Background: The mechanism by which 3-phosphoinositide-dependent kinase-1 (PDK1) is recruited to the plasma membrane is not fully understood.
Results: The PDK1 PH domain specifically binds phosphatidylserine, and the disruption of this binding abrogates membrane localization and signaling function of PDK1.
Conclusion: Phosphatidylserine binding is important for the membrane recruitment and signaling function of PDK1.
Significance: This study establishes an important signaling role of phosphatidylserine.
Keywords: Membrane Lipids, Phosphatidylinositol-dependent Kinase-1 (PDK1), Phosphatidylserine, PI 3-Kinase (PI3K), Plasma Membrane, PH Domain
Abstract
3-Phosphoinositide-dependent kinase-1 (PDK1) is a ubiquitously expressed serine/threonine kinase that functions downstream of phosphoinositide 3-kinase. Although binding of 3′-phosphoinositides, phosphatidylinositol 3,4,5-trisphosphate and phosphatidylinositol 3,4-bisphosphate, to the pleckstrin homology (PH) domain of PDK1 is known to be essential for its interaction with and activation of downstream kinases, the mechanism by which PDK1 is recruited to the plasma membrane remains controversial. Our surface plasmon resonance analysis of the PDK1 PH domain and selected mutants shows that the PH domain specifically binds phosphatidylserine using a site that is separate from the canonical phosphoinositide-binding site. Further cell studies show that this specific phosphatidylserine binding is important for the plasma membrane localization and signaling function of PDK1.
Introduction
The activation of phosphoinositide 3-kinase (PI3K) in response to growth factors and insulin stimulation leads to the generation of 3-phosphoinositides, phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3)2 and phosphatidylinositol 3,4-bisphosphate (PtdIns(3,4)P2), that act as second messengers triggering diverse metabolic, proliferative, and survival responses (1, 2). The 3-phosphoinositide-dependent kinase-1 (PDK1) is known as a master regulator of a subgroup of 3-phosphoinositide-responsive AGC protein kinase family members, including protein kinase B (PKB/Akt), p70 ribosomal S6 kinase, serum- and glucocorticoid-induced protein kinase, and atypical protein kinase C (PKC) (3, 4). Unlike other AGC kinases, PDK1 is found constitutively active in the cell (5) due to its intermolecular autophosphorylation activity (6). As a result, the signaling activity of PDK1 is thought to be regulated through agonist-induced transformation of downstream kinases into substrates for PDK1 (7). For example, in the PI3K-PDK1-Akt pathway, a transient increase in PtdIns(3,4,5)P3 and PtdIns(3,4)P2 in the plasma membrane (PM) by PI3K activation recruits Akt to the PM from the cytosol, where it can be phosphorylated at Thr308 by PDK1 (7). Akt and PDK1 both contain a pleckstrin homology (PH) domain capable of binding PtdIns(3,4,5)P3 and PtdIns(3,4)P2. It has been reported that binding of PtdIns(3,4,5)P3 to their PH domains promotes colocalization of the two proteins at the PM (5). For Akt, it also induces a conformational change of the protein, exposing Thr308 located in the T-loop (8) for phosphorylation by PDK1 (9, 10).
PtdIns(3,4,5)P3 is known to recruit a multitude of effectors, including Akt, to the PM. However, whether or not PtdIns(3,4,5)P3 recruits PDK1 to the PM remains controversial. Two studies reported that PDK1 translocated to the PM in response to PI3K activation (11, 12). However, another study found that a significant portion of the protein prelocalized at the PM and that growth factor stimulation had no effect on its localization (7). Also, Akt could be activated in cells expressing a PDK1 mutant, the PH domain of which lacks phosphoinositide binding activity (13). Collectively, these results suggest the presence of another factor besides 3′-phosphoinositides that is important for PM recruitment of PDK1.
Many cytosolic proteins are recruited to the PM by directly interacting with lipids present in the PM even when they have specific interaction partner proteins in the PM (14). The inner leaflet of the PM of mammalian cells is rich in anionic phospholipids, particularly phosphatidylserine (PS) (20–30 mol %) (15–17) and phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) (∼1 mol %) (18–21). Although much has been reported on the role of phosphoinositides, including PtdIns(4,5)P2, in PM recruitment of cytosolic proteins (19, 21), less has been known about the direct involvement of PS in their PM localization. Most lipid binding domains and proteins have cationic residues or patches on their membrane contact surfaces, and nonspecific electrostatic interactions between these residues and anionic lipids in cell membranes contribute significantly to the overall membrane binding of these proteins (22, 23). Because PS is the most abundant bulk anionic lipid in the PM, it is generally thought that PS is involved in this type of nonspecific interaction with cellular proteins. However, earlier in vitro membrane binding and cellular translocation studies of various proteins, including PKC (24) and sphingosine kinase (25), as well as their isolated lipid binding domains (26) have indicated that they are targeted to the PM through direct and specific interactions with PS in the PM. More recently, Yeung et al. (17) reported that PS binding is important for PM targeting of cytosolic proteins with polybasic motifs, including small G proteins. For the C2 domain of PKCα that can bind both PS (26, 27) and phosphoinositides (28, 29), it was shown that PS binding is essential for its PM recruitment, whereas phosphoinositide binding augments the PS-dependent membrane binding (30).
It has been reported that PS enhances the binding of PH domains to phosphoinositide-containing membranes. A single molecule study of the Grp1 PH domain showed that PS bound to an unidentified secondary binding site of the PH domain greatly enhanced its affinity for PtdIns(3,4,5)P3-containing vesicles and changed the diffusion behavior of the membrane-bound protein (31). More recently, binding of PS to basic residues near the PtdIns(3,4,5)P3-binding pocket of its PH domain was reported to be important for the activation of Akt (32). Here, we report that the PH domain of PDK1 specifically binds PS via a well defined site that is separate from its PtdIns(3,4,5)P3-binding pocket, and this specific PS binding is essential for its PM localization and signaling function in response to physiological stimuli.
