were used either directly or after further purification to assay NAD(P)HX dehydratase and epimerase activities.
HPLC Analysis of NAD(P)H Derivatives and Adenosine Nucleotides
The degradation products of NAD(P)H were analyzed by reverse-phase HPLC using a method adapted from Ref. 6 as described in the supplemental Experimental Procedures. AMP, ADP, and ATP were quantified by an anion-exchange HPLC method described previously (10).
Purification of (S)-NAD(P)HX and (R)-NAD(P)HX
The NAD(P)HX mixture for substrate purification was prepared as described above. Semipreparative HPLC was carried out on a Pursuit XRs C18 column (250 × 10.0 mm, 5 μm, Varian) at a flow rate of 5 ml/min using a linear gradient from 2 to 15.5% methanol over 30 min for NADH derivatives and from 0.5 to 5% methanol over 40 min for NADPH derivatives, and 0.5-ml fractions were collected. The aqueous phase consisted of sodium phosphate buffer (10 mm, pH 7.0). Compounds were detected by monitoring UV absorbance at 279 nm. The purest and most concentrated fractions for the two epimers of NAD(P)HX were adjusted to pH 8.0 with 1 m NaOH and lyophilized overnight. The dry residues were resuspended in Tris-HCl buffer (10 mm, pH 8.0), and NAD(P)HX concentration was determined by using ϵ290 = 13,500 m−1 cm−1 (4). The resulting (S)-NADHX, (R)-NADHX, (S)-NADPHX, and (R)-NADPHX solutions were 70–85% pure as determined by the analytical HPLC methods described above and were stored at −20 °C.
Purification and Molecular Identification of Yeast NADHX Dehydratase
NADHX dehydratase was purified from a dry bakers' yeast extract as described in the supplemental Experimental Procedures. A candidate protein band was excised from an SDS-PAGE gel loaded with the most purified enzyme fraction and analyzed by LC/MS/MS after trypsin digestion (11).
Cloning, Expression, and Purification of Recombinant NAD(P)HX Dehydratase and NAD(P)HX Epimerase from Mouse, Saccharomyces cerevisiae, and Escherichia coli
The coding sequences of mouse Carkd and mouse AIBP were amplified by PCR from full-length cDNA clones (IRAVp968B0872D and IRAVp968D07117D, respectively) of the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) library purchased from ImaGenes (Berlin, Germany). For Carkd and apolipoprotein A-1-binding protein (AIBP), the N-terminal sequences were predicted to correspond to mitochondrial targeting signals using subcellular localization programs (TargetP, MitoProt II). As for both of these proteins, the predicted mitochondrial propeptides were closely followed by another conserved and in-frame ATG codon, the forward primers were designed to anneal either to the first or to this second ATG codon to express long and short forms of Carkd and AIBP. The ORFs of yeast YKL151C and YNL200C and of E. coli yjeF were PCR-amplified from genomic DNA of boiled S. cerevisiae and E. coli cells, respectively. All of the primers used are listed in supplemental Table S1. The PCR products were cloned into the Champion pET100/D-Topo vector (Invitrogen) for isopropyl-1-thio-β-d-galactopyranoside-induced bacterial overexpression of N-terminally His-tagged proteins, and the DNA sequence of each insert was confirmed. Recombinant proteins were produced and purified as described in the supplemental Experimental Procedures.
