Background: Impaired oxidative phosphorylation (OXPHOS) is implicated in several metabolic disorders. Hence, molecules that stimulate OXPHOS may prove beneficial.
Results: LRP130, a protein involved in Leigh syndrome, activates mitochondrial transcription, which stimulates OXPHOS and oxidative metabolism in cells and mouse liver.
Conclusion: By activating mitochondrial transcription, LRP130 remodels mitochondria resulting in denser cristae.
Significance: An activator of OXPHOS, LRP130 may mitigate certain metabolic disorders.
Keywords: Energy Metabolism, Fatty Acid Oxidation, Mitochondria, Transcription, Transcription Co-activators, LRP130, Leucine-rich Pentatricopeptide Repeat Motif-containing, Cristae, Oxidative Phosphorylation
Abstract
Impaired oxidative phosphorylation (OXPHOS) is implicated in several metabolic disorders. Even though mitochondrial DNA encodes several subunits critical for OXPHOS, the metabolic consequence of activating mitochondrial transcription remains unclear. We show here that LRP130, a protein involved in Leigh syndrome, increases hepatic β-fatty acid oxidation. Using convergent genetic and biochemical approaches, we demonstrate LRP130 complexes with the mitochondrial RNA polymerase to activate mitochondrial transcription. Activation of mitochondrial transcription is associated with increased OXPHOS activity, increased supercomplexes, and denser cristae, independent of mitochondrial biogenesis. Consistent with increased oxidative phosphorylation, ATP levels are increased in both cells and mouse liver, whereas coupled respiration is increased in cells. We propose activation of mitochondrial transcription remodels mitochondria and enhances oxidative metabolism.
Introduction
Impaired mitochondrial function is implicated in neurodegenerative disease, diabetes, cancer, muscular dystrophy, heart failure, and aging (1–3). A primary mechanism of human mitochondrial diseases is dysfunctional oxidative phosphorylation (OXPHOS),3 which is carried out by five large enzymatic complexes in the mitochondrial inner membrane. Although mitochondria contain ∼1,200 proteins, only ∼90 of these proteins comprise OXPHOS complexes. Thirteen are encoded by mitochondrial DNA (mtDNA) (4, 5), whereas the remainder is encoded by nuclear DNA (nDNA) (6–8).
Mitochondrially encoded subunits are co-translationally inserted into the inner mitochondrial membrane. Serving as both a structural and functional scaffold for nuclearly encoded subunits, mtDNA-encoded subunits play a central role in the formation of functional OXPHOS complexes. Although highly orchestrated, the genetic determinants that increase OXPHOS complex formation and concomitant oxidative metabolism are poorly understood. In principle, this process could be regulated at the level of mitochondrial transcription, RNA processing, RNA stability, and/or translation. Indeed, it has been postulated that mitochondrial transcription might play a unique role in this regard (9). One impediment to deciphering this relationship is an incomplete understanding of regulators that activate mitochondrial transcription.
The first step in human mitochondrial gene expression is transcription initiation at the light strand promoter, heavy strand promoter 1 (HSP1), and HSP2 of mtDNA by a dedicated mitochondrial RNA polymerase (called POLRMT in humans). Like its yeast ortholog, human POLRMT is a single subunit, T7-like polymerase (10, 11), whose ability to efficiently initiate transcription depends on additional transcription factors (12). In humans, the basal transcriptional machinery consists of POLRMT and mitochondrial transcription factor B2, TFB2M (13). This core two-component system is activated differentially by the high mobility group box transcription factor, mitochondrial transcription factor A (TFAM or mtTFA) (13–18). TFAM binds at sites upstream of the promoters, and its C-terminal tail activates transcription via an interaction with TFB2M (19, 20). TFAM also facilitates packaging of mtDNA into nucleoids, regulates DNA copy number, and primers mtDNA during replication (12, 21–24). Once docked, TFB2M melts the promoter and interacts with POLRMT, culminating in transcription initiation by POLRMT (25). Although TFB2M and POLRMT clearly constitute the core transcription initiation complex (13) and TFAM is a well characterized transcriptional activator, the biological significance of activating mitochondrial transcription by other factors remains unclear and could have broad implications for understanding energy metabolism and metabolic disease.
LRP130 (official gene symbol, LRPPRC; leucine-rich pentatricopeptide repeat motif-containing) is the gene defective in Leigh syndrome French Canadian variant (26, 27). Various nuclear actions have been reported for LRP130 (28, 29); however, in vivo significance has not been established, and LRP130 protein is mostly present in mitochondria. Using knockdown cell lines, we and others previously reported that depletion of LRP130 attenuates mtDNA-encoded gene expression and, as predicted, impairs oxidative phosphorylation (30–34). Mechanisms underlying this regulation remain controversial and include mitochondrial RNA transcript stability, transcription termination, and RNA processing (32–35). Using mice transgenic for lrp130, cell-based assays, and in vitro transcription studies, we report here that LRP130 activates mitochondrial transcription, activation that is associated with remodeling of mitochondria, increased OXPHOS, and enhanced oxidative metabolism. Notably, increases in OXPHOS activity are independent of mitochondrial biogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cell lines HepG2, 293T, and Ad293 and mouse hepatoma cell line H2.35 were cultured at 37 °C in 5% CO2 in DMEM containing 10% FBS. Stable knockdown of LRP130 was achieved as described previously (31). Stable ectopic expression of LRP130, PORLMT, and TFB2M was achieved by infecting cells with a pMSCV-based retroviral vector. Greater than 50% of infected cells survived selection in puromycin or hygromycin. To block mitochondrial transcription, cells were treated with 0.4 μg/ml ethidium bromide for 0, 4, 8, or 16 h prior to harvest.
RNA Extraction and Gene Expression
RNA from cells or mouse liver were homogenized in TRIzol (Invitrogen) and prepared as described previously (31). For details see under supplemental “Experimental Procedures”.
In Organello Labeling of Mitochondrial Transcripts and Mitochondrial Run-off Transcription Assays
Labeling of mitochondrial RNA was performed as described previously (36). Mitochondrial run-off transcription assays were performed as described previously (13). For details see under supplemental “Experimental Procedures”.
Quantification of Cellular ATP and Measurement of Ketone Bodies
Ketone bodies were quantified using a β-hydroxybutyrate (ketone body) assay kit (catalog no. 700190, Cayman Chemical) per manufacturer's instructions. ATP in cells was measured using the CellTiter-Glo luminescent cell viability assay kit (Promega). Equivalent numbers of cells were trypsinized and incubated in 12 replicates in an opaque 96-well plate for 1 h. 100 μl of the reaction reagent was added directly to each well. After 2 min of shaking at room temperature and 10 minutes of stabilization in the dark, the luminescent signal was detected using a POLARstar Omega microplate reader (BMG Labtech).
Oxygen Consumption and Palmitate Oxidation Assays
Respiration in whole cells was measured using a Clark-type oxygen electrode (Hansatech) as described previously (30). Respiration in isolated mitochondria was performed as described previously (37). Palmitate oxidation was performed as described previously (38). For details see under supplemental “Experimental Procedures.”
Statistical and Microarray Analyses
Statistics were performed using the Prism software (GraphPad). Where indicated, two-way analysis of variance with or without post-hoc tests and Student's t test with Welch's correction were utilized. Global gene expression of shLrp130 and Lrp130 overexpression cells was examined by Affymetrix GeneChip Mouse Gene 1.0 ST array. cRNA was derived and hybridized to the chip using standard procedures recommended by Affymetrix. Statistical analysis of gene expression was performed using the Wilcoxon signed rank sum test (39).