EXPERIMENTAL PROCEDURES
Materials
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). PtdIns(3,4)P2, phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2), and PtdIns(3,4,5)P3 were purchased from Cayman. The concentrations of the phospholipids were determined by a modified Bartlett analysis. Fatty acid-free bovine serum albumin was from Bayer, Inc. (Kankakee, IL).
Expression Vector Construction and Mutagenesis
A construct of PDK1-PH (residues 409–556) was generated from murine cDNA using PCR. The amplified PCR product was subcloned as a BglII and EcoRI fragment into pGEX4T-1 (Novagen) vector at the BamHI and EcoRI restriction site. The PDK1 mutants, K465A, R466A, K467A, R466A/K467A, as well as corresponding Glu mutants were produced using overlap PCR. The plasmids were transformed into Top10F′ cells for high throughput DNA isolation, which was sequenced for verification. For cellular imaging work with NIH 3T3 cells, wild type and mutants were subcloned again as a BglII and EcoRI fragment between the EcoRI and BamHI site of pEGFP-C1 and mCherry-C1 vectors to generate PH domains tagged with an enhanced green fluorescence protein (EGFP) and a red fluorescence protein (mCherry), respectively, at their N termini. The N-terminal mCherry-tagged lactadherin C2 (Lact-C2) was prepared from the EGFP-tagged Lact-C2 (Addgene). mCerulean-tagged Btk-PH was prepared as described (33).
Protein Expression and Purification
The GST fusion proteins were expressed in BL21 RIL cells. For expression of the PDK1-PH wild type (WT) and mutants, 2 liters of Luria broth containing 100 μg/ml ampicillin were inoculated with BL21 RIL colonies containing the construct. Cells were allowed to grow in medium at 37 °C until an absorbance of 0.6 at 600 nm was reached. Protein expression was induced with the addition of 200 μm isopropyl 1-thio-β-d-galactopyranoside (Research Products, Mount Prospect, IL), at which point cells were moved to a 25 °C shaker for 14-h incubation. Cells were harvested through centrifugation (2500 × g for 10 min at 4 °C), and the pellet was resuspended in 20 ml of 20 mm Tris buffer, pH 8, with 160 mm KCl, 50 μm phenylmethylsulfonyl fluoride, and 2 mm dithiothreitol. The solution was sonicated for 8 min (30 s of sonication followed by a 30-s pause) and then centrifuged for 30 min (39,000 × g at 4 °C). When centrifugation was complete, the supernatant was filtered into a 50-ml Falcon tube and 500 μl of glutathione S-transferase tag resin was added (Novagen, Madison, WI). The supernatant was allowed to incubate with the resin for ∼30 min at 4 °C with moderate shaking. The supernatant was then poured onto a column, and the resin was washed with 20 ml of 20 mm Tris buffer, pH 8, 160 mm KCl to eliminate nonspecifically bound protein. After washing, the resin was resuspended in 1 ml of 20 mm Tris buffer, pH 8.4, containing 160 mm KCl, 25 mm CaCl2, and 1 μl of thrombin added to cleave the glutathione S-transferase tag, and the column was sealed and incubated at 4 °C for 12 h. The protein was eluted with three fractions of 1 ml of 20 mm Tris buffer, pH 8, 160 mm KCl. Protein purity was checked using a 18% polyacrylamide gel, and the protein concentration was determined using the bicinchoninic acid method. Protein was frozen in liquid nitrogen and stored at −80 °C.
SPR Measurements
All SPR measurements were performed at 23 °C using a lipid-coated L1 chip in a Biacore X system as described elsewhere (34). In brief, after washing the sensor chip with running buffer (20 mm Tris, pH 7.4, 160 mm KCl), 20 μl of lipid vesicles at a given composition (e.g. POPC/POPS = 8:2) were injected at 5 μl/min to give a response of ∼4000 resonance units. Similarly, a control surface was prepared by injecting 100% POPC onto a chip in a separate channel. The lipid layer was washed several times with 90 μl of 50 mm NaOH until the change in resonance units after each wash was less than 10. Once the signal was stabilized, equilibrium measurements were done at a flow rate of 5 μl/min. This allowed enough time for the R values of the association phase to reach near equilibrium levels (Req). A minimum of five different protein concentrations were injected to collect data for Kd determination. The Req values were plotted against the protein concentrations (Po), and the Kd was established by nonlinear least squares analysis of the binding isotherm using the equation, Req = Rmax/(1 + Kd/Po). The measurement was repeated at least three times to determine average and S.D. values. For kinetic measurements, the flow rate was changed to 30 μl/min.
Cell Studies, Microscopic Imaging, and Data Analysis
NIH 3T3 cells were seeded into eight wells of a sterile Nunc Lak-TeKIITM chambered coverglass plate, which was filled with 400 μl of Dulbecco's modified Eagle's medium (DMEM) and 10% (v/v) fetal bovine serum and incubated at 37 °C in a 5% CO2 environment for 24 h. For transfection, NIH 3T3 cells were incubated for 4 h with EGFP-C1 vector containing each PH domain (0.5 μg/ml) as well as mCerulean-C1 vector containing Btk-PH (1.0 μg/ml) in the presence of Lipofectamine reagent 2000 in DMEM. Cells were incubated overnight in DMEM with 10% fetal bovine serum, washed with DMEM, and then incubated in DMEM without serum for another 4 h. Prior to imaging, cells were washed twice with Hanks' balanced salt solutions. Subcellular localization of PDK1-PH WT and mutants was monitored at fixed time intervals (30 s) before and after stimulation with 50 ng/ml human platelet-derived growth factor (PDGF)-BB using a custom-built, two-photon microscope (24). For comparison, subcellular localization of a PtdIns(3,4,5)P3 sensor, mCerulean-tagged Btk-PH (33), was monitored under the same conditions. A minimum of three experiments were done for all reported results, with >80% of cells showing the same result as shown in each figure. All microscopic manipulation and data acquisition was controlled by the SimFCS program provided by Dr. Enrico Gratton. To calculate the time course of the degree of membrane localization of different proteins, cell images taken at regular intervals were analyzed using MATLAB. Lines were drawn in three places, crossing the PM and the cytosol, and photon count values were obtained along points on the lines. The values within the cytosol as well as at the PM were averaged. The relative PM distribution of a protein at a given time was then calculated by dividing the photon count average at PM by that at PM + cytosol.