Spectrophotometric Assays of NAD(P)HX Dehydratase and NAD(P)HX Epimerase
The spectrophotometric assays were based on the different absorption spectra displayed by NADH (or NADPH) and NADHX (or NADPHX) (4,7). NAD(P)H production was measured by monitoring A340 (wavelength at which the hydrated forms virtually do not absorb: ϵ340 = 200 m−1 cm−1), whereas NAD(P)HX consumption was determined by measuring A290 (wavelength of maximal difference between the absorption of NAD(P)HX and NAD(P)H). The NAD(P)HX dehydratase activities of Carkd, YKL151C, and YjeF were assayed in a reaction mixture at 30 °C containing, unless otherwise indicated, 25 mm Hepes, pH 7.1, or 25 mm Tris-HCl, pH 8.0, 2 mm MgCl2, 0.2–1 mm ATP (for Carkd and YKL151C) or ADP (for YjeF), 0.05–0.2 mg BSA/ml, and ∼20 μm (S)-NAD(P)HX. The NAD(P)HX epimerase activity of AIBP was measured in a reaction mixture at 30 °C containing 25 mm Tris-HCl, pH 8.0, 2 mm MgCl2, 0.2 mm ATP, 5 mm KCl, 0.05–0.1 mg of BSA/ml, an excess of Carkd, and ∼10 μm (R)-NAD(P)HX unless otherwise indicated. Activities were calculated based on previously published values for the extinction coefficients of NADH (ϵ340 = 6220 m−1 cm−1; ϵ290 = 2100 m−1 cm−1) and NADHX (ϵ290 = 13500 m−1 cm−1) (4). Given the higher spontaneous interconversion rates of the S and R forms of NAD(P)HX at pH 7 and below (6), the enzymatic assays in this study were preferably performed at pH 8 for both the NAD(P)HX dehydratases and epimerases.
RESULTS
Purification of Yeast NADHX Dehydratase and Identification of YKL151C as a Candidate Protein
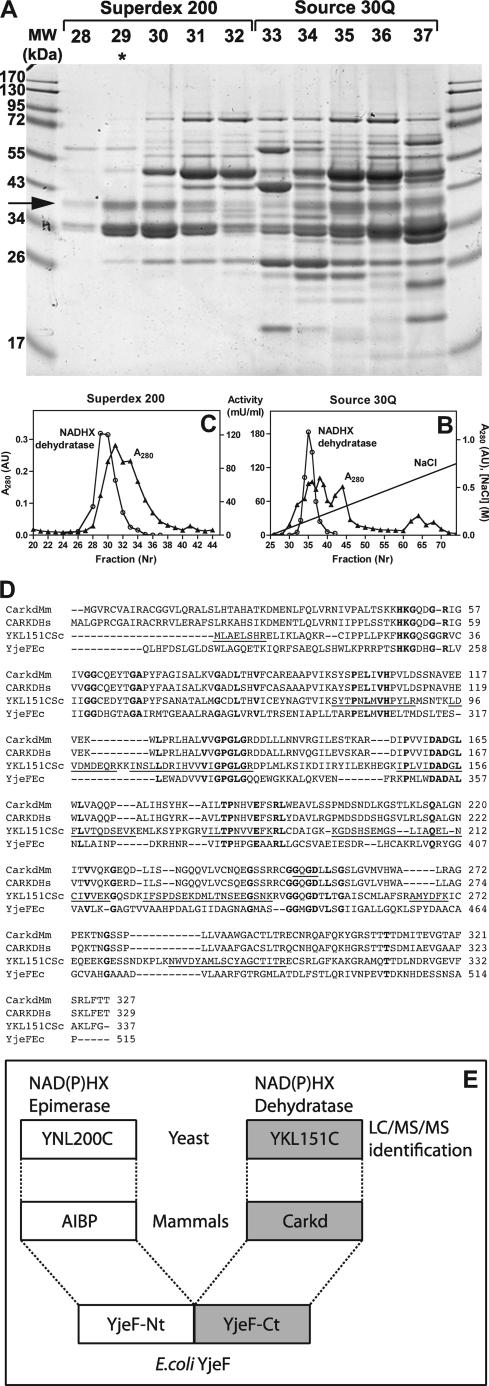
To identify the gene encoding the ATP-dependent NADHX dehydratase activity previously reported in yeast extracts (6, 9), we purified this enzyme from dry bakers' yeast. Yeast extract was obtained by autolysis in an ammonium phosphate buffer at pH 9 and passage through a French press apparatus. After precipitation of the enzyme by ammonium sulfate, it was further purified by three successive chromatographic steps on Blue Trisacryl, Source 30Q (Fig. 1B), and Superdex 200 (Fig. 1C) columns; this procedure allowed an ∼800-fold purification of the enzymatic activity with a yield of 7.5% (supplemental Table S2). A candidate protein band for NADHX dehydratase was identified by comparing its intensity on an SDS-PAGE gel stained with Coomassie Blue (Fig. 1A, arrow) with the elution profile of NADHX dehydratase activity from the Source 30Q and Superdex 200 columns. This band was excised from the gel lane that had been loaded with the most active fraction (fraction 29) eluted from the gel-filtration column. Trypsin digestion of the corresponding proteins followed by peptide analysis by LC/MS/MS suggested that yeast NADHX dehydratase corresponds to protein YKL151C, the sequence of which is shown in Fig. 1D. YKL151C was indeed the top candidate in terms of SEQUEST identification score in the list of proteins identified in the excised gel band. YKL151C belongs to the carbohydrate kinase family, which is part of the ribokinase superfamily (12). The latter comprises enzymes that phosphorylate a hydroxyl group of a substrate at the expense of ATP (e.g. ribokinase, adenosine kinase, hydroxyethylthiazole kinase) or ADP (ADP-glucokinase, ADP-phosphofructokinase).
