Background: HDV-like ribozymes map to several non-LTR retrotransposons, although their roles are not fully understood.
Results: Self-cleaving ribozymes are found widespread in retrotransposons and promote translation initiation in vitro and in vivo.
Conclusion: Ribozymes process many non-LTRs and facilitate translation of their ORFs.
Significance: These new roles further explain the retrotransposon cycle and expand the functions of catalytic RNA.
Keywords: Ribozyme, RNA, RNA Catalysis, RNA Processing, Translation, Translation Control, Cap-independent Translation Initiation, Non-LTR, Retrotransposon, Self-cleaving
Abstract
Many non-long terminal repeat (non-LTR) retrotransposons lack internal promoters and are co-transcribed with their host genes. These transcripts need to be liberated before inserting into new loci. Using structure-based bioinformatics, we show that several classes of retrotransposons in phyla-spanning arthropods, nematodes, and chordates utilize self-cleaving ribozymes of the hepatitis delta virus (HDV) family for processing their 5′ termini. Ribozyme-terminated retrotransposons include rDNA-specific R2, R4, and R6, telomere-specific SART, and Baggins and RTE. The self-scission of the R2 ribozyme is strongly modulated by the insertion site sequence in the rDNA, with the most common insertion sequences promoting faster processing. The ribozymes also promote translation initiation of downstream open reading frames in vitro and in vivo. In some organisms HDV-like and hammerhead ribozymes appear to be dedicated to processing long and short interspersed elements, respectively. HDV-like ribozymes serve several distinct functions in non-LTR retrotransposition, including 5′ processing, translation initiation, and potentially trans-templating.
Introduction
Retrotransposons are mobile genetic elements that utilize reverse transcriptase activity to propagate via an RNA intermediate in many genomes (1). The large fraction of eukaryotic genomes composed of retrotransposons coupled with their contributions to ectopic recombination and novel gene regulatory elements suggest potential for major evolutionary influence (2). Autonomous retrotransposons are divided into long terminal repeats (LTRs) and non-LTRs or long interspersed elements (LINEs).3 Short interspersed elements (SINEs) are non-autonomous elements thought to be mobilized primarily by non-LTR type elements (3). LTR elements have promoters within their LTRs (3); however, the promoters for non-LTR elements are less well conserved, and they can even be transcribed as introns of larger host transcripts (4). In such instances, the transcripts need to be processed so that the retrotransposons are liberated and propagated without the co-transcribed flanking sequences. Recently, the first suggestion that 5′ processing is performed by a ribozyme encoded in the 5′ UTR of the retrotransposon was proposed when an HDV-like ribozyme was discovered at the 5′ end of an Anopheles gambiae RTE (5). Subsequent analysis of the Drosophila R2 and Trypanosoma cruzi L1 elements showed that they also harbor HDV-like ribozyme at their 5′ termini (6, 7).
Among the most studied non-LTR retrotransposons are ribosomal DNA (rDNA) and telomeric elements that insert sequence-specifically into the host repeats (8–11). Perhaps the best characterized autonomous element is R2, which inserts site-specifically into the 28S rDNA of most insects (8). The R2 element was first identified as a Drosophila melanogaster insertion sequence co-transcribed with the 28S ribosomal RNA (rRNA) precursor by RNA polymerase I (4, 12). Transcripts of interrupted rRNA genes were shown to be processed co-transcriptionally at the boundary of the 28S rRNA transcript and 5′ and 3′ ends of the intervening sequence (13). The mechanism of R2 retrotransposition has been studied in depth, leading to a general model for non-LTR reverse transcription and integration, during which the endonuclease-catalyzed cleavage of the target DNA backbone is followed by target-primed reverse transcription and second strand synthesis by the RT, resulting in insertion of the newly synthesized cDNA into the genome (14, 15). Although much of this model has been validated in vitro, the mechanism of co-transcriptional processing of these elements by HDV-like ribozymes was not defined until recently (6).
The protein translated from the single ORF of the R2 element contains a restriction enzyme-like endonuclease domain, zinc finger and c-Myb-like DNA-binding motifs, and an RT domain (16, 17). Two subunits of the R2 protein are required to carry out the complete retrotransposition process, and they are thought to be tethered together by binding secondary structures in a single copy of its own RNA at the 5′ and 3′ ends (14, 18). The subunit that binds to the 3′ UTR RNA subsequently binds to DNA upstream of the target insertion site, and conversely, the remaining subunit binds the 5′ UTR RNA and DNA downstream of the target insertion site (18). Although much is known about the activity of the R2 protein, the mechanism by which it is translated remains to be defined. The N-terminal domain of the Drosophila R2 ORF is poorly conserved at the sequence level, and thus it is not surprising that the first AUG codon is variable. In many species an AUG codon is not found before the first conserved DNA-binding motif (4). Because the R2 RNA is co-transcribed with ribosomal RNA and is then processed, it is unlikely that R2 transcripts undergo canonical translation initiation that capped mRNAs do (19). The RNA structure in the R2 5′ UTR has been suggested to interact with translational machinery to initiate protein synthesis much like in internal ribosome entry sites (IRESes) seen in viruses and some cellular mRNAs (6, 19).
A model similar to the retrotransposition of R2 can be applied to other promoter-less retrotransposons, including other rDNA- and telomere-specific elements. For example, the SART retrotransposon inserts into the TTAGG telomeric repeats in a wide variety of insects via its protein encoded by a bicistronic RNA containing two ORFs that are thought to undergo a mechanism of translation similar to translational coupling seen in prokaryotes and viruses (20). The presence of a ribozyme capable of initiating translation might aid in this noncanonical translation mechanism. HDV-like ribozymes have previously been shown to cleave the pre-mRNA of the mammalian CPEB3 gene, and one potential function proposed for the ribozyme-terminated RNA was in translation initiation (21, 22). In another instance, an HDV-like ribozyme maps to the beginning of a viral mRNA in the insect iridescent virus 6 (also Chilo insect virus), where it forms the 5′ terminus of the RNA polymerase 5′ UTR (5) and may also serve to promote translation initiation.