Akt Activity Assay
NIH-3T3 cells were first differentiated as described previously (35, 36) and then co-transfected with Akt1 and PDK1 WT (or PDK1-R466A/K467A mutant). After a 16-h incubation to allow sufficient overexpression of proteins, cells were starved for a minimum of 3 h before they were stimulated with PDGF-BB (50 ng/ml) for 0, 10, or 20 min. Cell lysates were prepared using M-PER mammalian protein extraction reagent (Thermo Scientific) along with protease (Roche Applied Science) and phosphatase (Sigma-Aldrich) inhibitors, extracts were analyzed by SDS-PAGE, and the gel was transferred to polyvinylidene fluoride membranes (Millipore). Membranes were then incubated in blocking buffer TBS-T (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.05% Tween 20) with 5% (w/v) dry milk for 3 h. Immunoblots were washed, and probed with the following antibody: anti-Akt1, anti-Akt1-Thr(P)308, or anti-PDK1 at a 1:1000 ratio for each. Membranes were incubated with antibodies overnight and then washed with TBS-T with 1% milk a minimum of five times for 5 min. Blots were then incubated with anti-goat/anti-rabbit IgG secondary antibody for 2 h, washed five times, and prepared for development. Protein immunoblots were revealed using horseradish peroxidase. Intensities of protein bands were quantified using ImageJ software. The relative phosphorylation index was calculated by dividing the intensity of an Akt-Thr(P)308 band by that of its protein gel band, and this parameter was plotted as a function of time for both WT and the mutant.
Computer Modeling
A glycerol molecule in the crystal structure PDK1-PH (Protein Data Bank code 1W1D) (37) was replaced with a PS molecule using AutoDock (version 4.0), and the energy minimization was performed using the Lamarckian genetic algorithm. Among models with the lowest mean energy, the most plausible model was selected by considering the feasibility of embedding the hydrophobic tail of the PS molecule in the membrane.
RESULTS
Membrane Binding Properties of the PDK1 PH Domain
It was reported that PDK1-PH had high affinity for PtdIns(3,4,5)P3 and PtdIns(3,4)P2 and modest affinity for PtdIns(4,5)P2 (7). To explore the possibility that it binds other lipids, PS in particular, we reexamined the anionic lipid selectivity of PDK1-PH by kinetic SPR analysis. As shown in Fig. 1A, PDK1-PH has much higher affinity for POPC/PtdIns(3,4,5)P3 (97:3) and POPC/PtdIns(3,4)P2 (97:3) vesicles than for POPC/PtdIns(4,5)P2 (97:3) vesicles and vesicles containing other phosphoinositides. It has no detectable binding to pure POPC vesicles. Interestingly, PDK1-PH has high affinity for POPC/POPS (7:3) vesicles, which is comparable with its affinity for POPC/PtdIns(3,4,5)P3 (97:3) vesicles. Furthermore, its binding to POPC/POPS ((100 − x):x, where x = 0–30 mol %) vesicles shows a strong dependence on PS concentration (Fig. 1B). To determine whether this PS effect is due to specific binding to PS or nonspecific electrostatic interactions, we also measured the binding of PDK1-PH to POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (8:2) and POPC/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoinositol (8:2) vesicles. As shown in Fig. 1B, PDK1-PH had much lower affinity for these vesicles than POPC/POPS (8:2) vesicles, indicating that PDK1-PH specifically binds PS. To see whether PDK1-PH specifically binds PS using the PtdIns(3,4,5)P3-binding pocket or a separate site, we then measured the effect of adding PtdIns(3,4,5)P3 to POPC/POPS (8:2) vesicles on membrane affinity of PDK1-PH. Fig. 1C shows that PtdIns(3,4,5)P3 significantly enhances the affinity of PDK1-PH for POPC/POPS (8:2) vesicles in a concentration-dependent manner. Conversely, the addition of POPS to POPC/PtdIns(3,4,5)P3 (97:3) vesicles also increased the affinity of PDK1-PH (Fig. 1D). In both cases (i.e. Fig. 1, C and D), the addition of a second lipid, PS or PtdIns(3,4,5)P3, enhanced the membrane affinity of PDK1-PH, as expected from their individual effects (e.g. Fig. 1, compare B and D). These results indicate that PS and PtdIns(3,4,5)P3 bind to separate binding sites in PDK1-PH, and they work additively rather than synergistically in promoting membrane binding of PDK1-PH.
FIGURE 1.