FIGURE 1.
Molecular identification of yeast NADHX dehydratase as YKL151C, an extremely conserved protein among living species. A–C, NADHX dehydratase was purified from dry bakers' yeast (A), and the peak activity fractions of the Source 30Q (B) and the Superdex 200 (C) columns were analyzed by SDS-PAGE. A protein band co-eluting with enzyme activity (arrow) was excised from the gel (Superdex 200 fraction 29 indicated by an asterisk) and analyzed by LC/MS/MS. MW, molecular weight; AU, absorbance units. D, alignment of S. cerevisiae YKL151C (YKL151CSc; NP_012771.1) with mouse (CarkdMm; NP_001177286.1) and human (CARKDHs; NP_001229811.1) Carkd and with the C-terminal part of E. coli YjeF (YjeFEc; NP_418588.1; residues 211–515). Strictly conserved residues are in bold. The peptides identified in YKL151C are underlined. The GXGD motif, common to proteins of the ribokinase superfamily, with the catalytic aspartate is underlined in mouse Carkd. The human and mouse sequences starting at Met1 are predicted to be mitochondrial, and those starting at the second methionine are predicted to be cytosolic. For human CARKD, the long sequence is the most frequent one among expressed sequence tags. E, in E. coli, the Carkd domain represents the C-terminal part of the bifunctional YjeF protein. The N-terminal domain of YjeF is also very conserved, and the homologs in yeast and mammals are designated YNL200C and AIBP, respectively. Except in bacteria, AIBP and Carkd homologs are, however, encoded by separate genes.
YKL151C Is a Widely Conserved Protein That Is Associated to a Second, Uncharacterized Domain in Bacteria
Homologs of YKL151C are found in the overwhelming majority of eukaryotes and prokaryotes. The mouse and human homologs, known as Carkd, share ∼37% sequence identity with YKL151C (Fig. 1D). Interestingly, the bacterial homologs, including E. coli YjeF, are bidomain proteins comprising an ∼270 residue C-terminal domain, which in the case of YjeF shares ∼26% identity with YKL151C (Fig. 1D), and an ∼240-residue N-terminal domain, which is also highly conserved in prokaryotes and eukaryotes (Fig. 1E and supplemental Fig. S1). The eukaryotic homologs of YjeF-Nt, named AIBP in mammals and YNL200C in S. cerevisiae, are encoded by genes that are separate from the putative eukaryotic NADHX dehydratase genes (Fig. 1E). AIBP has been identified in a yeast two-hybrid screen using apolipoprotein A-1 as a bait (13). Intriguingly, however, its sequence does not comprise a signal peptide, but rather a mitochondrial propeptide as predicted by the TargetP program (14). Furthermore, its three-dimensional structure reveals the presence of a Rossmann-like fold (15), suggesting that this protein may bind a nicotinamide dinucleotide.
As bi- or multifunctional enzymes often catalyze functionally related reactions, we formulated the hypothesis that the reaction catalyzed by the N-terminal domain of E. coli YjeF (and presumably also by mammalian AIBP and yeast YNL200C) is physiologically related with the energy-dependent dehydration catalyzed by yeast YKL151C, mammalian Carkd, and the C-terminal domain of YjeF. This hypothesis was further supported by the highly correlated expression identified by the SPELL microarray search engine for the yeast proteins YKL151C (annotated as a heat-shock protein) and YNL200C. Because yeast NADHX dehydratase was previously shown to be specific for the S epimer of NADHX (6), an obvious possibility was that YjeF-Nt, AIBP, and YNL200C catalyze the isomerization of (R)- and (S)-NADHX (Fig. 2A). These hypotheses were confirmed by characterization of the recombinant proteins, as described below, first for the eukaryotic homologs and then for the bacterial enzyme.