HDV ribozymes fold into an intricate double-pseudoknot structure composed of five helical regions joined by single-stranded regions (23–25). We have previously used structure-based searches to identify the HDV family of self-cleaving ribozymes in many organisms (5), and we found that some of the sequences map to predicted RT genes in the purple sea urchin Strongylocentrotus purpuratus and the A. gambiae RTE retrotransposon (5). These results suggested that self-cleaving ribozymes might function in retrotransposition. However, the predicted secondary structure of these two sequences differs somewhat from the structures of canonical HDV-like ribozymes; some examples of the sea urchin ribozymes have shorter P1 regions than was previously observed, and the mosquito RTE ribozyme (drz-Agam-2) contains an unusually large peripheral domain in the J1/2 region (5). To test more broadly for the association of HDV-like ribozymes with retrotransposons, we performed motif searches (26) that allowed for variable P1 helices and large inserts in the J1/2 region of the ribozyme, and we analyzed their self-scission. Beyond processing, we examined the influence of the flanking insertion-site sequences on self-scission, and we tested the ability of ribozyme-terminated mRNAs to support efficient translation initiation.
EXPERIMENTAL PROCEDURES
Secondary Structure-based Searches
We used the RNABOB program (courtesy of S. Eddy, HHMI) to define the conserved sequence and structure elements of the HDV/CPEB3 ribozymes as described previously (5, 26, 27). Two changes were made to the descriptor as follows: P2 is 6 bp long and allows one insertion or a mispair in the middle (split between elements r3 and r4), and J1/4 is 3–7 nucleotides long.
Nomenclature
Naming of the ribozymes follows the rules proposed previously (5). Briefly, the name of the ribozyme was derived from the binomial nomenclature for the species (drz = delta-like ribozyme, Drosophila ananassae, Dana). Among the fruit fly ribozymes, family 1 (e.g. drz-Dmel-1) corresponds to R2 (or R2B) ribozymes, and family 2 (e.g. drz-Dmel-2) corresponds to Baggins ribozymes.
DNA Template Preparation
For in vitro cleavage kinetics, ribozyme constructs were either amplified from their respective genomes or synthesized by mutual priming using primers listed below and the DreamTaq PCR master mix (Fermentas). The constructs were designed to include a 40-nucleotide leader sequence, the full ribozyme, and a short (2–8 nucleotide) tail.
Cloning of Wild-type and Inactive Ribozyme Reporter Construct
Ribozyme DNA template was PCR-amplified with a Phusion high fidelity PCR master mix (New England Biolabs), forward primers to include the 300 bp upstream of the ribozyme and reverse primers with a BamHI site downstream of the ribozyme. The entire open reading frame of a firefly luciferase gene, including a BamHI site upstream of the AUG start codon, was amplified similarly from a plasmid gifted by the laboratory of Prof. Marian Waterman, University of California, Irvine. BamHI-digested ribozyme DNA was ligated to BamHI-digested luciferase DNA using T4 DNA ligase (New England Biolabs). Ligated constructs were PCR-amplified using Easy-A high fidelity PCR cloning enzyme, cloned into a pCR2.1-TOPO plasmid using a TOPO TA cloning kit (Invitrogen), and sequenced (GENEWIZ). Similar methods were followed for control constructs. To generate ribozymes with a uridine mutation at the active site cytidine, QuikChange mutagenesis (Stratagene) was used with oligonucleotides containing a T-A base pair where the wild-type construct contains a C-G base pair. All constructs were sequenced to check the ribozyme and luciferase sequence.
P1 Extension DNA Template Preparation
Mutants of drz-Dmel-1-1 were made via PCR from the cloned drz-Dmel-1-2 sequence using Phusion high fidelity PCR master mix (New England Biolabs), forward primers containing mutations only in the leader sequence, and a universal reverse primer. The constructs contained an ∼40-nucleotide leader sequence, the entire ribozyme sequence, and a 19-nucleotide tail. A second round of PCR was performed to add a T7 promoter upstream of the leader sequence.
RNA Transcription
For cleavage kinetics, α-32P-labeled RNA was transcribed at 37 °C for an hour in a 20-μl volume containing 40 mm Tris-HCl, 0.01% Triton X-100, 2 mm spermidine, 10 mm DTT, 2.5 mm each of GTP, UTP, and CTP, 250 μm ATP, 1.25 μCi of [α-32P]ATP (PerkinElmer Life Sciences), 7.75 mm MgCl2, 20 μm inhibitor oligonucleotide (specific for each construct), 1 unit of T7 RNA polymerase, and 0.5 pmol of DNA template. The desired RNA was purified from the reaction using denaturing PAGE. For in vitro and in vivo translation assays, nonradioactive RNA was transcribed at 37 °C for 3 h in a 50-μl volume containing 10 mm DTT, 2.5 mm each GTP, UTP, CTP, and ATP, 35 mm MgCl2, 40 units of RNasin Plus RNase inhibitor (Promega), 10 units of T7 RNA polymerase, and 1500 ng of restriction enzyme-digested plasmid DNA. After treatment with DNase I (RNase-free, Promega, 1 unit/μg DNA for 15 min at 37 °C), RNA was phenol/chloroform-extracted and purified on a Sephadex G-50 column (Sigma). Concentrations were determined by absorbance at 260 nm and adjusted based on densitometry measurements of full-length RNA from a SYBR Gold (Invitrogen)-stained 1% agarose gel.
Cleavage Kinetics
In vitro self-cleavage reactions were performed as described previously (5). Gel-purified RNA and 2× kinetics buffer (280 mm KCl, 20 mm NaCl, 100 mm Tris, pH 7.5, and twice the experimental concentration of divalent metal ions) were preincubated in separate tubes at the experimental temperature for 5 min. The zero time point was collected from the RNA stock solution. Self-scission was initiated by mixing the RNA with the 2× kinetics buffer, and aliquots were collected at indicated times and mixed with an equal volume of stop solution (8 m urea, 20 mm EDTA). The denaturing polyacrylamide gel of self-cleavage products was exposed to phosphorimager screens (GE Healthcare) and analyzed by using Typhoon phosphorimager and ImageQuant software (GE Healthcare). Fraction intact was modeled using a biexponential decay and uncleaved residuals function. The faster rate constant is reported in Table 1. Complete kinetic analyses are reported in the supplemental material.