Lipid selectivity of the PDK1 PH domain. A, phosphoinositide specificity of PDK1-PH. Kinetic sensorgrams show that it has the highest affinity for POPC/PtdIns(3,4,5)P3 (97:3) and POPC/PtdIns(3,4)P2 (97:3) vesicles while having much lower affinity for POPC/PtdIns(4,5)P2 (97:3) vesicles. It did not show any appreciable binding to vesicles containing other phosphoinositides. Note that PDK1-PH also has high affinity for POPC/POPS (7:3) vesicles. Protein concentration was 0.2 μm. B, PS-dependent membrane binding of PDK1-PH. PS concentration in POPC/POPS vesicles was varied from 0 to 30 mol %. Note that PDK1-PH has much lower affinity for vesicles containing 20% phosphatidylinositol (PI) or phosphatidylglycerol (PG) than for those containing 20% PS. Protein concentration was 0.2 μm. C, interplay of PS and PtdIns(3,4,5)P3 in membrane binding of PDK1-PH. Kinetic sensorgrams show binding to POPC/POPS/PtdIns(3,4,5)P3 ((80 − x):20:x, where x = 0–3 mol %) vesicles with fixed PS concentration and increasing PtdIns(3,4,5)P3 concentration. Protein concentration was 0.2 μm. D, interplay of PS and PtdIns(3,4,5)P3 in membrane binding of PDK1-PH. Kinetic sensorgrams show binding to POPC/POPS/PtdIns(3,4,5)P3 ((80 − x):x:3, where x = 0–30 mol %) vesicles with fixed PtdIns(3,4,5)P3 concentration and increasing PS concentration. Protein concentration was 0.2 μm. E, determination of Kd for binding of PDK1-PH to POPC/PtdIns(3,4,5)P3 (97:3) vesicles. PDK1-PH was injected at 5 μl/min at varying concentrations (0–200 nm) over the POPC/PtdIns(3,4,5)P3 (97:3) surface, and Req values were measured. A binding isotherm was generated from the Req (average of triplicate measurements) versus the concentration of PDK1-PH plot. A solid line represents a theoretical curve constructed from Rmax (= 1200 ± 40) and Kd (= 52 ± 4 nm) values determined by nonlinear least-squares analysis of the isotherm using the equation, Req = Rmax/(1 + Kd/C). All measurements were performed at 23 °C in 20 mm Tris-HCl buffer, pH 7.4, with 0.16 m KCl. Only association phases of sensorgrams were shown for clarity in A–D.
To quantitatively determine the lipid specificity of PDK1-PH, we performed equilibrium SPR measurements and determined Kd values for its binding to vesicles containing different anionic lipids (see Table 1). Consistent with kinetic SPR data shown in Fig. 1A, PDK1-PH has high affinity for POPC/PtdIns(3,4,5)P3 (97:3) (Kd = 52 nm) (see Fig. 1E) and POPC/PtdIns(3,4)P2 (97:3) vesicles (Kd = 70 nm) (data not shown). This affinity is comparable with that reported for other PtdIns(3,4,5)P3-binding PH domains (33). Kd for POPC/POPS (8:2) vesicles is 95 nm, confirming that PDK1-PH also has high affinity for PS-containing vesicles. The addition of 3 mol % PtdIns(3,4,5)P3 to POPC/POPS (8:2) vesicles causes a modest 3-fold increase in the membrane affinity of PDK1-PH, corroborating the additive effect of the two lipids in driving membrane binding of PDK1-PH.
TABLE 1.
Membrane binding properties of the PDK1 PH domain WT and mutants
Equilibrium SPR experiments were performed in 20 mm Tris-HCl, pH 7.4, containing 0.16 m KCl, and Kd values were determined as shown in Fig. 1E.
| PDK1 PH domains |
Kd |
||
|---|---|---|---|
| POPC/PtdIns(3,4,5)P3 (97:3) | POPC/POPS (80:20) | POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) | |
| nm | |||
| WT | 52 ± 4 | 95 ± 17 | 30 ± 5 |
| K465A | 1700 ± 160 | 160 ± 50 | 150 ± 25 |
| R466A | 58 ± 9 | 390 ± 180 | 65 ± 20 |
| K467A | 59 ± 8 | 200 ± 25 | 65 ± 10 |
| R466A/K467A | 50 ± 15 | 560 ± 200 | 220 ± 20 |
Identification of the PS-binding Site of the PDK1 PH Domain
To help determine the location of a PS-specific binding site in the PDK1-PH, we examined the crystal structure (37) and the surface electrostatic distribution of PDK1-PH and searched for cationic grooves on or near the putative membrane binding surface surrounding the PtdIns(3,4,5)P3-binding pocket. Interestingly, we identified near the PtdIns(3,4,5)P3-binding pocket a cationic groove comprising Arg466 and Lys467. In the crystal structure, these residues coordinate a glycerol molecule (Fig. 2A). This suggested that the two basic residues may be directly involved in PS binding. Molecular modeling also suggested that the cationic groove could accommodate a PS headgroup (Fig. 2B). We thus mutated these residues to Ala individually or in combination and measured the effects on binding to vesicles with various compositions. We also mutated Lys465, which is known to directly interact with the PtdIns(3,4,5)P3 headgroup (see Fig. 2A) (37). Results are summarized in Table 1.
FIGURE 2.
Identification of the PS-binding site of the PDK1 PH domain. A, the crystal structure of PDK1-PH complexed with inositol 1,3,4,5-tetrakisphsphate (IP4) and glycerol (Protein Data Bank code 1W1D) (37). The PDK1-PH structure is shown in a ribbon representation with the Connolly molecular surface. IP4 and glycerol are shown in space-filling representations. PtdIns(3,4,5)P3 and PS-binding residues are shown in stick representations and labeled. The molecule is orientated with its top part facing the membrane. B, a model structure of a PDK1-PH-IP4-PS complex. The PDK1-PH molecule is shown in a surface representation in the same orientation as in Fig. 1A. Red and blue colors qualitatively indicate negative and positive potentials, respectively. IP4 and PS molecules are shown in space-filling representations.
As expected, R465A shows drastically lower binding to POPC/PtdIns(3,4,5)P3 (97:3) vesicles than WT but only modestly reduced affinity for POPC/POPS (8:2) vesicles. Also, this mutant did not show increased binding to POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) over POPC/POPS (8:2) vesicles. Thus, Lys465 is clearly involved in PtdIns(3,4,5)P3 binding but not in PS binding. In contrast, R466A and K467A had significantly lower affinity for POPC/POPS (8:2) vesicles than WT but not for POPC/PtdIns(3,4,5)P3 (97:3) vesicles. Likewise double-site mutation of Arg466 and Lys467 to Ala caused ∼6-fold reduction in affinity for POPC/POPS (8:2) vesicles but did not decrease the affinity for POPC/PtdIns(3,4,5)P3 (97:3) vesicles. All single- and double-site mutations of Arg466 and Lys467 also significantly lowered the affinity for POPC/POPS/PtdIns(3,4,5)P3 (77:20:3) vesicles. Collectively, these mutational data indicate that Arg466 and Lys467 constitute the PS binding site but are not involved in PtdIns(3,4,5)P3 interaction. This in turn corroborates the notion that separate binding sites exist for PS and PtdIns(3,4,5)P3.