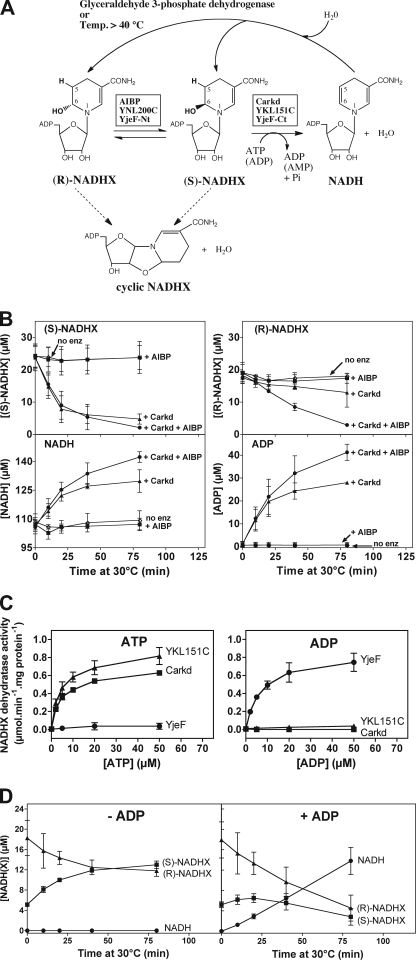
FIGURE 2.
Characterization of the enzymatic reactions catalyzed by mammalian Carkd and AIBP and E. coli YjeF to repair hydrated NAD(P)H. A, scheme illustrating the reactions leading to NADHX formation and those leading to NADHX repair by the enzymes identified in this study. Although only NADHX repair is represented, the same reactions are catalyzed by the same enzymes on NADPHX. YjeF-Ct, C-terminal domain of YjeF. B, time course showing the stereospecific (S)-NADHX conversion to NADH by Carkd accompanied by the formation of stoichiometric amounts of ADP from ATP. Significant (R)-NADHX consumption is observed only when both Carkd and the AIBP epimerase are present. Carkd and/or AIBP were added at final concentrations of 2 and 8 μg/ml, respectively, to a reaction mixture at pH 8 containing (S)- and (R)-NADHX and 0.1 mm ATP-Mg. Reactions were stopped at the indicated times by protein denaturation through the addition of an equal volume of 8 m urea. Control reactions without added enzyme (no enz) were also performed. The concentrations of the indicated compounds were determined by HPLC analysis. The values shown are means ± S.D. (n = 3). C, dependence of the (S)-NADHX dehydratase activity of Carkd and YKL151C on ATP and of YjeF on ADP. Enzyme activities were determined spectrophotometrically in a reaction mixture at pH 7.1 containing ∼20 μm (S)-NADHX. D, time course demonstrating the NADHX epimerase activity and the ADP-dependent NADHX dehydratase activity of the bifunctional YjeF enzyme. YjeF, at a final concentration of 2.1 μg/ml, was incubated at pH 8 in the presence of an NADHX solution enriched in (R)-NADHX, 5 mm KCl, and with or without 0.1 mm ADP-Mg for the times indicated. Reactions were stopped by the addition of an equal volume of a mixture containing 8 m urea and 10 mm EDTA. The concentrations of the indicated compounds were determined by reverse-phase HPLC. Control reactions without added enzyme were also performed in the presence of ADP-Mg; little change in concentration for the various compounds analyzed was observed under these conditions. The values shown represent means ± S.D. (n = 3).