Quantification of Hepatic ATP
After euthanasia, the left lower lobe of the liver was immediately frozen in liquid nitrogen. Three pieces weighing 50–100 mg were assayed separately for each animal. Each sample of liver was transferred to 1 ml of ice-cold 5% perchloric acid in DMEM and homogenized at 30 Hz for 3 min in a Tissue Lyser II apparatus (Qiagen). After vortexing, 75 μl of homogenate was transferred to a new tube for protein quantification (outlined below). The remaining sample was centrifuged at 16,000 × g for 10 min at 4 °C. An aliquot of 200 μl of cleared lysate, taken below the fatty layer, was transferred to a new tube for neutralization. Exactly 0.083 volumes (16.6 μl) of 10 n NaOH was added to the aliquot and immediately vortexed, followed by addition of exactly 1/9 volume (22 μl) of 1 m Tris-Cl, pH 7.4, and immediate vortexing. The neutralized sample was diluted 1:300 with water, and 50 μl was assayed for ATP content, using the Cell Titer Glo kit (Promega catalog no. G7571).
For protein quantification, the 75-μl aliquot of lysate saved from above was neutralized with 6.23 μl of 10 n NaOH and 8.3 μl of 1 m Tris-Cl, pH 7.5, with immediate vortexing after each addition. The sample was raised to 750 μl with 660.47 μl of PBS containing 1.137% SDS. The lysate was boiled for 30 min and briefly vortexed every 10 min. Insoluble debris was pelleted at 12,000 rpm for 5 min. After diluting the cleared lysate by 1:2 with water, 50 μl was assayed for protein content using a BCA protein assay kit (catalog no. 23227, Pierce). ATP content was normalized to protein content.
Generation of Liver-specific lrp130 Transgenic Mice
Liver-specific expression was driven by a transthyretin enhancer and promoter, using the pTTR1ExV3 vector (a gift from Dr. Van Dyke) (40). To facilitate cloning, the EcoRI site in intron 1 of the transthyretin gene was eliminated by site-directed mutagenesis. Containing a C-terminal Myc tag, murine lrp130 cDNA was blunt-ligated into the StuI site of the pTTR1ExV3 vector. After EcoRI and SgrA1 digestion, the gel-purified construct was microinjected into fertilized C57BL/6 embryos by the transgenic core at the University of Massachusetts Medical School. The mice were hemizygous for lrp130 in liver.
Mitochondria Volume and Cristae Density
60–90-nm sections of cells were sectioned for transmission electron microscopy after conventional fixation and staining. Two sets of images were taken using a Philips CM10 electron microscope as follows: ×5800 images for measuring mitochondrial volume using ImageJ software (National Institutes of Health), and ×19,000 images for cristae density measurement using standard stereological measurements.
Assessment of Mitochondrial Complex Activity
The activity of each mitochondrial complex was performed as described previously (41). The activities of complex I (NADH-ubiquinone oxidoreductase, EC 1.6.99.3), complex II (succinate dehydrogenase, EC 1.3.5.1), complex III (ubiquinol-cytochrome c reductase, EC 1.10.2.2), complex V (ATPase, EC 3.6.1.3), and citrate synthase (EC 4.1.3.7) were assessed using crude mitochondria. Mitochondria were suspended in 25 mm potassium phosphate buffer, lysed by three cycles of freeze-thawing, and centrifuged at 11,000 × g, 4 °C for 10 min. Pellets were resuspended in 25 mm potassium phosphate buffer. 50 μl of the reaction was assayed using a half-area, UV-visible 96-well plate (Corning Costar). All chemicals were purchased from Sigma
For complex I, isolated mitochondria were suspended in distilled water to a final concentration of 0.05–0.1 mg/ml for 1–2 min at 37 °C. 40 μl of the mitochondrial solution and 10 μl of complex I assay medium (50 mm Tris-Cl, pH 8.0, 5 mg/ml BSA, 0.8 mm NADH (an electron donor), 240 μm KCN, and 4 μm antimycin A) were mixed together in a well. Complex activity was then measured at 340 nm by adding 50 μm decylubiquinone, an electron acceptor.
For complex II, the reaction was set up using 10 mm succinate as electron donor in 1 ml, containing 10 mm potassium phosphate buffer, pH 7.8, 1 mg/ml BSA, 2 mm EDTA, 4 μm rotenone, 0.2 mm ATP, and 80 μm 2,6-dichlorophenol-indophenol. Two to 4 μg of mitochondrial protein was added into the medium and incubated for 10 min at 25 °C. The assay was started by adding decylubiquinone (80 μm) and then measured at 600 nm.
For complex III, 2–4 μg of mitochondrial protein was suspended in 1-ml final volume containing 20 mm potassium phosphate buffer, pH 7.8, 4 mm EDTA, 2 mg/ml BSA, 1.2 mm n-dodecyl-β-d-maltoside, 160 μm decylubiquinol, 480 μm KCN, 8 μm rotenone, and 400 μm ATP. The medium was incubated for 5–10 min at 25 °C. After the addition of oxidized cytochrome c (40 μm), activity was measured at 550 nm.
For complex V, assay medium (25 mm Tris-Cl, pH 8.0, 2.5 mg/ml BSA, 10 mm MgCl2, 25 mm KCl, 7.5 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone, 2.5 μm antimycin A, 5 mm phosphoenolpyruvate, 1.25 mm ATP, 10 units of lactate dehydrogenase, 10 units of pyruvate kinase, and 0.5 mm NADH) was incubated for 5 min at 37 °C. Isolated mitochondria were suspended in distilled water to a final concentration of 0.05–0.1 mg/ml for 30 s at 37 °C. 30 μl of the mitochondrial solution and 20 μl of assay medium were mixed together in a well. Complex V activity was measured by following the time kinetics over 3 min at an absorbance of 340 nm. Representative of complex V activity, the resulting slope was calculated.
The activity of complex IV (cytochrome c oxidase, EC 1.9.3.1) was assayed using a kit (CYTOCOX1, Sigma). Absorbance measurements were recorded every 12 s using a plate reader operating in plate mode (POLARstar Omega plate reader manufactured by BMG Labtech). Relative activities for complex I, II, and V and citrate synthase were derived after normalization (nmol/min/mg protein). Relative activity for complex III and IV was derived after calculating the first order rate constant each reaction (k/min/mg protein).
Blue-native PAGE Analysis
Blue-native PAGE was performed as described previously (42) with some modifications. Isolated mitochondria were suspended with 1× Native PAGE Sample Buffer (Invitrogen), and the protein concentration was measured. Mitochondria were incubated with digitonin (1.6 digitonin/g of protein) for 15 min on ice and centrifuged at 16,000 × g for 40 min in 4 °C. Supernatants were collected, and digitonin was increased to a final concentration of 4 g/g of protein. The supernatant was mixed with G-250 CBB solution (Invitrogen), and 10 μg (for complex II, III, and V) or 25 μg (complex I and IV) of protein was applied to a 3–12% Native polyacrylamide gel (Invitrogen). After electrophoresis at 150 V for 60 min, the voltage was increased to 250 V for 90 min. The gel was soaked for 30 min in MOPS SDS running buffer (Invitrogen) pre-warmed to 70 °C, containing 1% fresh β-mercaptoethanol. The gel was transferred to PVDF membrane and immunoblotted. For Coomassie Brilliant Blue staining, the gel was stained immediately following electrophoresis, using the NOVEX Colloidal Blue Staining kit from Invitrogen.
Mitochondrial ChIP Assay, Immunoblotting, and Purification of LRP130 Protein
Mitochondrial ChIP was performed as described previously (43). The latter two assays were performed using commercially available reagents per manufacturer's instructions. For details see under supplemental “Experimental Procedures”.
Animal Models
Male mice ages 12–16 weeks on C57BL6 background were used for studies. All studies on mice were performed in accordance with guidelines of the IACUC committee at the University of Massachusetts Medical School. Mice were maintained in a 12-h light/12 h-dark cycle.
RESULTS
LRP130 Stimulates Oxidative Phosphorylation
LRP130 is primarily a mitochondrial protein, localized to the mitochondrial matrix (31, 35, 44, 45). Prior studies have established that cells deficient for LRP130 have impaired OXPHOS (31, 33). Molecules that stimulate OXPHOS, however, number in the few but may have important therapeutic consequences for metabolic disorders characterized by impaired OXPHOS. We therefore were interested if ectopic expression of LRP130 could reciprocally stimulate OXPHOS and, if true, how might LRP130 influence metabolism.