Effects of PS Binding on PM Localization of PDK1-PH
To determine the importance of PS binding of PDK1-PH on its subcellular localization, we transfected NIH 3T3 cells with EGFP-tagged PDK1-PH WT and mutations and monitored their subcellular localization under different conditions. Cells expressing similar levels of EGFP-tagged protein were selected and used for translocation measurements. We found that a large proportion of PDK1-PH WT was prelocalized to the PM in unstimulated NIH 3T3 cells (Fig. 3A), consistent with the previous report (7). In contrast to the WT, the R466A/K467A mutant, which has much reduced PS affinity but has intact PtdIns(3,4,5)P3 binding, exhibited largely cytosolic distribution under the same conditions (Fig. 3A). Interestingly, the K465A mutant, which is deficient in PtdIns(3,4,5)P3 binding, still showed significant PM prelocalization (Fig. 3A). The same pattern was observed for >80% of 50 or more cells investigated. These results suggest that PS binding is necessary for the PM localization of PDK1-PH in quiescent NIH 3T3 cells. Although we primarily used NIH 3T3 cells here because they showed robust response to PDGF stimulation (33), experiments with other mammalian cell lines, including HeLa and HEK293 cells, showed essentially the same trend (data not shown).
FIGURE 3.
Membrane translocation of EGFP-tagged PH domains in NIH 3T3 cells upon PDGF treatment. A, the time courses of subcellular localization of EGFP-tagged PDK1-PH WT and mutants in response to 50 ng/ml PDGF in NIH 3T3 cells. The arrow indicates the timing of PDGF addition. For comparison, subcellular localization of a PtdIns(3,4,5)P3 sensor, mCerulean-tagged Btk-PH was monitored under the same conditions. B, the time course in the relative PM distribution (i.e. photon count average at PM/photon count average at (PM + cytosol)) of PDK1-PH WT, PDK1-PH-K465A, PDK1-PH-R466A/K467A, and Btk-PH calculated from Fig. 3A. Bars, 5 μm.
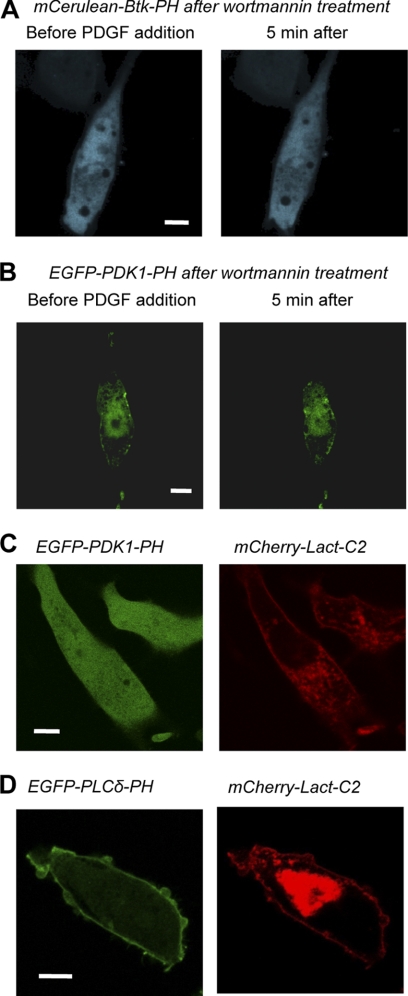
In order to verify that PM localization of PDK1-PH in unstimulated NIH 3T3 cells is due not to PtdIns(3,4,5)P3 binding but to PS binding, we measured the effects of suppressing PtdIns(3,4,5)P3 and PS levels in NIH 3T3 cells. It has been suggested that the background PtdIns(3,4,5)P3 level in the PM of transformed cell lines may be higher than that of normal cells (21). We thus treated NIH 3T3 with a PI3K inhibitor, wortmannin, and measured the effect on the PM localization of PDK1-PH WT. Treatment of NIH 3T3 cells with 50 nm wortmannin for 30 min abrogated PtdIns(3,4,5)P3 generation by PI3K, as evidenced by the cytosolic distribution of the mCerulean-tagged Btk-PH domain that has been used as cellular PtdIns(3,4,5)P3 sensor (33) even after cells were stimulated by PDGF (Fig. 4A). However, PDK1-PH still showed a significant degree of PM prelocalization pattern after the same wortmannin treatment (Fig. 4B). This control experiment thus precludes the possibility that PM prelocalization of PDK1-PH is due to its binding to residual PtdIns(3,4,5)P3 at the PM of quiescent NIH 3T3 cells. Due to the lack of a potent inhibitor for PS biosynthesis, we overexpressed the N-terminal mCherry-tagged Lact-C2 domain, which is known to bind PS with high affinity and specificity (17), to sequester PS in the PM. When NIH 3T3 cells were co-transfected with PDK1-PH WT and Lact-C2, PDK1-PH was fully cytosolic (Fig. 4C). This PS-masking effect of the Lact-C2 is specific to PS-binding proteins because the overexpression of Lact-C2 did not alter the PM localization of the PtdIns(4,5)P2-binding phospholipase Cδ (PLCδ) PH domain (Fig. 4D). These results further support the notion that PS binding, but not PtdIns(3,4,5)P3 binding, is important for PM localization of PDK1-PH in unstimulated NIH 3T3 cells.
FIGURE 4.