Characterization of Carkd/YKL151C and AIBP/YNL200C as (S)-NAD(P)HX Dehydratases and NAD(P)HX Epimerases, Respectively
The coding sequences of the putative mouse and yeast NADHX dehydratases (Carkd and YKL151C) and epimerases (AIBP and YNL200C) were cloned into TOPO vectors for bacterial isopropyl-1-thio-β-d-galactopyranoside-induced expression of N-terminally His-tagged recombinant proteins. For Carkd and AIBP, long and short forms of the proteins were produced that contained or did not contain the N-terminal mitochondrial propeptide. For Carkd, sufficient amounts of purified protein could be obtained for either form and, as similar kinetic properties were found for both forms, they were used indiscriminately in the experiments described below. For AIBP, much higher expression levels were obtained using the short construct, and the protein lacking the mitochondrial propeptide was used for enzyme characterization. All of the recombinant proteins were purified by at least one metal affinity chromatography step to obtain homogeneous enzyme preparations.
NADHX (or NADPHX) can be produced by incubating NADH (or NADPH) in the presence of elevated concentrations of Pi and slightly acidic pH (6, 7). The resulting mixture contains, in addition to (S)-NAD(P)HX and (R)-NAD(P)HX (in a 60:40 ratio), non-reacted NAD(P)H and cyclic forms derived from NAD(P)HX (Fig. 2A) (5–7, 16). All of these compounds can be separated by reverse-phase HPLC (6, 17), which allowed us to functionally characterize our recombinant proteins. As shown in Fig. 2B, during incubation of such an NADHX mixture with Carkd and ATP, (S)-NADHX was rapidly depleted, whereas the (R)-NADHX levels decreased only slightly over the same time period. This (S)-NADHX consumption was accompanied by the formation of stoichiometric amounts of NADH and ADP, indicating that the conversion of 1 mol of (S)-NADHX to NADH is accompanied by the hydrolysis of ∼1 mol of ATP to ADP. This confirms the identity of Carkd as an ATP-dependent and stereospecific (S)-NADHX dehydratase. By contrast, when both Carkd and AIBP were present in the incubation mixture, both the S and the R forms of NADHX were entirely converted to NADH (Fig. 2B), supporting the hypothesis that AIBP acts as an NADHX epimerase, catalyzing the conversion of (R)-NADHX to the S epimer used by the NADHX dehydratase. In these time-course experiments, we also monitored the concentrations of the cyclic forms of NADHX (contained in the NADHX mixture added to the reaction medium); these concentrations remained stable throughout the incubation time, showing that neither Carkd nor AIBP acts on those cyclic forms of NADHX in the assay conditions used (data not shown). Even more direct evidence for the NAD(P)HX epimerase activity of AIBP was obtained by incubating purified solutions enriched in either the R epimer or the S epimer of NAD(P)HX with recombinant AIBP and by showing that the latter greatly accelerates the re-equilibration of both epimers to a 60:40 ratio (shown for NADPHX in supplemental Fig. S2).
Using a spectrophotometric assay, we confirmed that Carkd and also YKL151C convert (S)-NADHX to NADH in an ATP-dependent manner (Fig. 2C). In those assays, Carkd and YKL151C indeed caused a decrease in A290 (NADHX consumption) as well as an increase in A340 (NADH production) only when ATP was present. End point assays in which Carkd and AIBP (or YNL200C) were successively added to a mixture of (S)- and (R)-NAD(P)HX (60:40 ratio) showed that the initial decrease in A290 (or increase in A340) caused by the dehydratase activity of Carkd was followed by a second decrease in A290 (or increase in A340) of similar magnitude upon the addition of AIBP or YNL200C (supplemental Fig. S3). This observation further supported that Carkd is stereospecific for its NAD(P)HX substrate and that AIBP (and YNL200C) functions as an NAD(P)HX epimerase, thereby allowing the complete reconversion of NAD(P)HX to NAD(P)H in the presence of the NAD(P)HX dehydratase.
The addition of lactate dehydrogenase (and pyruvate) to a mixture in which (S)-NADHX had been completely converted to NADH by Carkd or YKL151C (as evidenced by a plateau reached while monitoring A340) led to a decrease in A340 approximately equaling the increase in A340 caused by the recombinant NADHX dehydratases (not shown). This strongly indicated that NADHX dehydratase reconverts (S)-NADHX to the β-anomer of NADH. All of the findings obtained using the spectrophotometric assay and NADHX as a substrate could be repeated with similar results when replacing NADHX by NADPHX.