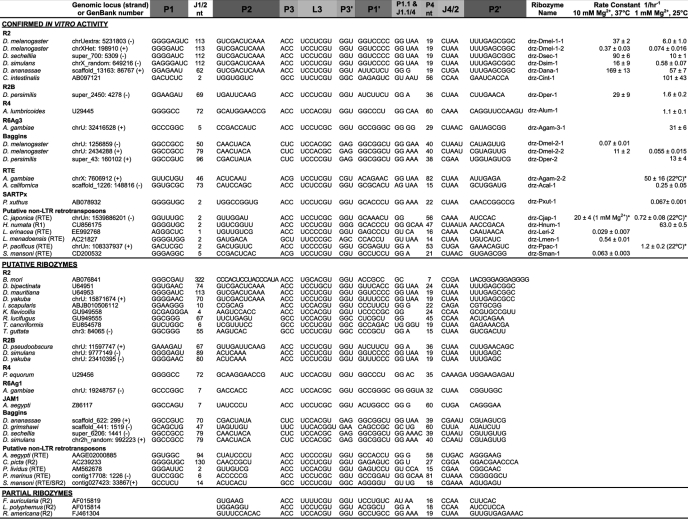
TABLE 1.
Segmental alignment and activity of HDV-like ribozymes mapping to 5′ termini of retrotransposons
Rate constants are average values ± average deviations.
*Data are from Ref. 4.
In Vitro Translation
The Retic Lysate IVT kit (Ambion) was used for 10 μl in vitro translation reactions containing 68% (v/v) rabbit reticulocyte lysate (RRL), 125 mm KOAc, 0.5 mm Mg(OAc)2, 10 mm creatine phosphate, 50 μm amino acids, 13 units of RNasin Plus RNase inhibitor (Promega), and 12 ng of uncapped RNA. Reactions were incubated at 30 °C for 90 min, stopped on ice, immediately examined for luciferase activity by mixing with 100 μl of luciferase assay reagent (Promega) in black 96-well plates (Falcon), and imaged using the IVIS Lumina II system (Caliper Life Sciences). Translation efficiency was measured based on integrated density of luminescence from each reaction and reported as the mean of four independent translation experiments from two independent transcriptions.
Cell Culture and Transfection
Drosophila S2 cells were maintained at 26 °C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and 50 units of penicillin G and 50 μg of streptomycin sulfate per ml of medium. Cells were seeded at 0.65 × 106 cells per well in 24-well plates 24 h before transfecting 2 μg of purified RNA per well using TransMessenger transfection reagent (Qiagen). Luciferase activity of cell lysates collected using Passive Lysis Buffer (Promega) 24 h after transfection was measured as described above.
RESULTS
Structure-based Searches Reveal Many Retrotransposon-associated Ribozymes
To establish the distribution of HDV-like retrotransposon-associated ribozymes, we searched expressed sequence tags, RT-containing mRNAs, and genomic regions defined as repeats by RepeatMasker for sequences capable of assuming the ribozyme double-pseudoknot secondary structure (26). The expanded search criteria allowed identification of many ribozyme candidates, a subset of which mapped to the 5′ termini of known non-LTR retrotransposons (Table 1). These included additional A. gambiae RTEs, the JAM1/RTE (28) in the yellow fever mosquito Aedes aegypti, the telomeric R1-like repeat element SARTPx2 (10, 11) in the Asian swallowtail butterfly Papilio xuthus, and Baggins retrotransposons in a number of Drosophila species. Ribozymes were also found at 5′ termini of many rDNA-targeting retrotransposons as follows: the R2 element in Drosophila; the termites Kalotermes flavicollis and Reticulitermes lucifugus (29), the sea squirt Ciona intestinalis (30), the black-legged tick Ixodes scapularis (GenBank accession number ABJB010506112), the horseshoe shrimp Triops cancriformis (GenBank accession number ABJB010506112), and the Zebra finch Taeniopygia guttata (20); the R4 elements (9) in the human intestinal roundworm Ascaris lumbricoides and the horse intestinal roundworm Parascaris equorum; and R6Ag1 and R6Ag3 elements (11) in A. gambiae. Partial ribozymes, missing only the 5′ strand of P1, were found in R2 elements of the European earwig Forficula auricularia (16), the Atlantic horseshoe crab Limulus polyphemus (16, 31) and the fly Rhynchosciara americana (32). In the silkworm Bombyx mori R2 element, the structure-based search uncovered the core of the HDV-like ribozyme several hundred nucleotides downstream of the retrotransposon 5′ terminus and the putative cleavage site precisely at the 5′ terminus of the retrotransposon, making the J1/2 region the largest found to date (Table 1). Interestingly, various ovarian small antisense RNAs implicated in regulation of retrotransposon activity (33) are complementary to the J1/2 and core sequences of this putative ribozyme.
Ribozyme candidates also map to the 5′ termini of putative non-LTR elements, classified by the similarity of the RT-coding downstream regions, in the mosquito A. aegypti (RTE), the brush-footed butterfly Heliconius numata (R1-like), the flatworm Schistosoma mansoni (RTE), the sea slug Aplysia californica (RTE), the common sea urchin Paracentrotus lividus (RTE), the Indonesian coelacanth Latimeria menadoesnsis (RTE), the sea lamprey Petromyzon marinus (RTE), the little skate Leucoraja erinacea (RTE), and the painted turtle Chrysemys picta (R2) (Table 1).
Finally, analysis of sequences downstream of HDV-like ribozymes previously identified in nematodes Caenorhabditis japonica and Pristionchus pacificus (5) revealed that they potentially code for proteins similar to the RTE RT (Table 1), suggesting that these are also ribozyme-terminated retrotransposons. The discovery of HDV-like self-cleaving ribozymes in a number of different retrotransposons and species suggests a widespread use of these catalytic RNAs in retrotransposition.