Effect of PtdIns(3,4,5)P3 depletion and PS sequestration on the subcellular localization of PDK1-PH in NIH-3T3 cells. A, subcellular localization of EGFP-PDK1-PH when NIH 3T3 cells were pretreated with 50 nm wortmannin and then stimulated with 50 ng/ml PDGF. B, subcellular localization of mCerulean-Btk-PH when NIH 3T3 cells were pretreated with 50 nm wortmannin and then stimulated with 50 ng/ml PDGF. C, subcellular localization of EGFP-PDK1-PH (left) and mCherry-Lact-C2 (right) when co-expressed in NIH 3T3 cells. D, subcellular localization of EGFP-PLCδ-PH (left) and mCherry-Lact-C2 (right) when co-expressed in NIH 3T3 cells. >80% of 20–30 cells investigated for each experiment showed similar patterns, and a representative cell image was taken for illustration from these cell populations. Fluorescence proteins are tagged at the N termini of the proteins. Bars, 5 μm.
We then measured the effect of PI3K activation on the subcellular localization of PDK1-PH WT and mutants. The time courses of membrane localization of these proteins in response to PDGF stimulation, which is known to activate PI3K, are illustrated in Fig. 3B. Consistent with their largely PM-prelocalized behaviors in the resting cells, WT and PtdIns(3,4,5)P3 binding-deficient K465A showed little movement in response to PDGF stimulation. However, R466A/K467A, which showed the cytosolic distribution in the resting state, spontaneously and reversibly translocated to the PM in response to PDGF stimulation. Its time course of PM localization was similar to that of the mCerulean-tagged Btk-PH domain. These results again show that under normal conditions, the PM localization of PDK1-PH does not depend on the PtdIns(3,4,5)P3 increase in PM.
Effects of PS Binding on Cellular Function of PDK1-PH
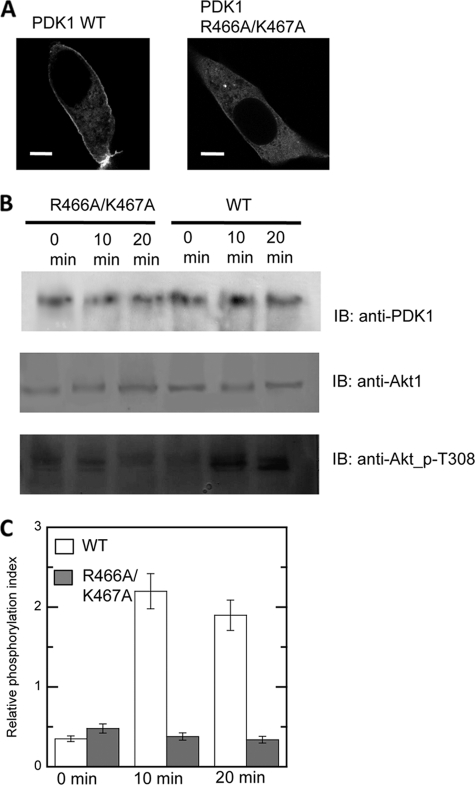
To demonstrate the physiological significance of PS-dependent PM localization of PDK1-PH, we measured the effect of the PS site mutation on the Akt phosphorylation activity of PDK1. PDK1 is known to phosphorylate Thr308 of Akt1 in response to PI3K activation (9, 10). First, we transfected NIH 3T3 cells with the EGFP-tagged full-length PDK1 WT and R466A/K467A mutant and monitored their localization. As was the case with the isolated PH domains, PDK1 WT showed a significant degree of PM localization in unstimulated NIH 3T3 cells, whereas R466A/K467A was primarily cytosolic (Fig. 5A). This indicates that the membrane localization behavior of its PH domain largely governs that of the intact PDK1. We then measured the phosphorylation of Thr308 of Akt1 in the cells that were co-transfected with Akt1 and PDK1 (WT or R466A/K467A) according to an established protocol (38). For all cells co-transfected with PDK1 (WT or R466A/K467A) and Akt1, a similar level of background Akt1-Thr308 phosphorylation was observed before stimulation by PDGF, presumably due to transient colocalization of PDK1 and Akt1 (Fig. 5, B and C), as reported previously (38). After PDGF stimulation, however, a dramatic difference in Akt1 phosphorylation among these cells was clearly visible (i.e. 10-min PDGF stimulation caused a >4-fold increase in Akt1 phosphorylation over background in cells expressing PDK1 WT while having little to no effect on Akt1 phosphorylation in cells expressing PDK1 R466A/K467A). Also, PDK1 R465A did not cause increased Akt1 phosphorylation in response to PDGF stimulation, confirming that PDGF stimulation proceeds through PtdIns(3,4,5)P3 generation at the PM (data not shown). These results show that PS binding activity of PDK1-PH is essential for the cellular localization and the signaling activity of PDK1 under physiological conditions.
FIGURE 5.
Akt phosphorylation activity of PDK1 WT and R466A/K467A mutant. A, subcellular localization of EGFP-tagged full-length PDK1 WT and R466A/K467A in unstimulated NIH 3T3 cells. Bars, 5 μm. B, NIH3T3-L1 fibroblasts co-transfected with Akt1 and PDK1 (WT or R466A/K467A) were stimulated for a given period of time with 50 ng/ml PDGF, and the whole cell lysates were separated on the gel and immunoblotted (IB) using anti-PDK1 (top), anti-Akt1 (middle), and anti-Akt1-Thr(P)308 (bottom) antibodies, respectively. C, intensities of protein bands were quantified using ImageJ software. Relative phosphorylation index was calculated by dividing the intensity of an Akt-Thr(P)308 band (bottom) by that of its corresponding Akt protein band (middle), and this parameter was plotted as a function of time for PDK1 WT and R466A/K467A.
DISCUSSION
Because PDK1 plays a central role in various PI3K signaling pathways, the mechanisms by which it activates a wide variety of downstream kinases have been intensely investigated. Given the fact that PDK1 is constitutively active in mammalian cells, it has been generally considered that its PM recruitment and co-localization with downstream kinases are key regulatory steps for PDK1. Evidence suggests that PtdIns(3,4,5)P3 is important for the co-localization of PDK1 with its substrates (5). However, it still remains unknown how PDK1 is recruited to the PM. The present study provides strong evidence for the notion that specific binding of PS to its PH domain is essential for the PM localization and hence signaling function of PDK1.