Using a Clark-type oxygen electrode, we quantified cellular respiration in cells replete with LRP130 and found that LRP130 increases cellular respiration (Fig. 1, A–C). This was accompanied by an increase in ATP, observed in both cells and mouse liver transgenic for lrp130 (Fig. 1, D and E) (details of transgenic mice are discussed below). Because oxygen consumption and ATP content were both increased and there was no change in proton leak (which permits oxygen consumption without ATP synthesis), these data argue that LRP130 increases oxidative phosphorylation. In cells deficient for LRP130, reciprocal results were observed, a diminution in OXPHOS consistent with prior observations (supplemental Fig. S1) (30, 33). In addition to coupled respiration (OXPHOS), we measured maximally stimulated cellular respiration (also called uncoupled respiration) and individual respiratory complex activity. Again, LRP130 influenced both, providing additional evidence in support of global OXPHOS control (supplemental Fig. S2).
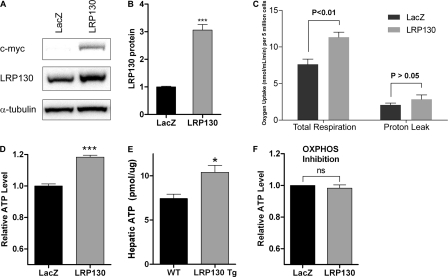
FIGURE 1.
LRP130 stimulates oxidative phosphorylation. A, stable retroviral expression of C-terminally tagged LRP130 protein in H2.35 cells. B, quantification of LRP130 protein normalized to loading control. Endogenous LRP130 was assigned a value of 1. C, measured in a Clark-type electrode, oxygen consumption was increased in cells replete with LRP130. D and E, ectopic expression of LRP130 protein increases ATP levels in cells (D, n = 4) as well as mouse liver (E, n = 4), indicative of coupled respiration (i.e. oxidative phosphorylation). F, in H2.35 cells, inhibition of OXPHOS with inhibitors of respiration, rotenone (2 μm), myxothiazol (4 μm), and oligomycin (5 μm), abrogate differences in ATP, indicating LRP130 influences ATP via OXPHOS. Cells were treated for 7 h and then assayed for ATP. Error bars indicate S.E. *, p < 0.05; ***, p < 0.001. ns = not statistically significant.
Because ATP content is determined by many variables, including OXPHOS, we evaluated if LRP130-mediated increases in ATP were attributable to increased OXPHOS. We inhibited OXPHOS using inhibitors of the respiratory chain, rotenone and myxothiazol, as well as an inhibitor of complex V, oligomycin. We then measured ATP content to assess the non-OXPHOS pathways. Inhibition of OXPHOS abrogated differences in ATP (Fig. 1F), indicating that LRP130-mediated increases in ATP require an increase in OXPHOS. Although we did not pursue this further, an increase ATP could entail an imbalance of ATP homeostasis (ATP synthesis that exceeds ATP consumption) or an expansion of the adenylate pool due to increased metabolic demand (46). Even so, our results are consistent with prior reports showing that activation of OXPHOS is sufficient to increase ATP (47, 48).
Next, we evaluated mitochondrial function directly by measuring oxygen consumption in isolated mitochondria. Using mitochondria isolated from cells or liver, forced expression of LRP130 increased State 3 respiration, active respiration driven by substrate. We noted an increase of 12 and 15% for cell and liver mitochondria, respectively. Maximally stimulated respiration, achieved by chemically uncoupling mitochondria with carbonyl cyanide p-trifluoromethoxyphenylhydrazone, was increased ∼25% for both cell and liver mitochondria (Tables 1 and 2). Forced expression of LRP130 had no effect on resting respiration (State 4). Within cells, mitochondria exist in a state near State 3, an active respiratory state that likely explains increased respiration observed in cells replete with LRP130. Because equal mitochondrial content was used for each comparison, these studies further signify that LRP130 confers greater oxidative capacity per mitochondrion.
TABLE 1.
Relative respiration in mitochondria isolated from H2.35 cells
Values represent relative means ± S.E. (n = 3). Each sample was measured in triplicate for n = 3 samples (i.e. nine measurements per group). For both groups, the respiratory control ratio (RCR) averaged 1.86 ± 0.04 and was insignificant, indicating similar isolation quality. Outer mitochondrial membrane integrity was greater than 70%. p values were calculated using Student's t test. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; NS, means not statistically significant.
| Respiration | LRP130 vs. LacZ | p value |
|---|---|---|
| State 3 | 1.12 ± 0.02 | <0.01 |
| State 4 | 1.11 ± 0.05 | NS |
| FCCP | 1.26 ± 0.05 | <0.05 |
TABLE 2.
Relative respiration in isolated liver mitochondria
Values represent relative means ± S.E. (n = 3). Each sample was measured in triplicate for n = 3 samples (i.e. nine measurements per group). For both groups, the respiratory control ratio (RCR) averaged 3.56 ± 0.04 and was insignificant, indicating similar isolation quality. Outer mitochondrial membrane integrity was greater than 80%. p values were calculated using Student's t test. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; NS, means not statistically significant.
| Respiration | LRP130 Tg vs. WT | p value |
|---|---|---|
| State 3 | 1.15 ± 0.01 | <0.01 |
| State 4 | 1.02 ± 0.13 | NS |
| FCCP | 1.25 ± 0.16 | <0.05 |
Induction of LRP130 in Liver Increases Ketogenesis and Fatty Acid Oxidation
Based on the aforementioned results, LRP130 increases OXPHOS. In vivo, OXPHOS exerts critical control over several metabolic pathways. Prior studies suggest an increase in OXPHOS may influence metabolic pathways linked to oxidative phosphorylation (49–51). We therefore hypothesized that if LRP130 activates OXPHOS in vivo, it might accelerate oxidative metabolism. For this purpose, we generated transgenic mice that ectopically express LRP130 in liver (supplemental Fig. S3). To query the in vivo consequences of activating OXPHOS, we evaluated ketogenesis/fatty acid oxidation (FAO), metabolic pathways linked to OXPHOS. Redox coenzymes (NAD and FAD) cycle between OXPHOS and FAO/ketogenic pathways, inextricably linking the two. Because ketogenesis is abundant in fasting liver and accounts for 70% of FAO (52), it is a crude yet simple measure of hepatic OXPHOS in vivo (Fig. 2A).
FIGURE 2.
Fatty acid oxidation and ketogenesis are increased by LRP130. A, schematic of β-fatty acid oxidation (β-FAO) and ketogenesis. Rate controlling enzymes are shown in bold. Ketogenesis accounts for 70% of fasting β-FAO. Dependent on oxidized co-enzymes, NAD and FAD, β-FAO in mitochondria is inextricably linked to OXPHOS. In fasting liver, only a small portion of acetyl-CoA enters Krebs cycle for complete oxidation (dotted arrow). B and C, following a 16-h fast, serum β-hydroxybutyrate, the most abundant ketone body in serum, was quantified (n = 8) in wild-type and LRP130 transgenic (LRP130 Tg) mice. B, relationship to time. C, area under the curve (AUC) quantification. D, quantification of fasted serum free fatty acids (FFA) at 16 h (n ≥ 4). E–I, hepatic action of LRP130 is cell autonomous. Isolated primary hepatocytes were transduced with adenovirus, GFP versus LRP130. E, quantification of β-hydroxybutyrate in hepatocyte media (n = 6). F, immunoblot of LRP130 protein in GFP and LRP130-infected hepatocytes. Numbers below the blot indicate quantification of LRP130 protein normalized to loading control, performed in triplicate. G, quantification of [14C]palmitate oxidation (n = 5), measuring acid soluble material (ASM) (mostly ketones and acetyl-CoA); H, CO2 (complete oxidation in Krebs cycles). To facilitate reading of the y axis, scale starts at values >0. I, gene expression of rate controlling enzymes involved in β-FAO and ketogenesis in isolated primary hepatocytes (n = 3). Abbreviations used are as follows: PRC (peroxisome proliferator-activated receptor γ coactivator-related 1), ERRa (estrogen-related receptor α), ACSM (acyl-CoA synthetase medium chain family member), ACSL1 (acyl-CoA synthetase long chain family member 1), CPT-1a (carnitine palmitoyltransferase 1A), MCAD (acyl-CoA dehydrogenase, medium chain), LCAD (acyl-CoA dehydrogenase, long chain), HMGCS2 (3-hydroxy-3-methylglutaryl-coenzyme A synthase 2). Error bars represent S.E. **, p < 0.01. ns = not statistically significant.