Retrotransposon Ribozymes Self-cleave in Vitro with Variable Kinetic Rates
To investigate the activity of the ribozymes, we amplified the genomic regions containing the 5′ termini of a representative set of the putative ribozymes, transcribed the products in vitro, and tested the Mg2+-dependent self-scission of the purified RNAs. As in the case of the A. gambiae RTE ribozyme (5), these RNAs showed robust self-cleavage kinetics at ambient temperature and physiological Mg2+, K+, and Na+ concentrations (Fig. 1, Table 1, and supplemental Table S1), demonstrating that these structures are bona fide ribozymes. No self-scission was observed in physiological monovalent salts alone (data not shown).
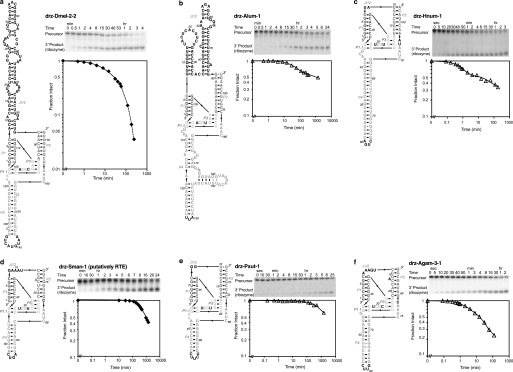
FIGURE 1.
Secondary structures and in vitro activity of retrotransposon HDV-like ribozymes. a, Drosophila Baggins drz-Dmel-2-2; b, A. lumbricoides R4 drz-Alum-1; c, H. numata R1 drz-Hnum-1; d, S. mansoni RTE drz-Sman-1; e, P. xuthus SARTPx-2 drz-Pxut-1; and f, A. gambiae R6Ag3 drz-Agam-3 ribozymes. Conditions for in vitro self-cleavage assays are as follows: 10 mm MgCl2 at 37 °C (♦) and 1 mm MgCl2 at 25 °C (△). All graphs are logarithmic with vertical axes on the same scale; b, d, and e span longer time periods. The data were fit to mono- or bi-exponential decay. Rate constants and amplitudes are reported in Table 1 and supplemental Table S1.
The D. melanogaster Baggins, A. lumbricoides R4, H. numata putative R1, S. mansoni putative RTE, P. xuthus SARTPx-2, and A. gambiae R6Ag3 retrotransposons all harbor active ribozyme sequences at their 5′ termini (Fig. 1, a–e, and supplemental Table S1). As in the case of other HDV-like ribozymes, these sequences have variable peripheral domains but a highly conserved core structure. Notable among these is the A. lumbricoides R4 ribozyme (drz-Alum-1), in which the J1/2 region is predicted to fold into two helices that may form a tertiary contact between the 5′ stem-loop and the purine-rich internal loop of the 3′ stem (Fig. 1b). Such tertiary interactions are known to stabilize hammerhead ribozymes (34–37) but have not been proposed to form in HDV-like ribozymes.
In the R2 elements, the conserved regions at the 5′ termini (4) form the catalytic core of the putative ribozyme double-pseudoknot structure (Fig. 2a), whereas the less conserved sequences map primarily to the peripheral domains (J1/2 and P4, supplemental Fig. S2), which are known to be variable in HDV-like ribozymes (5, 38). In drz-Dmel-1-1, we observed similar rates of self-scission in the presence of 1 mm Ca2+ and Mn2+ (13 ± 2 and 8 ± 4 h−1, respectively, at 25 °C, see supplemental Table S2). Recently, Eickbush and Eickbush (6) showed similar in vitro activity in the Drosophila R2 retrotransposons.
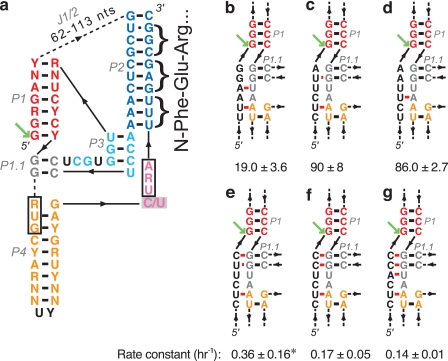
FIGURE 2.
Drosophila R2 ribozymes. a, Drosophila R2 ribozyme consensus secondary structure. Core elements are colored by region corresponding to the HDV ribozyme (23). Boxed nucleotides are start and stop codons found upstream of the R2 coding region (4). Green arrow points to the cleavage site. Consensus is based on in vitro active R2 ribozymes (Table 1). The inhibitory active site C/U mutation used in translation experiments is shown by a gray box. The first three amino acids of the R2 protein are shown to the right of nucleotides that code for them. b–g, potential extension of the P1 helix with the leader sequence of various wild-type and mutant constructs. b–d, common 28S rDNA insertion sequences that promote self-scission. e–g, mutations in the insertion sequence that allow extension of the P1 helix. Potential alternative pairing between the leader sequence (black) and the P1.1 region (gray) is shown in red. Rate constants are reported with units of h−1 and were measured in 1 mm MgCl2 at 25 °C. All data are average values ± average deviations. *, faster self-cleavage also occurs, see supplemental Table S3.
The Baggins retrotransposons and their 5′-ribozymes were found in only a subset of the Drosophila species (Table 1). These ribozymes have predicted overall structures similar to the Drosophila R2 and Anopheles RTE RNAs, including large peripheral domains in the J1/2 region (Table 1, Fig. 1a, and supplemental Fig. S1). Their in vitro self-scission is also variable, and the sequences we tested had similar cleavage rate constants compared with the R2 ribozymes (Table 1, Fig. 1a, and supplemental Table S1) and, as in the case of other HDV-like ribozymes (5, 21), showed temperature and/or Mg2+ dependence (Table 1 and supplemental Table S1).