Our SPR studies of PDK1-PH WT clearly show that this domain can specifically bind PS using a site that does not overlap with the canonical phosphoinositide binding pocket. Molecular modeling and mutational analysis identified two basic residues, Arg466 and Lys467, constituting a PS-specific binding pocket. Mutations of these residues significantly lower the affinity of PDK1-PH for PS-containing vesicles but not for POPC/PtdIns(3,4,5)P3 (97:3) vesicles. In contrast, the mutation of a PtdIns(3,4,5)P3 ligand, Lys465, reduces the affinity for PtdIns(3,4,5)P3-containing vesicles without affecting affinity for PS-containing vesicles. Arg466 and Lys467 are located in the loop connecting β-strands. Intriguingly, multiple sequence alignment (Fig. 6) shows that many PtdIns(3,4,5)P3-binding PH domains have basic residues in this loop region, suggesting that these residues may also form similar cationic grooves and accommodate a PS headgroup. In fact, Arg10 and Lys15 of Akt1-PH were found to be involved in its PS binding (32). In contrast, the PtdIns(4,5)P2-selective PLCδ-PH lacks a basic residue in the same region. Thus, it would seem that specific PS binding is a common property of PtdIns(3,4,5)P3-binding PH domains.
FIGURE 6.
Partial sequence alignment of PH domains. N-terminal amino acid sequences of selected PtdIns(3,4,5)P2-binding PH domains are shown. Basic residues in the canonical phosphoinositide-binding pocket are shown in boldface characters. Putative PS-binding cationic residues in the loop region (box) are shown in italic, boldface type. Note that PLCδ-PH that selectively binds PtdIns(4,5)P2 does not have a cationic residue in this region.
What are the physiological consequences of specific PS binding to PtdIns(3,4,5)P3-binding PH domains? In the study of Grp1-PH, PS binding was shown to enhance its affinity for PtdIns(3,4,5)P3-containing vesicles and slow the lateral diffusion of the membrane-bound domain; however, the physiological significance of PS binding of this PH domain was not investigated (31). For Akt-PH, PS binding not only augments the domain's affinity for PtdIns(3,4,5)P3-containing membranes but also promotes a conformational change and activation of the full-length Akt (32). For both PH domains, it was speculated that PS binding is not strong enough to drive the PM recruitment of the domains (or intact proteins) and that the coincident interaction of the PH domain with PS and PtdIns(3,4,5)P3 is necessary for effective PM localization (32, 39). In contrast, our SPR data (Fig. 1A) show that PDK1-PH has comparable affinity for POPC/POPS (7:3) and POPC/PtdIns(3,4,5)P3 (97:3) vesicles. Given that the PS level in the inner leaflet of PM is estimated to be in the range of 20–30 mol %, PDK1-PH-PS binding may be strong enough to drive the PM translocation of the domain (hence the intact protein), at least partially, in the absence of PtdIns(3,4,5)P3. Consistent with this notion, a significant portion of PDK1-PH (and intact PDK1) is prelocalized to the PM of unstimulated NIH 3T3 cells, and this PM prelocalization is disrupted either by mutation of PS-binding residues or by PS sequestration by another PS-binding protein but not by mutation of a PtdIns(3,4,5)P3-binding residue or by PtdIns(3,4,5)P3 depletion. One may argue that the mutation of Arg466 and Lys467 may interfere with the interaction of PDK1-PH with other proteins present in the PM. Although we cannot completely rule out this possibility, the fact that the PS-binding Lact-C2 domain selectively blocks the PM localization of PDK1-PH but not the PtdIns(4,5)P2-binding phospholipase Cδ-PH, strongly suggests that PS binding is mainly responsible for the prelocalization of PDK1-PH in the PM of quiescent cells.
In agreement with the critical role of PS binding in PM localization of PDK1-PH, a signature signaling activity of PDK1 (i.e. phosphorylation of Thr308 of Akt) depends on the PS binding activity of PDK1-PH. It should be noted that this activity also requires the presence of PtdIns(3,4,5)P3 in the PM because it is necessary for colocalization of two proteins and the conformational change of Akt. In view of a recent report showing the role of PS in the membrane binding and the conformational change of Akt (32), it would seem that PS plays a more direct and crucial role in regulation of the PI3K-PDK1-Akt signaling pathway than previously thought.
Collectively, the present study establishes that PS plays a key role in PM localization of PDK1. As such, it adds to the growing evidence for the critical role of PS as a PM-specific marker that recruits many cytosolic proteins through specific interaction. Mammalian cells use complex mechanisms to maintain a high concentration of PS selectively in the inner leaflet of the PM (16), and it is therefore expected that membrane-binding cytosolic proteins should take advantage of its local abundance in their specific and reversible PM targeting. Genome-wide identification and characterization of the proteins that are targeted to the PM through specific interaction with PS would be necessary to fully assess the significance of PS as a site-specific marker at the PM.
Acknowledgments
We thank Dr. Hui Lu and Morten Kallberg for help in molecular modeling. We thank Dr. Richard Ye and Ni Cheng for assisting the Akt phosphorylation assay. We also thank Dr. Hee-Yong Kim for helpful discussion on PS binding of the Akt PH domain.
This work was supported, in whole or in part, by National Institutes of Health Grant GM68849.
- PtdIns(3,4,5)P2
- phosphatidylinositol 3,4,5-trisphosphate
- EGFP
- enhanced green fluorescence protein
- Lact-C2
- lactadherin C2
- PH
- pleckstrin homology
- PLCδ
- phospholipase Cδ
- PM
- plasma membrane
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine
- PS
- 1-phosphatidylserine
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- SPR
- surface plasmon resonance
- IP4
- inositol 1,3,4,5-tetrakisphsphate.