In mice transgenic for liver-specific lrp130 (LRP130 Tg), we quantified β-hydroxybutyrate, the most abundant ketone body generated by ketogenesis (Fig. 2, B and C). Animals were of similar weight (supplemental Fig. S3), and their fasting serum glucose was not significantly different (data not shown). Transgenic expression of LRP130 in mouse liver increased serum β-hydroxybutyrate. In contrast, ketogenesis was blunted in mouse liver deficient for LRP130 (supplemental Figs. S4 and S5). Although it is tempting to ascribe these changes to altered OXPHOS, several other variables govern ketogenesis. Key variables include serum free fatty acids, peripheral consumption of ketone bodies, and rate-controlling enzymes (Fig. 2A). We therefore evaluated the effect of ectopic LRP130 on several of these variables.
Serum free fatty acids were unchanged (Fig. 2D). Altered peripheral consumption of ketone bodies seemed less likely, because ectopic expression of LRP130 was limited to liver (data not shown). Moreover, ketogenesis and β-FAO were increased in isolated primary hepatocytes, indicating cell autonomous action of LRP130 (Fig. 2, E–H). An increase in FAO/ketogenesis could be the result of increased mitochondrial enzymes that regulate the FAO/ketogenic axis. To exclude this, we measured the mRNA of rate-controlling enzymes, which are largely regulated at the level of transcription. In primary hepatocytes replete with LRP130, changes in the mRNA of key FAO/ketogenic genes proved insignificant (Fig. 2I). Even in H2.35 cells replete with LRP130, which express LRP130 at a higher level, differences in mRNA of FAO/ketogenic genes were still insignificant (supplemental Fig. S3E). We also measured FAO/ketogenic gene expression in LRP130-deficient hepatocytes. Deficiency of LRP130, which impairs OXPHOS, was accompanied by an increase in PGC-1 co-activators (peroxisome proliferator-activated receptor γ, co-activator 1) and several PGC-1 target genes, although changes in several other genes were insignificant (supplemental Fig. S5D). In sum, these data strongly argue that LRP130 does not directly control FAO/ketogenic gene expression. Of the key variables we investigated none except OXPHOS correlated with increased oxidative metabolism.
Based on this analysis, we conclude LRP130 increases OXPHOS, which likely stimulates FAO/ketogenesis. We cannot exclude an unaccounted variable, which incidentally regulates FAO/ketogenesis. Given known pathways and mechanisms of FAO/ketogenesis, we believe our conclusion is reasonable. In support of our conclusion, studies using isolated perfused liver suggest OXPHOS exerts strong regulatory control over several metabolic pathways (49).
LRP130 Remodels Mitochondria by Increasing Cristae Density
Initial observations indicate that LRP130 increases OXPHOS and that activation of OXPHOS in vivo increases oxidative metabolism. Based on respiration studies in isolated mitochondria, LRP130 increases OXPHOS per mitochondrion (remodeling). These studies however do not take account of mitochondrial biogenesis. Increased mitochondrial number, in addition to greater OXPHOS per mitochondrion, could have jointly increased OXPHOS. We therefore sought to determine the contribution of mitochondrial biogenesis versus mitochondrial remodeling. Using electron microscopy to quantify mitochondrial biogenesis, we observed that LRP130 had no impact on mitochondrial biogenesis (Fig. 3A). If true, then LRP130 functions mainly by remodeling mitochondria, changes that should be apparent at the ultrastructural level. We therefore evaluated cristae density, the ultrastructural basis for OXPHOS, using electron microscopy. Indeed, forced expression of LRP130 increased cristae density (Fig. 3, B and C). Interestingly, the dense pattern of cristae in cells replete with LRP130 resembles those of brown fat mitochondria, which are known to have dense cristae.
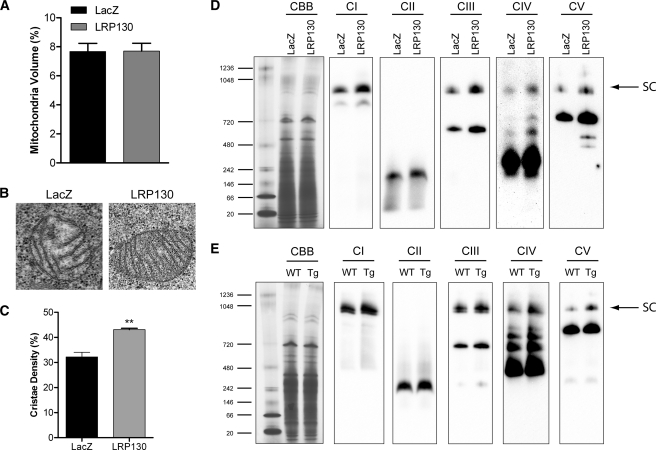
FIGURE 3.
LRP130 remodels mitochondria by increasing cristae density. A, mitochondrial volume, a measure of mitochondrial biogenesis, is not affected by LRP130 (p = ns). B, representative electron micrographs showing that LRP130 increases cristae density. C, quantified from electron micrographs at ×19,000 magnification, cristae density was increased by LRP130. D and E, Coomassie Blue (CBB) staining of Blue Native polyacrylamide gel shows equal protein loading. Immunoblot of Blue Native polyacrylamide gel indicates that mitochondria replete with LRP130 have increased respiratory supercomplexes (SC) designated by the arrow as well as lower molecular weight complexes below the arrow in cells (D) and mouse liver (E). Abbreviations used are as follows: CI (complex I), CII (complex II), CIII (complex III), CIV (complex IV), and CV (complex V). Antibodies used for immunoblotting detect nuclearly encoded subunits: Tg (LRP130 Tg), CI (NDUFA9), CII (70-kDa subunit), CIII (UQCRC2, also called Core2 antigen), CIV (Cox5a), and CV (ATPase α subunit). Images shown are representative of three or more independent experiments. Error bars indicate S.E. ***, p < 0.001
Individual respiratory complexes are assembled into higher order structures termed respiratory supercomplexes (42, 53, 54). Because respiratory supercomplexes constitute the structural and functional underpinning of cristae (54, 55), we evaluated respiratory supercomplexes using Blue Native gel electrophoresis to provide further evidence for remodeling by LRP130. Consistent with increased cristae density, supercomplexes were increased in mitochondria replete with LRP130 whether derived from H2.35 cells or mouse liver (Fig. 3, D and E). Likely because of higher LRP130 expression, greater changes were noted in mitochondria from cultured cells. In contrast, cristae density and supercomplexes were decreased in cells deficient for LRP130 (supplemental Fig. S6), reinforcing the role of LRP130 in mitochondrial remodeling. Of note, depletion of LPR130 also failed to alter mitochondrial biogenesis (supplemental Fig. S6). In conclusion, these data indicate that LRP130 stimulates OXPHOS by promoting denser cristae, independent of mitochondrial biogenesis.
LRP130 Stimulates mtDNA Gene Expression
Our data indicate that LRP130 stimulates OXPHOS by promoting supercomplex formation, which increases cristae density. Intuitively, this observation requires an increase in respiratory subunits. The OXPHOS system is encoded by two genomes. The nucleus encodes ∼75 polypeptides, although mtDNA encodes 13 polypeptides, in addition to 22 tRNAs and 2 ribosomal RNAs (rRNA). Using Affymetrix gene arrays, we queried if LRP130 regulates OXPHOS gene expression. As illustrated in supplemental Fig. S7A, LRP130 failed to increase OXPHOS subunits encoded by nuclear DNA (nDNA), but it did increase respiratory subunits encoded by mtDNA (supplemental Fig. S7B). Conversely, in cells deficient for LRP130, mitochondrially encoded transcripts were decreased (supplemental Fig. S8, A and B), consistent with prior reports (30–33). Perturbation in mitochondrial genome copy number could not explain changes in mtDNA gene expression, because mtDNA copy number was unchanged, irrespective of LRP130 status (supplemental Figs. S7C and S8C). Because mtDNA is also a marker of mitochondrial biogenesis, these data bolster the observation that LRP130 does not influence mitochondrial biogenesis.