R2 Ribozyme Self-scission Is Modulated by Upstream Insertion Sequences
In the Drosophila R2 ribozymes, the cleavage rate constants varied among isolates, which differ mostly in composition of the leader sequence just upstream of the cleavage site and the J1/2 peripheral region, whereas the cores of the ribozymes are conserved (Table 1). This observation suggests that folding of the sequences surrounding the cleavage site and in the peripheral domains affect the formation of active ribozymes. In HDV ribozymes, the sequence upstream of the cleavage site has previously been shown to influence proper folding of the ribozymes into active conformations (39–41). When the leader sequence has the capacity to extend the P1 helix of the ribozyme, it may prevent formation of the P1.1 region of the active site, resulting in slower self-scission. Our data correlate well with this hypothesis. We observe significantly faster self-cleavage kinetics in identical ribozymes preceded by leader sequences that cannot extend the P1 helix and slower kinetics in constructs that can form the longer P1. To investigate this trend, we made several wild-type and mutant constructs of drz-Dmel-1-1 with variable leader sequences and measured their self-scission. The insertion site for the R2 element in D. melanogaster is reported as CTCTTAAG(G/T)AGC (Fig. 2b), although the three nucleotides upstream of the insertion site are often deleted upon insertion of the R2 element (8), leaving CTCTTA upstream and making this another possible common leader sequence (Fig. 2c). The deletion of AGG from the leader sequence results in a substantial increase in the rate of self-scission (19 ± 3.6 to 90 ± 8.1 h−1, see supplemental Table S3), which may be caused by the elimination of an A-U base pair between the leader and J1.1/4. A third wild-type leader sequence, TCTTAA, is found in the 28S insertion site in the reference D. melanogaster genome (BDGP Release 5), and this construct displayed similar self-cleavage activity to the GAA deletion construct (Fig. 2d and supplemental Table S3). Mutating this leader sequence to allow 1–3 bp to extend the P1 helix (Fig. 2, e–g) results in a 250-, 530-, and 640-fold reduction in the cleavage rate constant, respectively (supplemental Table S3). Depending on the preparation of the RNA, however, the leader sequence ending in CTCTTC (Fig. 2e) can sustain fast self-cleavage (supplemental Table S3). This variability may result from an alternate folding event that occurs in the leader sequence that prevents P1 extension and allows the correct ribozyme structure to form. Overall, the most common (wild-type) insertion sites for the R2 element in the 28S rRNA gene (8, 42) create leader sequences that support fast ribozyme self-scission, suggesting that the selection of the insertion sequence, although unique for each class of R elements, may be influenced by its ability to promote efficient 5′ processing by self-cleaving ribozymes.
Dedicated Processing of SINEs and LINEs by Hammerhead and HDV-like Ribozymes
The S. mansoni ribozyme drz-Sman-1 (Fig. 1d) maps upstream of an RTE-like ORF (43), and a different HDV-like sequence maps to the SR2 element (Table 1). S. mansoni has previously been shown to harbor hammerhead ribozymes, which process the Smα SINEs (44). Thus, in S. mansoni two different types of self-cleaving ribozymes, HDV-like and hammerhead, process distinct types of mobile elements, LINEs and SINEs, respectively. Our data (Table 1), in conjunction with the recently published mapping of the hammerhead ribozymes (37, 45, 46), suggest that two Drosophila species (persimilis and pseudoobscura), the mosquito A. aegypti, the black-legged tick I. scapularis, and the sea lamprey P. marinus also harbor retrotransposon-associated HDV-like and hammerhead ribozymes. In I. scapularis, the hammerhead ribozymes appear associated with satellite repeats (e.g. in seven copies within an ∼700-nucleotide region of sequence ABJB010761563) and HDV-like ribozyme upstream of an R2 LINE. In one locus in the sea lamprey genome (contig 90380), a hammerhead sequence is followed by the Iun10 (47) and Pmaμ6 (48) microsatellites, a putative HDV-like ribozyme, and an RTE-like RT coding region. This finding suggests that in the tick and lamprey genomes, like schistosomes, the hammerhead and HDV-like ribozymes are dedicated to processing SINEs and LINEs, respectively.
HDV-like Ribozymes Near LTR Retrotransposons
Analysis of sequences downstream of previously reported HDV-like ribozymes (5) revealed two examples of gag/pol/env polyprotein gene typical of LTR retrotransposons, but no coding regions of non-LTR proteins. In the fungus Ajellomyces capsulatus and the triatomine Rhodnius prolixus, ribozymes are found near putative LTR retrotransposons similar to Tf2 and BEL12-AG elements, respectively. LTR retrotransposons are flanked by sequences that contain promoters for transcription by RNA polymerase II (3); therefore, they presumably do not require processing from a longer transcript. Interestingly, we find these ribozymes in an antisense orientation with respect to the LTR ORF, suggesting that these ribozymes do not act to liberate a retroelement from the upstream sequence but may participate in an as-yet unknown process associated with the LTR retrotransposons.
Ribozymes in Retrotransposons Promote Translation Initiation
Our results show that structure-based searches identify active HDV-like ribozymes distributed through multiple types of retrotransposons in species spanning most of the animal phyla, including arthropods, molluscs, platyhelminths, nematodes, echinoderms, and chordates. Taken in conjunction with previous results (6, 13, 49, 50), we show that the terminal ribozymes of the R2 retrotransposons are active in vitro and in vivo, leading to a model of R2 RNA production involving co-transcriptional cleavage by an HDV-like ribozyme (Fig. 3a). This model explains how a promoter-less retrotransposon that inserts into other genes, sequence-specifically or not, can liberate its 5′ terminus from the upstream transcript. Once ribozyme cleavage occurs, the retrotransposon transcript must be translated for the cycle to continue. The mechanism by which many retrotransposons are translated has not been explicitly defined, but it has been suggested to occur in a noncanonical fashion (4, 19). The presence of an HDV-like ribozyme at the 5′ terminus of the retrotransposon supports this hypothesis. The HDV-like ribozymes found in retrotransposons contain a complex pseudoknot structure, which is integral to the activity of some IRESes (51, 52). In contrast to most eukaryotic mRNAs, ribozyme self-scission yields a transcript with a 5′-hydroxyl group that cannot be capped. In some cases, notably the Drosophila R2 elements, the ribozymes constitute the entire 5′ UTR of the element. These observations led us to hypothesize that the ribozyme is directly involved in noncanonical translation initiation of the downstream retrotransposon ORF.