REFERENCES
- 1. Cantley L. C. (2002) Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 2. Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010) Nat. Rev. Mol. Cell Biol. 11, 329–341 [DOI] [PubMed] [Google Scholar]
- 3. Mora A., Komander D., van Aalten D. M., Alessi D. R. (2004) Semin. Cell Dev. Biol. 15, 161–170 [DOI] [PubMed] [Google Scholar]
- 4. Bayascas J. R. (2008) Cell Cycle 7, 2978–2982 [DOI] [PubMed] [Google Scholar]
- 5. Alessi D. R., Deak M., Casamayor A., Caudwell F. B., Morrice N., Norman D. G., Gaffney P., Reese C. B., MacDougall C. N., Harbison D., Ashworth A., Bownes M. (1997) Curr. Biol. 7, 776–789 [DOI] [PubMed] [Google Scholar]
- 6. Wick M. J., Ramos F. J., Chen H., Quon M. J., Dong L. Q., Liu F. (2003) J. Biol. Chem. 278, 42913–42919 [DOI] [PubMed] [Google Scholar]
- 7. Currie R. A., Walker K. S., Gray A., Deak M., Casamayor A., Downes C. P., Cohen P., Alessi D. R., Lucocq J. (1999) Biochem. J. 337, 575–583 [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas C. C., Deak M., Alessi D. R., van Aalten D. M. (2002) Curr. Biol. 12, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 9. Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R., Reese C. B., Cohen P. (1997) Curr. Biol. 7, 261–269 [DOI] [PubMed] [Google Scholar]
- 10. Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B., Painter G. F., Holmes A. B., McCormick F., Hawkins P. T. (1997) Science 277, 567–570 [DOI] [PubMed] [Google Scholar]
- 11. Anderson K. E., Coadwell J., Stephens L. R., Hawkins P. T. (1998) Curr. Biol. 8, 684–691 [DOI] [PubMed] [Google Scholar]
- 12. Filippa N., Sable C. L., Hemmings B. A., Van Obberghen E. (2000) Mol. Cell. Biol. 20, 5712–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bayascas J. R., Wullschleger S., Sakamoto K., García-Martínez J. M., Clacher C., Komander D., van Aalten D. M., Boini K. M., Lang F., Lipina C., Logie L., Sutherland C., Chudek J. A., van Diepen J. A., Voshol P. J., Lucocq J. M., Alessi D. R. (2008) Mol. Cell. Biol. 28, 3258–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cho W. (2006) Sci. STKE 2006, pe7. [DOI] [PubMed] [Google Scholar]
- 15. Vance J. E., Steenbergen R. (2005) Prog. Lipid Res. 44, 207–234 [DOI] [PubMed] [Google Scholar]
- 16. Leventis P. A., Grinstein S. (2010) Annu. Rev. Biophys. 39, 407–427 [DOI] [PubMed] [Google Scholar]
- 17. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 18. Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 19. McLaughlin S., Murray D. (2005) Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 20. McLaughlin S., Wang J., Gambhir A., Murray D. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 [DOI] [PubMed] [Google Scholar]
- 21. Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho W., Stahelin R. V. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 119–151 [DOI] [PubMed] [Google Scholar]
- 23. Mulgrew-Nesbitt A., Diraviyam K., Wang J., Singh S., Murray P., Li Z., Rogers L., Mirkovic N., Murray D. (2006) Biochim. Biophys. Acta 1761, 812–826 [DOI] [PubMed] [Google Scholar]
- 24. Stahelin R. V., Digman M. A., Medkova M., Ananthanarayanan B., Rafter J. D., Melowic H. R., Cho W. (2004) J. Biol. Chem. 279, 29501–29512 [DOI] [PubMed] [Google Scholar]
- 25. Stahelin R. V., Hwang J. H., Kim J. H., Park Z. Y., Johnson K. R., Obeid L. M., Cho W. (2005) J. Biol. Chem. 280, 43030–43038 [DOI] [PubMed] [Google Scholar]
- 26. Stahelin R. V., Rafter J. D., Das S., Cho W. (2003) J. Biol. Chem. 278, 12452–12460 [DOI] [PubMed] [Google Scholar]
- 27. Verdaguer N., Corbalan-Garcia S., Ochoa W. F., Fita I., Gómez-Fernández J. C. (1999) EMBO J. 18, 6329–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sánchez-Bautista S., Marín-Vicente C., Gómez-Fernández J. C., Corbalán-García S. (2006) J. Mol. Biol. 362, 901–914 [DOI] [PubMed] [Google Scholar]
- 29. Evans J. H., Murray D., Leslie C. C., Falke J. J. (2006) Mol. Biol. Cell 17, 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. (2008) J. Biol. Chem. 283, 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knight J. D., Falke J. J. (2009) Biophys. J. 96, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang B. X., Akbar M., Kevala K., Kim H. Y. (2011) J. Cell Biol. 192, 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Manna D., Albanese A., Park W. S., Cho W. (2007) J. Biol. Chem. 282, 32093–32105 [DOI] [PubMed] [Google Scholar]
- 34. Stahelin R. V., Cho W. (2001) Biochemistry 40, 4672–4678 [DOI] [PubMed] [Google Scholar]
- 35. Garza L. A., Birnbaum M. J. (2000) J. Biol. Chem. 275, 2560–2567 [DOI] [PubMed] [Google Scholar]
- 36. Calleja V., Alcor D., Laguerre M., Park J., Vojnovic B., Hemmings B. A., Downward J., Parker P. J., Larijani B. (2007) PLoS Biol. 5, e95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Komander D., Fairservice A., Deak M., Kular G. S., Prescott A. R., Peter Downes C., Safrany S. T., Alessi D. R., van Aalten D. M. (2004) EMBO J. 23, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whiteman E. L., Chen J. J., Birnbaum M. J. (2003) Endocrinology 144, 3811–3820 [DOI] [PubMed] [Google Scholar]
- 39. Corbin J. A., Dirkx R. A., Falke J. J. (2004) Biochemistry 43, 16161–16173 [DOI] [PMC free article] [PubMed] [Google Scholar]