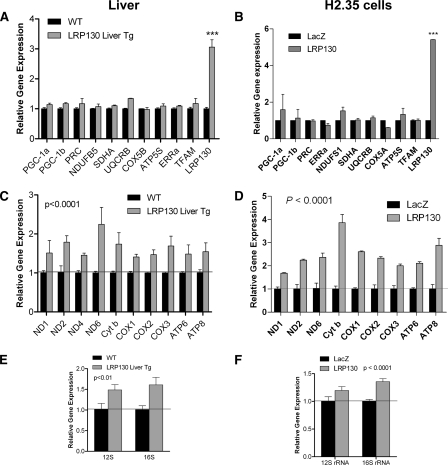
Gene array data were confirmed in cells and mouse liver using RT-qPCR. In addition to several OXPHOS subunits, several key regulators of mitochondrial biogenesis were quantified (Fig. 4 and supplemental Fig. S8). Immunoblotting of a few subunits revealed that protein expression correlated with mRNA levels (supplemental Figs. S6D and S7D). Thus, LRP130 induces the core mitochondrially encoded subunits but fails to regulate nuclearly encoded OXPHOS subunits. As mentioned earlier, mitochondrial DNA also encodes several nonpolypeptide transcripts, including 22 tRNAs and 2 rRNAs. We observed a small increase in rRNA (Fig. 4, E and F), which also was confirmed using in organello labeling (discussed below). Regarding tRNA, cycle numbers were too high, making RT-qPCR unsuitable for quantifying tRNA. tRNA was therefore quantified using in organello labeling, which is discussed below. Parallel loss-of-function studies in mouse liver and cells revealed that LRP130 is required for mitochondrial gene expression (supplemental Fig. S8, F–I), consistent with prior reports in cultured cells.
FIGURE 4.
LRP130 induces mitochondrially encoded gene expression. A and B, RT-qPCR of several nuclearly encoded subunits in mouse liver (A) and cells (B). Two-way analysis of variance with Bonferroni post tests. C and D, RT-qPCR of several mitochondrially encoded genes for mouse liver (C) and cells (D). E and F, RT-qPCR of rRNA in mouse liver (E) and cells (F) (n = 3). Error bars indicate S.E. ***, p < 0.001.
In summary, these data indicate that LRP130 increases gene expression of mitochondrially encoded genes, 13 of which encode OXPHOS subunits. LRP130 does not regulate the remaining ∼75 subunits, encoded by the nucleus. Because subunits encoded by the mitochondrion direct nuclearly encoded subcomplexes from the matrix to mature membrane-bound supercomplexes (56), one consequence of increasing mtDNA gene expression may be more supercomplexes and denser cristae, as we have observed. Given a certain level of nuclearly encoded subunits, these data imply induction of mitochondrially encoded genes is sufficient to increase OXPHOS, which in turn increases the capacity for oxidative metabolism.
LRP130 Activates Mitochondrial Transcription
We sought to understand how LRP130 increases mitochondrial gene expression. In mitochondria, steady-state RNA levels are influenced by several variables, including mtDNA copy number, RNA processing (cleavage), RNA stability, and transcription of mtDNA. As stated earlier, mtDNA copy number was unchanged, irrespective of LRP130 status (supplemental Figs. S7C and S8C). Therefore, we focused our attention on RNA metabolism.
It has been reported that LRP130 regulates transcript stability (32). To assess transcript stability, we treated cells with ethidium bromide, an inhibitor of mitochondrial transcription (supplemental Fig. S9, A and B). Decay rates shown are not identical to transcript half-lives reported for human cells, because mouse cells respond less effectively to ethidium bromide (57, 58). The decay rates shown are not dissimilar from a prior study using mouse cells (57). Irrespective of LRP130 status, mitochondrial transcript decay was unchanged. Without evidence to support a role in RNA stability, we evaluated the effect of LRP130 on mitochondrial transcription.
We monitored de novo transcription in isolated mitochondria, using in organello labeling. In support of transcription, mitochondria replete with LRP130 had increased de novo transcription, following short or long labeling (Fig. 5A). Quantification of total de novo transcripts was consistent with gene expression data, signifying that increases in steady-state gene expression were in part due to increased mitochondrial transcription (Fig. 5B). Indicative of global effects on mitochondrial transcription, rRNA and tRNA were both increased as well. Moreover, an incompletely processed precursor RNA (pc-RNA) was increased, again buttressing evidence in favor of mitochondrial transcription (Fig. 5A). These data also argue against enhanced RNA processing (cleavage), because there was an increase in the precursor RNA. Collectively, these data indicate that LRP130 stimulates mitochondrial transcription.
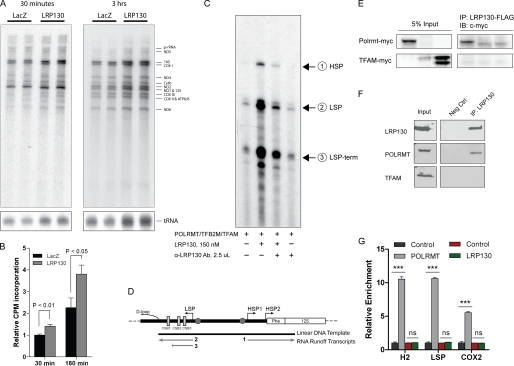
FIGURE 5.
LRP130 is a transcriptional activator of mitochondrial transcription. A, in organello labeling of mitochondria with [32P]UTP for 30 min or 3 h shows increased de novo mitochondrial transcripts for all RNA species in cells replete with LRP130, indicating LRP130 globally stimulates mitochondrial transcription. Image shown is representative of n = 3. B, quantification of de novo transcripts shown in A, n = 3. C, in vitro mitochondrial transcription using purified factors: POLRMT, TFAM, TFB2M, and LRP130 protein. Addition of an LRP130-specific antibody abrogates its stimulatory effect. D, schematic of DNA template and expected in vitro transcripts. E, ectopically expressed proteins, LRP130 and POLRMT, interact. 1st and 3rd lanes contain POLRMT and TFAM, respectively. IP, immunoprecipitate; IB, immunoblot. F, endogenous LRP130 and POLRMT proteins interact. Neg Ctrl, negative control. G, mitochondrial ChIP of POLRMT and LRP130 indicates LRP130 does not directly dock DNA. Representative graph of n = 3–4 experiments performed in triplicate. Designations are as follows: H2 (HSP2 promoter), LSP (light strand promoter), and COX2 (cox2), a polypeptide gene situated ∼8 kb from the promoters. Although not demarcated on the graph, enrichment of POLRMT at either promoter was significant compared with its enrichment at Cox2 (p < 0.001). Two-way analysis of variance with Bonferroni post-tests: ***, p < 0.001, ns = not statistically significant.
Prior studies have demonstrated that increases in mitochondrial ATP can stimulate mitochondrial transcription (59, 60). For in organello labeling studies, we isolated mitochondria from cells replete with LRP130, cells which have increased ATP. Therefore, it is conceivable that in organello labeling studies simply reflect energetically superior mitochondria, not enhanced transcriptional control. We therefore evaluated mitochondrial transcription in vitro, using purified POLRMT, TFAM, and TFB2M, the basal transcription machinery, as well as purified LRP130 protein (supplemental Fig. S10A). LRP130 protein activated in vitro mitochondrial transcription in a dose-dependent manner (supplemental Fig. S10B). As shown in Fig. 5C, activation of mitochondrial transcription by LRP130 proved specific, because its stimulatory action was abrogated by an antibody directed against LRP130 protein. A map of the template used for transcription and the expected products are illustrated in Fig. 5D. In this assay ATP levels are not a confounding variable, and RNA processing is inoperable. We therefore conclude that LRP130 functions as a bona fide activator of mitochondrial transcription, activation we propose increases mitochondrial gene expression in mitochondria.