Bacterial YjeF Is a Bifunctional and ADP-dependent NAD(P)HX Repair Enzyme
With recombinant E. coli YjeF, the protein comprising a C-terminal domain homologous to Carkd and an N-terminal domain homologous to AIBP, we also detected an NAD(P)HX dehydratase activity when performing the spectrophotometric assay in the presence of ATP. However, the incomplete substrate (NAD(P)HX) consumptions observed during those assays, especially at low ATP concentrations, led us to include a P-enolpyruvate/pyruvate kinase-based regeneration system for ATP in the reaction mixture. Contrary to our expectations, the enzymatic reaction was completely inactive under those conditions, which suggested that ADP is involved in the reaction. This led us to test whether ADP rather than ATP is the co-substrate used by the bacterial enzyme, a hypothesis that was indeed confirmed as shown in Fig. 2C.
Furthermore, we found that the addition of YjeF to 60:40 mixtures of (S)- and (R)-NADHX or (S)- and (R)-NADPHX (in the presence of ADP) caused an increase in A340 that was ∼2-fold higher than the increase in A340 caused by the addition of Carkd or YKL151C to the same reaction mixtures (except for the presence of ATP instead of ADP) (shown for NADPHX in supplemental Fig. S4). These observations supported the hypothesis that YjeF not only dehydrates (S)-NAD(P)HX, but also interconverts (R)- and (S)-NAD(P)HX.
We confirmed the bifunctionality and ADP dependence of E. coli YjeF by using the HPLC-based assays. The addition of recombinant YjeF to an incubation mixture containing a purified, (R)-NADHX-enriched solution, but no ADP, only accelerated the conversion of (R)-NADHX to (S)-NADHX (Fig. 2D). In the presence of ADP, however, both the R and the S forms of NADHX were completely converted to NADH (Fig. 2D), and this conversion was accompanied by the hydrolysis of a stoichiometric amount of ADP to AMP (not shown).
Kinetic Properties of the NAD(P)HX Dehydratase and NAD(P)HX Epimerase Activities
Both Carkd and YjeF displayed high affinities for their (S)-NADHX and (S)-NADPHX substrates, as well as for their respective co-substrates (ATP or ADP) with low micromolar Km values (supplemental Table S3). For Carkd, we found an ∼4-fold higher Vmax value when (S)-NADPHX was used as a substrate instead of (S)-NADHX. For YjeF, the difference was not as pronounced, but (S)-NADPHX was also a better substrate for this enzyme than (S)-NADHX, as indicated by the 2-fold higher catalytic efficiency with (S)-NADPHX. Similar results were obtained with yeast YKL151C for (S)-NADHX and ATP (not shown).
The kinetic properties of AIBP for (R)-NADHX and (R)-NADPHX were determined spectrophotometrically by coupling the epimerization reaction to the dehydration reaction catalyzed by Carkd. We found that KCl stimulates the NADHX epimerase activity of AIBP by ∼14-fold (supplemental Fig. S5), whereas this salt did not have any effect on the activity of Carkd. Similar effects on AIBP activity were obtained when using potassium acetate instead of the chloride salt, indicating that K+ exerts the stimulatory effect. NH4Cl also had a similar stimulatory effect on AIBP activity, whereas NaCl did not affect this activity up to a concentration of 5 mm (supplemental Fig. S5). The concentrations of KCl and NH4Cl needed for half-maximal activation of AIBP activity were remarkably low (<150 μm). AIBP also displayed high affinities for its NAD(P)HX substrates and a higher catalytic efficiency for the hydrated nicotinamide phosphate nucleotide (supplemental Table S3).
DISCUSSION
The molecular identification of the NAD(P)HX repair enzymes provides critical new information on this system. First, it consists of two distinct enzymatic activities, an ATP- or ADP-dependent dehydratase and an as yet undescribed epimerase. Together these enzymes are able to repair the two epimers of NADHX and NADPHX. This identification also leads to a reappraisal of the function of AIBP, which has previously been claimed to be secreted and to interact with lipoproteins (13). Preliminary data from our laboratory indicate that AIBP (like Carkd) is essentially mitochondrial, cytosolic, and nuclear,5 supporting its role in NAD(P)HX repair.