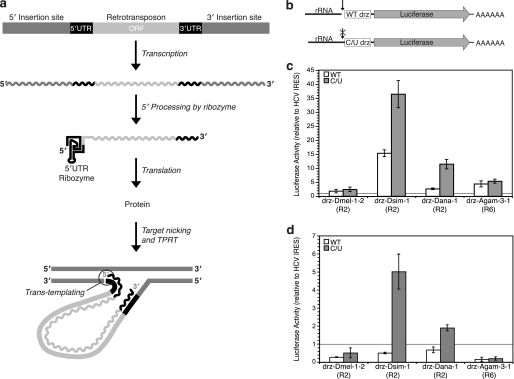
FIGURE 3.
Proposed model of retrotransposon RNA processing and translation initiation. a, retrotransposon is co-transcribed with its host DNA, and the 5′ terminus is generated by the self-cleaving ribozyme. The ribozyme forms the 5′ UTR of the retrotransposon ORF and aids in the efficiency of the trans-templating event that occurs after target nicking and target-primed reverse transcription (TPRT) by the protein of the retroelement. b, reporter constructs used in translation assays. Wild-type and inactive (C/U) ribozymes were cloned upstream of a luciferase gene and downstream of a 300-nucleotide rRNA leader sequence. A T7 RNA polymerase promoter was used to drive in vitro transcription. c, translation activity of wild-type and inactive ribozymes relative to HCV IRES from in vitro experiments with RRL. d, translation of luciferase mRNA terminated with wild-type and inactive ribozymes relative to HCV IRES in transfected insect S2 cells. The horizontal lines in the graphs represent the activity of the HCV IRES. All data are average values ± average deviations.
To test this hypothesis, we cloned wild-type and inactive mutant ribozymes and their respective leader sequences upstream of a luciferase reporter gene (Fig. 3b), and these constructs were transcribed in vitro using T7 RNA polymerase. Mutant ribozymes contained an active-site cytosine-to-uracil (C/U) mutation (Fig. 2a) that prevents self-cleavage, while maintaining the global structure of the ribozyme (24). The RNA constructs were either translated in vitro using RRL or transfected into insect S2 cells to measure their translation in vivo. The luciferase activity of the ribozymes was normalized to the activity of a construct initiated by the hepatitis C virus (HCV) IRES. All ribozymes tested had activity equal to or greater than the HCV IRES, with the D. simulans R2 ribozyme (drz-Dsim-1) exhibiting the highest translation in RRL. Surprisingly, the inactive (C/U) ribozymes yielded higher in vitro translation activity than their wild-type counterparts, although the relative translation efficiency of the self-cleaved and inhibited ribozyme constructs varied among the R2 and R6 elements tested (Fig. 3c). Transfection of these RNAs into Drosophila S2 cells and subsequent measurement of the luciferase activity showed similar results (Fig. 3d). Notably, the difference in translation initiation between wild-type and mutant ribozymes in vivo was less dramatic than in vitro. As a control experiment, we annealed a DNA oligonucleotide that binds across the initiator codon in the luciferase ORF to the drz-Dsim-1 RNA. This oligonucleotide inhibited translation in vitro and in vivo by about 75% (data not shown). These data support a novel role for HDV-like self-cleaving ribozymes in initiating translation of retrotransposon ORFs. In addition to liberating the 5′ terminus of the retrotransposon transcript via self-scission, the ribozyme acts similarly to an IRES and presumably binds the translation machinery, allowing translation initiation to occur. This mechanism bypasses the need for a 5′-methylguanosine cap on the RNA and explains the absence of a conserved AUG codon in the R2 elements.
DISCUSSION
Over the past 30 years, ribozymes have been shown to have many biological roles, including self-splicing, processing of rolling-circle transcripts, and metabolite-dependent gene regulation (53–55). Our work, and that of Eickbush and Eickbush (6) and Thomas and coworkers (7), expands these biological roles to the processing of 5′ termini of non-LTR retrotransposons by self-cleaving ribozymes. These RNAs exhibit self-scission rates in physiological conditions consistent with a model in which they are responsible for the co-transcriptional production of the retrotransposon 5′ termini.
A number of identified ribozyme-terminated retrotransposons insert sequence-specifically into rDNA (R2, R4, and R6) or telomeric repeats (SART). In the Drosophila R2 elements, this upstream insertion sequence was found to strongly influence the activity of the ribozyme, and the typical rDNA insertion sequences promote the fastest processing of the retrotransposon terminus (Fig. 2, b–g). Similar bias toward upstream sequences that minimize misfolding is observed in other sequence-specific retrotransposon ribozymes, indicating that efficient co-transcriptional 5′ processing of the elements is evolutionarily advantageous to their propagation. Because the endonuclease domain of the protein of the retrotransposon determines the exact sequence for insertion, we hypothesize that the preferred upstream sequence for the ribozyme influenced the evolution of the endonuclease domain. Alternatively, the inherent specificity of the endonuclease domain may have provided a sequence constraint as the ribozyme fold evolved. In either case, the seemingly arbitrary distribution of insertion sites, particularly among R elements, which insert within the ∼50-bp region in the large rRNA genes of their respective organisms, may be more predetermined than previously thought.