With convergent evidence supporting mitochondrial transcription, we investigated the mechanism of transcriptional activation in greater detail. The initiation complex consists of POLRMT and TFB2M, which is activated by TFAM. We evaluated if LRP130 complexes with the aforementioned factors. Although ectopically expressed LRP130 failed to co-immunoprecipitate with TFAM, LRP130 did co-immunoprecipitate with POLRMT (Fig. 5E). Implicated in nucleic acid binding, LRP130 and POLRMT might co-immunoprecipitate via tethering to nucleic acid. To exclude this possibility, RNase A and DNase were added to the co-immunoprecipitation reaction. In fact, interactions with POLRMT remained stable even after washing in detergent and high salt (see “Experimental Procedures”). Interaction with POLRMT was also confirmed using endogenous proteins (Fig. 5F). Of note, LRP130 did interact with ectopically expressed TFB2M, however, we could not detect an interaction with endogenous TFB2M (data not shown). Because polyclonal antibodies directed against endogenous proteins may have disrupted an endogenous interaction, we cannot exclude an interaction between TFB2M and LRP130.
Based on the above data, LRP130 activates mitochondrial transcription and complexes with POLRMT, a component of the initiation complex. LRP130 might function as a transcription factor or a transcriptional co-activator. Transcription factors dock DNA directly and activate transcription. In contrast, co-activators do not directly dock DNA, even though they interact with the transcription apparatus and activate transcription (61). Using mitochondrial ChIP assays, we evaluated whether LRP130 directly docks the mitochondrial HSP2 and light strand promoters. Primers designed for the HSP1 promoter did not work; therefore, the HSP1 promoter could not be evaluated. Formaldehyde, a short distance cross-linker, cross-links protein to DNA. Proteins situated far from DNA, like co-activators, cannot be efficiently cross-linked to DNA by formaldehyde. Serving as a control DNA-binding protein, POLRMT was enriched at promoters, validating our experimental conditions (Fig. 5G). Under identical conditions, LRP130 failed to dock mitochondrial DNA promoters, indicating it is situated far from DNA (Fig. 5G). We conclude that LRP130 activates mitochondrial transcription and interacts with the transcriptional machinery, but it does not directly dock mitochondrial DNA promoters. By definition this profile classifies LRP130 as a co-activator of mitochondrial transcription.
In summary, LRP130 is a novel co-activator of mitochondrial transcription that stimulates mitochondrial gene expression, cristae density, and oxidative phosphorylation. Genetic, structural and functional alterations collectively remodel mitochondria, permitting enhanced oxidative metabolism.
DISCUSSION
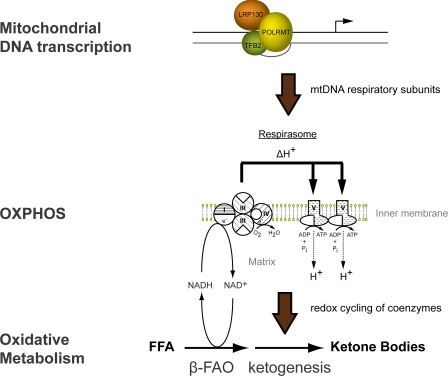
In this report, we conclude LRP130 activates mitochondrial transcription, activation that is coupled with mitochondrial remodeling and enhanced energy metabolism. We propose that mitochondrial transcription induced by LRP130 drives biogenesis of OXPHOS complexes, manifested as increased cristae density and supercomplex formation. With enhanced respiratory capacity, cells and animal liver replete with LRP130 exhibit increased oxidative metabolism, most notably β-fatty acid oxidation and ketogenesis (Fig. 6).
FIGURE 6.
Model of LRP130 action and effect on energy metabolism. LRP130 interacts with POLRMT, a component of the initiation complex, to activate mitochondrial transcription. Polypeptides encoded by mtDNA are increased, promoting supercomplexes and OXPHOS. Metabolic pathways dependent on cycling of coenzymes by the electron transport chain are stimulated by increased OXPHOS. In summary, increases in mitochondrial transcription by LRP130 are associated with remodeling of mitochondria and enhanced oxidative metabolism, notably hepatic FAO and ketogenesis.
LRP130 increases mitochondrially encoded subunits by activating mitochondrial transcription, but it fails to increase nuclearly encoded subunits. Functional OXPHOS complexes, however, require nuclearly encoded subunits. How then does LRP130 increase functional OXPHOS complexes? We propose two mechanisms. One, prior reports indicate post-transcriptional mechanisms regulate many nuclearly encoded subunits. In a comparison of tissues with cultured cells, variability in OXPHOS subunits was limited mainly to mitochondrially encoded genes, changes that correlated with OXPHOS (62). Even in tissues with low expression of nuclearly encoded genes, OXPHOS still proved robust (63), provided mtDNA expression was intact. These reports indicate stoichiometric gene expression (mitochondrial DNA-encoded versus nuclearly encoded subunits), poorly predicts OXPHOS activity, reinforcing the importance of post-transcriptional control of nuclearly encoded subunits (62–65). It can be hypothesized that post-transcriptional control (of nuclearly encoded subunits) paired with transcriptional control of mitochondrial subunits via LRP130 permits functional OXPHOS subunits. In support of this notion, our data show that LRP130 activates mitochondrially encoded gene expression and stimulates OXPHOS.
Two, mitochondrially encoded subunits influence redistribution of nuclearly encoded subcomplexes from the matrix to the inner mitochondrial membrane (56). Many nuclearly encoded subunits form subcomplexes in the matrix (66, 67), and their distribution depends on the level of mitochondrially encoded subunits. Unlike hydrophobic mitochondrially encoded subunits, which are co-translationally inserted into the inner mitochondrial membrane, water-soluble nuclearly encoded subunits assemble as subcomplexes in the matrix. Formation of matrical subcomplexes precedes their incorporation into mature membrane complexes. In cells deficient for ND4 or ND5, which are mitochondrially encoded subunits, or in rho0 cells, which lack all mitochondrially encoded transcripts, subcomplexes composed of nuclearly encoded subunits accumulate in the matrix (56). In cells replete with mitochondrially encoded subunits, however, fewer nuclearly encoded subcomplexes form in the matrix. If true, then LRP130-mediated increases in mitochondrial proteins would obviate the need for increased gene expression of nuclearly encoded subunits by altering the distribution of nuclearly encoded subcomplexes. Although we did not experimentally address this concept, it can be hypothesized that LRP130 increases mitochondrially encoded subunits, which accelerate redistribution of matrical subcomplexes into mature membrane-bound complexes.
The role of mtDNA gene expression in mitochondrial function has largely been inferred from human subjects with point mutations or deletions of mtDNA (3, 68). Similarly, mouse and Drosophila models with reduced mtDNA copy number or deficient mitochondrial transcription have been studied in this regard (22, 69–73). Predictably, attenuated mitochondrial gene expression reduces mitochondrial transcripts and polypeptides, impairing mitochondrial function and physiology. In principle, increases in mtDNA gene expression might increase mitochondrial function and alter physiology. Experimentally, this has proven challenging, because activators of mitochondrial transcription number in few. In cells, TFAM increases mtDNA transcripts (74), but interpretation is confounded by concomitant increases in mtDNA copy number (22). TFB2M and TFB1M cooperate to induce mitochondrial gene expression and mtDNA copy number in cells (75). Functionally, these cells exhibit higher oxygen consumption and increased ATP (76), but this is accompanied by an increase in mtDNA content and mitochondrial biogenesis (75). Although not explored, mitochondrial biogenesis mediated by TFB2M could entail induction of PGC-1 co-activators. Like TFAM, TFB2M and TFB1M present challenges in delineating the effect of mtDNA transcription on mtDNA gene expression from concurrent changes in mitochondrial biogenesis and/or mtDNA copy number. Our results show that LRP130 stimulates mitochondrial transcription independent of mtDNA content or mitochondrial biogenesis. In this regard, LRP130 is unique as an activator of mitochondrial transcription and energy metabolism.