A second new piece of information is that homologs of the NAD(P)HX repair enzymes are found in the overwhelming majority of eukaryotes, prokaryotes, and archaea, indicating that NAD(P)HX repair is a very ancient, and presumably important, process for many species. This is not surprising if one considers the central role played by NAD and NADP in intermediary metabolism, as well as the fact that NAD(P)HX is known to inhibit several dehydrogenases (7,8). The NAD(P)HX repair enzymes are not essential, as indicated by the finding that the E. coli (yjeF) and S. cerevisiae (YKL151C and YNL200C) knock-out mutants are viable (18). This may be related to the capacity of these organisms to carry out de novo synthesis of nicotinamide nucleotides. It should be stressed, however, that the yeast YKL151C and YNL200C mutants show a modest, yet significant competitive growth defect (19), which is likely to be enhanced under conditions where nicotinamide nucleotide synthesis is compromised.
The fact that the yeast YKL151C gene is up-regulated under heat shock (20) has to be put in relation with the heat lability of NAD(P)H. Preliminary data from this laboratory indicate first order rates of conversion of NADPH to NADPHX of 0.002 and 0.05 min−1 at 37 and 72 °C, respectively, and ∼5-fold lower rates for NADH.6 These findings suggest that the NAD(P)HX repair system is critical for thermophiles. However, this repair system is present even in psychrophiles, pleading in favor of enzyme-catalyzed hydration of NAD(P)H being an important contributor of NAD(P)HX formation in many species. These findings appeal to the search of NADHX- and NADPHX-producing enzymes.
The NAD(P)HX dehydratase is a new, but atypical, member of the ribokinase superfamily, not only because it is not a kinase, unlike the other members of this superfamily, but also because it represents an unprecedented example of an enzyme whose activity has evolved from being ADP-dependent to being ATP-dependent. The identification of the NAD(P)HX dehydratase as a member of a superfamily of enzymes that catalyze the phosphorylation of substrates, however, is an important argument in favor of the mechanism proposed by Acheson et al. (6), in which dehydration involves the transfer of a phosphoryl group onto the C6-hydroxyl of the nicotinamide ring followed by phosphate elimination. The availability of recombinant protein will allow the further exploration of this unique reaction mechanism.
A remarkable feature of the repair system, which has been found for all enzymes investigated here, is that it works on derivatives of both NADH and NADPH, yet it works significantly better on hydrated forms of the latter than of the former nucleotide. This is logical if one considers that the intracellular NADPH/NADP+ ratio is much higher than the NADH/NAD+ ratio as well as the higher intrinsic reactivity of NADPH as compared with NADH.
While this work was almost completed, several structures of YjeF bacterial homologs appeared in the Protein Data Bank (PDB), including one annotated as “a fusion of a domain of unknown function and ADP-/ATP-dependent NAD(P)H-hydrate dehydratase” (PDB ID 3rs8). This structure shows NADHX and ADP in the catalytic site of the C-terminal domain of YjeF (YjeF-Ct), consistent with this enzyme catalyzing the ADP-dependent dehydration of NADHX as described in the present study with E. coli YjeF. The N-terminal domain comprises an analog of NADHX (ADP-ribosylamine) in the putative catalytic site of YjeF-Nt (AIBP domain), as well as a nearby potassium ion forming seven bonds with neighboring atoms. These structural features are also consistent with the function identified in the present study for the AIBP domain and its dependence on K+.
In conclusion, we have identified a widely distributed NAD(P)HX repair system consisting of two enzymatic activities. This system is likely to serve both as a proofreading system and to repair heat-damaged pyridine nucleotides. This work appeals to the search for other enzymes that repair metabolites and coenzymes.
Note Added in Proof
Following a suggestion of Dr. Kenneth Rudd (University of Miami), we propose to designate the E. coli yjeF gene “nnr” for nicotinamide nucleotide repair. Stand-alone bacterial genes encoding NAD(P)HX dehydratase or NAD(P)HX epimerase could be named nnrD and nnrE, respectively.