The R2 element of the silkworm B. mori has been shown to possess a 5′ UTR that folds into a pseudoknot similar to that of the HDV ribozymes (19), but this element lacks sufficient nucleotides to form the second nested pseudoknot (P1.1). We tested this element for in vitro self-scission but did not observe any activity, even at elevated Mg2+ concentrations and temperatures. However, further upstream of the pseudoknot described by Kierzek et al. (19), we identified another potential ribozyme in the 5′ UTR of the B. mori R2 element. This RNA does not form a canonical HDV-like ribozyme because the lower base pair of the P1.1 region is formed by a CG base pair, as opposed to a GC or a mispair observed in other HDV-like ribozymes, but this potentially stable P1.1, together with a P2 helix that is 14 bp long, may lead to a stable catalytic core capable of supporting self-cleavage of a substrate strand 300 nucleotides upstream. Future self-scission experiments with the entire 5′ UTR of the B. mori R2 element will test whether there is a genuine HDV-like ribozyme.
Ribozymes were also found terminating a number of sequence-independent retrotransposons, including the Drosophila Baggins and Anopheles RTEs. Although these retrotransposons lack conserved insertion sequences, analysis of the Baggins and RTE sequences in reference genomes shows that most of the insertion sites map to introns or immediately downstream of 3′ UTRs or LTR retrotransposons, frequently of the Gypsy and Pao families. This observation suggests a model in which the Baggins and RTEs, like the rDNA and SART elements, ensure expression through co-transcription with other genetic elements and use self-cleaving ribozymes for 5′ processing.
The role of self-cleaving ribozymes beyond 5′ terminal processing has been speculative; however, the ribozymes have been proposed to influence other steps in the retrotransposition cycle (6). In the R2 elements, the ribozyme appears to act in a manner similar to a viral IRES (56, 57). Self-scission yields an uncapped R2 transcript with a 5′-hydroxyl that lacks a conserved initiation codon downstream of the ribozyme sequence. However, a short coding region just upstream of the RT coding region (4), complete with start and stop codons, maps to the P4 helix and J4/2 strand of the ribozyme, respectively (Fig. 2a). The role of this region in translation is unknown; however, efficient production of the downstream ORFs for the R2 element is observed both in vitro and in vivo, and thus it may serve simply to melt the P4 stem-loop to allow for efficient translation of the downstream P2 sequence, which is highly conserved among the Drosophila R2 ribozymes. Surprisingly, uncleaved ribozymes initiated translation at least as well as self-cleaved ones, suggesting that the ribozyme structure, rather than the state of its 5′ terminus, is responsible for translation initiation. Whereas the presence of an upstream rRNA sequence does appear to increase translation yield in vitro, the effect is much smaller in vivo; we therefore do not expect a significant contribution of rRNA-ribozyme fusions toward translation. The translation initiation role of the catalytic RNA in the R2 and R6 elements may extend to other retrotransposons, including those that do not insert into ribosomal DNA, and perhaps to other HDV-like ribozymes.
The R2 ribozymes may also be playing a role in the insertion of R2 cDNA. Upon completing cDNA production, the R2 RT enzyme efficiently switches from its RNA template to the newly made DNA strand to facilitate insertion of the element at new loci (Fig. 3a) (58). This trans-templating reaction is significantly more efficient when the 5′ end of an RNA template has a 5′-hydroxyl group as opposed to a methylguanosine cap analog or a triphosphate (59). As the downstream self-cleavage product of HDV-like ribozymes is a 5′-hydroxyl, ribozyme-processed RNA provides a superior substrate for trans-templating by the protein and may be beneficial for overall retrotransposon insertion efficiency.
Finally, mRNA beginning with a self-cleaved ribozyme is resistant to degradation. Self-cleaved ribozymes lack a 5′-triphosphate and so will avoid detection by the RIG-I pathway (60), and the stable fold (61) formed by HDV-like ribozymes confers resistance to the 5′ end of the retrotransposon transcript from 5′-exonucleases. Higher levels of ribozyme-terminated mRNA relative to HCV IRES-terminated mRNA were observed via quantitative RT-PCR analysis of transfected S2 cell lysates 24 h after equal amounts of RNA were transfected into the cells (supplemental Fig. S3). Although protection from 5′-exonuclease activity is common with transcripts beginning in structured RNAs, the exceptional stability of the HDV-like fold likely better protects the 5′ terminus than a self-cleaved hammerhead ribozyme, for example, which also yields a 5′-hydroxyl product but has comparatively less thermodynamic stability.
Together, these findings point to a model wherein transcription, translation, and retrotransposition are linked in such a way that templates with ribozyme-terminated 5′ ends are highly efficient retrotransposons. The mechanism through which the 5′ end is retained in these non-LTRs is distinct from LTR elements and may provide protection from 5′ end erosion in actively propagating elements.
Recent analysis of the Drosophila R2 ribozymes interpreted the diversity of the sequences as evidence of convergent evolution (6); however, several features of the HDV ribozyme fold suggest otherwise. HDV-like ribozymes are characterized by a highly constrained core comprised of an intricate nested double-pseudoknot with six invariant positions (5, 26, 27). This complex structure yields an information content of at least 55 bits, making it unlikely that the fold has evolved multiple times in eukaryotic genomes, because the likelihood of its appearance in a random sequence is less than one in 4 × 1016. Experimentally, HDV-like ribozymes have never been found during in vitro selections (62, 63) and possess limited accessible sequence space, as observed from reselections of HDV ribozymes (64, 65). Furthermore, the retrotransposon-associated ribozymes presented here (Table 1) exhibit a greater level of sequence conservation than has been observed among other HDV-like ribozymes (5, 21, 27). The P3 region in particular is highly conserved among the rDNA elements, which, combined with the high information content of the fold, indicates a common origin for these sequences. Finally, phylogenetic analysis of various non-LTR reverse transcriptase domains point to vertical transfer of these elements (20, 66–68). As the P2 helix of the R2 ribozymes is thought to code the N terminus of its RT protein, it likely evolved in a similar manner.