As a proof-of-principle, this study provides evidence that activation of mitochondrial transcription is associated with increased cristae density and enhanced energy metabolism. It is not apparent that isolated increases in mitochondrial transcription mediated via LRP130 occur in nature, a concern that pertains to the regulation of lrp130 mRNA. Previously, we demonstrated that PGC-1s regulate lrp130 gene expression during certain developmental processes (30). Given LRP130 is a mitochondrial protein, this observation is fully anticipated. Even so, PGC-1 co-activators do not entirely predict lrp130 expression. Although Northern blotting of pgc-1 co-activators and lrp130 across various tissues showed strong correlation in heart, muscle, and brown fat; in other tissues (testis, brain, and liver), this correlation was lacking. This could mean regulation of lrp130 in certain tissues is not entirely dependent on PGC-1 co-activator expression, and perhaps in some instances isolated increases of LRP130 are feasible. Even without a complete understanding of lrp130 regulation, this study provides a rationale for pharmacologically targeting LRP130 action and predicts that LRP130 may mitigate metabolic disorders caused by impaired energy metabolism.
The mechanism of LRP130 action has proven controversial. Some studies reported that LRP130 binds to RNA (32, 44, 77), stabilizes Cox I (cytochrome c oxidase I) and Cox III (cytochrome c oxidase III) mtRNA in fibroblasts (35, 78), interacts with SLIRP (SRA stem-loop interacting RNA-binding protein) to stabilize mitochondrial RNAs that encode polypeptides (32), participates in post-transcriptional processing (33), or is involved in transcription termination (34). The single report relating to transcription termination made use of crude lysates, which are unable to distinguish between direct versus indirect control. Those implying binding or RNA processing are supported by the fact that LRP130 contains 21 pentatricopeptide repeat motifs, domains implicated in RNA processing and protein-protein interactions in chloroplasts and mitochondria. Additionally, LRP130 protein shares some homology with yeast protein, PET309, which is implicated in RNA processing and/or Cox1 translation (79, 80). If LRP130 has a major role in RNA stability and/or processing, then the status of LRP130 should dictate RNA decay. We observed no change in RNA stability, and thus, our results are not consistent with this action. It is also possible that LRP130 has different roles in various tissues and/or states of cellular differentiation. Furthermore, our results do not preclude some role in RNA processing directly coupled to mitochondrial transcription. The coupling of transcription and RNA processing is well appreciated in the nucleus and may occur in mammalian mitochondria. In yeast, Nam1p regulates both RNA processing and translation but also binds directly to the mitochondrial RNA polymerase (81, 82), a relationship that suggests tight coordination among mitochondrial transcription, processing, and translation. In particular, mitochondrially encoded Cox transcripts exhibit a notable coordination (82). If LRP130 functions analogously, disruption of transcript processing or translation might attenuate transcription and vice versa, potentially unifying disparate observations made across various studies (32–35) and account for COX-specific defects observed in fibroblasts from patients with Leigh syndrome French Canadian variant (26). The fact that LRP130 interacts with POLRMT and directly stimulates mitochondrial transcription strongly argues that activation of mitochondrial transcription constitutes an important function of LRP130.
In conclusion, we have identified that LRP130 directly activates mitochondrial transcription, which is accompanied by remodeling of mitochondria, increased energy expenditure, and enhanced oxidative metabolism. By elucidating that LRP130 effects changes in energy metabolism, therapeutic targeting of LRP130, and/or other regulators of mitochondrial transcription may mitigate certain metabolic disorders.
Acknowledgments
Core resources were supported by National Institutes of Health Diabetes Endocrinology Research Center Grant DK32520 from NIDDK.
This work was supported, in whole or in part, by National Institutes of Health Grants K08DK071017, R03DK081564, and R01DK089185 (to M. P. C.) and R01HL059655 (to G. S. S.). This work was also supported by a James Hudson Brown-Alexander Brown Coxe Fellowship (to Y. V. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S10, Table 1, “Experimental Procedures,” and additional references.
- OXPHOS
- oxidative phosphorylation
- POLMRT
- mitochondrial RNA polymerase
- TFAM
- mitochondrial transcription factor A
- FAO
- fatty acid oxidation
- qPCR
- quantitative PCR.
REFERENCES
- 1. Luft R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 8731–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rimoin D. L., Connor J. M., Pyeritz R. E., Korf B. R. (eds) (2006) in Principles and Practice of Medical Genetics, 5th Ed., Churchill Livingstone, New York [Google Scholar]
- 3. Wallace D. C., Fan W. (2010) Mitochondrion 10, 12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., Young I. G. (1981) Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 5. Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. (1981) Cell 26, 167–180 [DOI] [PubMed] [Google Scholar]
- 6. Falkenberg M., Larsson N. G., Gustafsson C. M. (2007) Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 7. Shadel G. S. (2008) Am. J. Pathol. 172, 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wallace D. C., Fan W. (2009) Genes Dev. 23, 1714–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Allen J. F. (1993) J. Theor. Biol. 165, 609–631 [DOI] [PubMed] [Google Scholar]
- 10. Tiranti V., Savoia A., Forti F., D'Apolito M. F., Centra M., Rocchi M., Zeviani M. (1997) Hum. Mol. Genet. 6, 615–625 [DOI] [PubMed] [Google Scholar]
- 11. Masters B. S., Stohl L. L., Clayton D. A. (1987) Cell 51, 89–99 [DOI] [PubMed] [Google Scholar]
- 12. Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 13. Shutt T. E., Lodeiro M. F., Cotney J., Cameron C. E., Shadel G. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12133–12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher R. P., Clayton D. A. (1985) J. Biol. Chem. 260, 11330–11338 [PubMed] [Google Scholar]
- 15. Fisher R. P., Clayton D. A. (1988) Mol. Cell. Biol. 8, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parisi M. A., Clayton D. A. (1991) Science 252, 965–969 [DOI] [PubMed] [Google Scholar]
- 17. Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 18. Gaspari M., Falkenberg M., Larsson N. G., Gustafsson C. M. (2004) EMBO J. 23, 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dairaghi D. J., Shadel G. S., Clayton D. A. (1995) J. Mol. Biol. 249, 11–28 [DOI] [PubMed] [Google Scholar]
- 20. McCulloch V., Shadel G. S. (2003) Mol. Cell. Biol. 23, 5816–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. (1992) J. Biol. Chem. 267, 3358–3367 [PubMed] [Google Scholar]
- 22. Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., Hultenby K., Rustin P., Gustafsson C. M., Larsson N. G. (2004) Hum. Mol. Genet. 13, 935–944 [DOI] [PubMed] [Google Scholar]
- 23. Kanki T., Ohgaki K., Gaspari M., Gustafsson C. M., Fukuoh A., Sasaki N., Hamasaki N., Kang D. (2004) Mol. Cell. Biol. 24, 9823–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufman B. A., Durisic N., Mativetsky J. M., Costantino S., Hancock M. A., Grutter P., Shoubridge E. A. (2007) Mol. Biol. Cell 18, 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. (2009) Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morin C., Mitchell G., Larochelle J., Lambert M., Ogier H., Robinson B. H., De Braekeleer M. (1993) Am. J. Hum. Genet. 53, 488–496 [PMC free article] [PubMed] [Google Scholar]