This work was supported by the Fonds de la Recherche Scientifique (FRS-FNRS), by the European Union Seventh Framework Programme (Grant FP7/2007-2013) under Grant Agreement 276814, and by Welbio (Walloon region).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables S1–S3, and Figs. S1–S5.
A. Marbaix and C. Linster, unpublished results.
C. Linster and A. Aklé, unpublished results.
- NADHX
- hydrated form of NADH
- NADPHX
- hydrated form of NADPH
- AIBP
- apolipoprotein A-1-binding protein
- LC/MS/MS
- liquid chromatography coupled to tandem mass spectrometry
- YjeF-Nt
- N-terminal domain of YjeF.
REFERENCES
- 1. Van Schaftingen E., Rzem R., Veiga-da-Cunha M. (2009) J. Inherit. Metab. Dis. 32, 135–142 [DOI] [PubMed] [Google Scholar]
- 2. Adler L. N., Gomez T. A., Clarke S. G., Linster C. L. (2011) J. Biol. Chem. 286, 21511–21523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rafter G. W., Chaykin S., Krebs E. G. (1954) J. Biol. Chem. 208, 799–811 [PubMed] [Google Scholar]
- 4. Chaykin S., Meinhart J., Krebs E. G. (1956) J. Biol. Chem. 220, 811–820 [PubMed] [Google Scholar]
- 5. Oppenheimer N. J., Kaplan N. O. (1974) Biochemistry 13, 4685–4694 [DOI] [PubMed] [Google Scholar]
- 6. Acheson S. A., Kirkman H. N., Wolfenden R. (1988) Biochemistry 27, 7371–7375 [DOI] [PubMed] [Google Scholar]
- 7. Yoshida A., Dave V. (1975) Arch. Biochem. Biophys. 169, 298–303 [DOI] [PubMed] [Google Scholar]
- 8. Prabhakar P., Laboy J. I., Wang J., Budker T., Din Z. Z., Chobanian M., Fahien L. A. (1998) Arch. Biochem. Biophys. 360, 195–205 [DOI] [PubMed] [Google Scholar]
- 9. Meinhart J. O., Chaykin S., Krebs E. G. (1956) J. Biol. Chem. 220, 821–829 [PubMed] [Google Scholar]
- 10. Linster C. L., Van Schaftingen E. (2003) J. Biol. Chem. 278, 36328–36333 [DOI] [PubMed] [Google Scholar]
- 11. Drozak J., Veiga-da-Cunha M., Vertommen D., Stroobant V., Van Schaftingen E. (2010) J. Biol. Chem. 285, 9346–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guixé V., Merino F. (2009) IUBMB Life 61, 753–761 [DOI] [PubMed] [Google Scholar]
- 13. Ritter M., Buechler C., Boettcher A., Barlage S., Schmitz-Madry A., Orsó E., Bared S. M., Schmiedeknecht G., Baehr C. H., Fricker G., Schmitz G. (2002) Genomics 79, 693–702 [DOI] [PubMed] [Google Scholar]
- 14. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 15. Jha K. N., Shumilin I. A., Digilio L. C., Chertihin O., Zheng H., Schmitz G., Visconti P. E., Flickinger C. J., Minor W., Herr J. C. (2008) Endocrinology 149, 2108–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oppenheimer N. J., Kaplan N. O. (1974) Biochemistry 13, 4675–4685 [DOI] [PubMed] [Google Scholar]
- 17. Miksic J. R., Brown P. R. (1978) Biochemistry 17, 2234–2238 [DOI] [PubMed] [Google Scholar]
- 18. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 19. Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., Newman H. W., Braun S., Madhani H. D., Krogan N. J., Weissman J. S. (2008) Nat. Methods 5, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Treger J. M., Schmitt A. P., Simon J. R., McEntee K. (1998) J. Biol. Chem. 273, 26875–26879 [DOI] [PubMed] [Google Scholar]
- 21. Linster C. L., Noël G., Stroobant V., Vertommen D., Vincent M. F., Boomer G. T., Veiga-da-Cunha M., Van Schaftingen E. (2011) J. Biol. Chem. 10.1074/jbc.M111.281527 [DOI] [PMC free article] [PubMed] [Google Scholar]