- 27. Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Mitchell G. A., Morin C., Mann M., Hudson T. J., Robinson B., Rioux J. D., Lander E. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Labialle S., Dayan G., Gayet L., Rigal D., Gambrelle J., Baggetto L. G. (2004) Nucleic Acids Res. 32, 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Topisirovic I., Siddiqui N., Lapointe V. L., Trost M., Thibault P., Bangeranye C., Piñol-Roma S., Borden K. L. (2009) EMBO J. 28, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper M. P., Uldry M., Kajimura S., Arany Z., Spiegelman B. M. (2008) J. Biol. Chem. 283, 31960–31967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cooper M. P., Qu L., Rohas L. M., Lin J., Yang W., Erdjument-Bromage H., Tempst P., Spiegelman B. M. (2006) Genes Dev. 20, 2996–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E. A., Consortium L. (2010) Mol. Biol. Cell 21, 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gohil V. M., Nilsson R., Belcher-Timme C. A., Luo B., Root D. E., Mootha V. K. (2010) J. Biol. Chem. 285, 13742–13747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sondheimer N., Fang J. K., Polyak E., Falk M. J., Avadhani N. G. (2010) Biochemistry 49, 7467–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu F., Morin C., Mitchell G., Ackerley C., Robinson B. H. (2004) Biochem. J. 382, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Enríquez J. A., Fernández-Silva P., Pérez-Martos A., López-Pérez M. J., Montoya J. (1996) Eur. J. Biochem. 237, 601–610 [DOI] [PubMed] [Google Scholar]
- 37. Frezza C., Cipolat S., Scorrano L. (2007) Nat. Protoc. 2, 287–295 [DOI] [PubMed] [Google Scholar]
- 38. Spurway T. D., Sherratt H. A., Pogson C. I., Agius L. (1997) Biochem. J. 323, 119–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilcoxon F. (1946) J. Econ. Entomol. 39, 269 [DOI] [PubMed] [Google Scholar]
- 40. Wu H., Wade M., Krall L., Grisham J., Xiong Y., Van Dyke T. (1996) Genes Dev. 10, 245–260 [DOI] [PubMed] [Google Scholar]
- 41. Barrientos A., Fontanesi F., Diaz F. (2009) Curr. Protoc. Hum. Genet. Chapter 19, Unit 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., Enriquez J. A. (2008) Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 43. Kucej M., Kucejova B., Subramanian R., Chen X. J., Butow R. A. (2008) J. Cell Sci. 121, 1861–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mili S., Piñol-Roma S. (2003) Mol. Cell. Biol. 23, 4972–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sterky F. H., Ruzzenente B., Gustafsson C. M., Samuelsson T., Larsson N. G. (2010) Biochem. Biophys. Res. Commun. 398, 759–764 [DOI] [PubMed] [Google Scholar]
- 46. Ataullakhanov F. I., Vitvitsky V. M. (2002) Biosci. Rep. 22, 501–511 [DOI] [PubMed] [Google Scholar]
- 47. Liang H., Bai Y., Li Y., Richardson A., Ward W. F. (2007) Ann. N.Y. Acad. Sci. 1100, 264–279 [DOI] [PubMed] [Google Scholar]
- 48. Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49. Soboll S., Oh M. H., Brown G. C. (1998) Eur. J. Biochem. 254, 194–201 [DOI] [PubMed] [Google Scholar]
- 50. Hafner R. P., Brown G. C., Brand M. D. (1990) Eur. J. Biochem. 188, 313–319 [DOI] [PubMed] [Google Scholar]
- 51. Ainscow E. K., Brand M. D. (1999) Eur. J. Biochem. 266, 737–749 [DOI] [PubMed] [Google Scholar]
- 52. Sunny N. E., Satapati S., Fu X., He T., Mehdibeigi R., Spring-Robinson C., Duarte J., Potthoff M. J., Browning J. D., Burgess S. C. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E1226–E1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schäfer E., Seelert H., Reifschneider N. H., Krause F., Dencher N. A., Vonck J. (2006) J. Biol. Chem. 281, 15370–15375 [DOI] [PubMed] [Google Scholar]
- 54. Vonck J., Schäfer E. (2009) Biochim. Biophys. Acta 1793, 117–124 [DOI] [PubMed] [Google Scholar]
- 55. Minauro-Sanmiguel F., Wilkens S., García J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12356–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bourges I., Ramus C., Mousson de Camaret B., Beugnot R., Remacle C., Cardol P., Hofhaus G., Issartel J. P. (2004) Biochem. J. 383, 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hayashi J., Tanaka M., Sato W., Ozawa T., Yonekawa H., Kagawa Y., Ohta S. (1990) Biochem. Biophys. Res. Commun. 167, 216–221 [DOI] [PubMed] [Google Scholar]
- 58. Inoue K., Takai D., Hosaka H., Ito S., Shitara H., Isobe K., LePecq J. B., Segal-Bendirdjian E., Hayashi J. (1997) Biochem. Biophys. Res. Commun. 239, 257–260 [DOI] [PubMed] [Google Scholar]
- 59. Amiott E. A., Jaehning J. A. (2006) Mol. Cell 22, 329–338 [DOI] [PubMed] [Google Scholar]
- 60. das Neves R. P., Jones N. S., Andreu L., Gupta R., Enver T., Iborra F. J. (2010) PLoS Biol. 8, e1000560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Näär A. M., Lemon B. D., Tjian R. (2001) Annu. Rev. Biochem. 70, 475–501 [DOI] [PubMed] [Google Scholar]
- 62. Duborjal H., Beugnot R., Mousson de Camaret B., Issartel J. P. (2002) Genome Res. 12, 1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Duggan A. T., Kocha K. M., Monk C. T., Bremer K., Moyes C. D. (2011) J. Exp. Biol. 214, 1880–1887 [DOI] [PubMed] [Google Scholar]
- 64. Giegé P., Sweetlove L. J., Cognat V., Leaver C. J. (2005) Plant Cell 17, 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Garbian Y., Ovadia O., Dadon S., Mishmar D. (2010) PLoS One 5, e9985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Antonicka H., Ogilvie I., Taivassalo T., Anitori R. P., Haller R. G., Vissing J., Kennaway N. G., Shoubridge E. A. (2003) J. Biol. Chem. 278, 43081–43088 [DOI] [PubMed] [Google Scholar]
- 67. Lenaz G., Genova M. L. (2010) Antioxid. Redox. Signal. 12, 961–1008 [DOI] [PubMed] [Google Scholar]
- 68. Trifunovic A., Hansson A., Wredenberg A., Rovio A. T., Dufour E., Khvorostov I., Spelbrink J. N., Wibom R., Jacobs H. T., Larsson N. G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17993–17998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ikeuchi M., Matsusaka H., Kang D., Matsushima S., Ide T., Kubota T., Fujiwara T., Hamasaki N., Takeshita A., Sunagawa K., Tsutsui H. (2005) Circulation 112, 683–690 [DOI] [PubMed] [Google Scholar]
- 70. Park C. B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C. M., Larsson N. G. (2007) Cell 130, 273–285 [DOI] [PubMed] [Google Scholar]
- 71. Martínez-Azorín F., Calleja M., Hernández-Sierra R., Farr C. L., Kaguni L. S., Garesse R. (2008) J. Neurochem. 105, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wenz T., Luca C., Torraco A., Moraes C. T. (2009) Cell Metab. 9, 499–511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 73. Hokari M., Kuroda S., Kinugawa S., Ide T., Tsutsui H., Iwasaki Y. (2010) Neuropathology 30, 401–407 [DOI] [PubMed] [Google Scholar]
- 74. Montoya J., Perez-Martos A., Garstka H. L., Wiesner R. J. (1997) Mol. Cell. Biochem. 174, 227–230 [PubMed] [Google Scholar]
- 75. Cotney J., Wang Z., Shadel G. S. (2007) Nucleic Acids Res. 35, 4042–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cotney J., McKay S. E., Shadel G. S. (2009) Hum. Mol. Genet. 18, 2670–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tsuchiya N., Fukuda H., Nakashima K., Nagao M., Sugimura T., Nakagama H. (2004) Biochem. Biophys. Res. Commun. 317, 736–743 [DOI] [PubMed] [Google Scholar]
- 78. Xu F., Addis J. B., Cameron J. M., Robinson B. H. (2011) Biochem. J. in press [Google Scholar]
- 79. Manthey G. M., McEwen J. E. (1995) EMBO J. 14, 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tavares-Carreón F., Camacho-Villasana Y., Zamudio-Ochoa A., Shingú-Vázquez M., Torres-Larios A., Pérez-Martínez X. (2008) J. Biol. Chem. 283, 1472–1479 [DOI] [PubMed] [Google Scholar]
- 81. Rodeheffer M. S., Boone B. E., Bryan A. C., Shadel G. S. (2001) J. Biol. Chem. 276, 8616–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Naithani S., Saracco S. A., Butler C. A., Fox T. D. (2003) Mol. Biol. Cell 14, 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]