Our results show that multiple non-LTR retrotransposons harbor self-cleaving ribozymes that are used to liberate their 5′ termini from upstream transcripts. This work helps explain observations made over 25 years ago that showed that the Drosophila R2 retrotransposons are processed co-transcriptionally and points to multiple roles of self-cleaving ribozymes in retrotransposition. Our findings support a general trend indicating that self-cleaving ribozymes are associated with many retrotransposons, including satellites in newts (69), fungi (70), salamanders (71), schistosomes (44), and crickets (72) and the human L1PA8 non-LTR retrotransposon (21). Recent results (37, 46) and our study suggest that in several organisms, including sea lamprey, schistosomes, and ticks, the hammerhead and HDV-like ribozymes are dedicated to processing of SINEs and LINEs, respectively. The function of the human L1 ribozyme (21) is likely different from the HDV-like ribozymes presented in this work and those associated with SINEs, because the L1 ribozyme maps downstream of the retrotransposon 5′ terminus (typically between the predicted RUNX3 and SRY-binding sites in the 5′ UTR). The 5′ UTR of L1PA8, which was active in primates about 40 Myr ago, has diverged substantially from other L1 UTRs, including the currently active L1Hs (73). The fact that the ribozyme was not retained by the L1PA lineage suggests that the ribozyme was not beneficial for the retrotransposon and raises the possibility that it disrupted the 5′ UTR, suppressing the activity of the retrotransposon. This function would stand in stark contrast to the proposed function of the HDV-like ribozymes presented here. Our finding that self-cleaving ribozymes process the 5′ termini of many different non-LTR retrotransposons and promote translation initiation expands the roles of catalytic RNAs, originally established for self-splicing group I and group II introns, in mobile DNA elements (74).
Supplementary Material
Acknowledgments
We thank M. Jafari for the W1118 strain of D. melanogaster, J. Prescher for use of the IVIS Spectrum system, and S. Sandmeyer, R. Martin, and members of the Lupták laboratory for critical evaluation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM094929. This work was also supported by the University of California, Irvine, and the Pew Trusts.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1–S3, and “Experimental Procedures.”
- LINE
- long interspersed element
- SINE
- short interspersed element
- HDV
- hepatitis delta virus
- IRES
- internal ribosomal entry site
- RRL
- rabbit reticulocyte lysate
- HCV
- hepatitis C virus
- RRL
- rabbit reticulocyte lysate
- RTE
- retrotransposable element.
REFERENCES
- 1. Han J. S. (2010) Mob. DNA 1, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bourque G. (2009) Curr. Opin. Genet. Dev. 19, 607–612 [DOI] [PubMed] [Google Scholar]
- 3. Mourier T., Willerslev E. (2009) Brief. Funct. Genomic Proteomic 8, 493–501 [DOI] [PubMed] [Google Scholar]
- 4. George J. A., Eickbush T. H. (1999) Insect Mol. Biol. 8, 3–10 [DOI] [PubMed] [Google Scholar]
- 5. Webb C. H., Riccitelli N. J., Ruminski D. J., Lupták A. (2009) Science 326, 953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eickbush D. G., Eickbush T. H. (2010) Mol. Cell. Biol. 30, 3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanchez-Luque F. J., Lopez M. C., Macias F., Alonso C., Thomas M. C. (2011) Nucleic Acids Res. 39, 8065–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jakubczak J. L., Burke W. D., Eickbush T. H. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3295–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burke W. D., Müller F., Eickbush T. H. (1995) Nucleic Acids Res. 23, 4628–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kubo Y., Okazaki S., Anzai T., Fujiwara H. (2001) Mol. Biol. Evol. 18, 848–857 [DOI] [PubMed] [Google Scholar]
- 11. Kojima K. K., Fujiwara H. (2003) Mol. Biol. Evol. 20, 351–361 [DOI] [PubMed] [Google Scholar]
- 12. Roiha H., Miller J. R., Woods L. C., Glover D. M. (1981) Nature 290, 749–753 [DOI] [PubMed] [Google Scholar]
- 13. Jamrich M., Miller O. L., Jr. (1984) EMBO J. 3, 1541–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luan D. D., Korman M. H., Jakubczak J. L., Eickbush T. H. (1993) Cell 72, 595–605 [DOI] [PubMed] [Google Scholar]
- 15. Kurzynska-Kokorniak A., Jamburuthugoda V. K., Bibillo A., Eickbush T. H. (2007) J. Mol. Biol. 374, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burke W. D., Malik H. S., Jones J. P., Eickbush T. H. (1999) Mol. Biol. Evol. 16, 502–511 [DOI] [PubMed] [Google Scholar]
- 17. Yang J., Malik H. S., Eickbush T. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7847–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Christensen S. M., Ye J., Eickbush T. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17602–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kierzek E., Christensen S. M., Eickbush T. H., Kierzek R., Turner D. H., Moss W. N. (2009) J. Mol. Biol. 390, 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kojima K. K., Fujiwara H. (2005) Mol. Biol. Evol. 22, 2157–2165 [DOI] [PubMed] [Google Scholar]
- 21. Salehi-Ashtiani K., Lupták A., Litovchick A., Szostak J. W. (2006) Science 313, 1788–1792 [DOI] [PubMed] [Google Scholar]
- 22. Luptak A., Szostak J. W. (2007) in Ribozymes and RNA Catalysis (Lilley D. M. J., Eckstein F. eds) pp. 130–131, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 23. Ferré-D'Amaré A. R., Zhou K., Doudna J. A. (1998) Nature 395, 567–574 [DOI] [PubMed] [Google Scholar]
- 24. Ke A., Zhou K., Ding F., Cate J. H., Doudna J. A. (2004) Nature 429, 201–205 [DOI] [PubMed] [Google Scholar]
- 25. Chen J. H., Yajima R., Chadalavada D. M., Chase E., Bevilacqua P. C., Golden B. L. (2010) Biochemistry 49, 6508–6518 [DOI] [PubMed] [Google Scholar]
- 26. Riccitelli N. J., Lupták A. (2010) Methods 52, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Webb C. H., Luptak A. (2011) RNA Biol. 8, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warren A. M., Hughes M. A., Crampton J. M. (1997) Mol. Gen. Genet. 254, 505–513 [DOI] [PubMed] [Google Scholar]
- 29. Ghesini S., Luchetti A., Marini M., Mantovani B. (2011) J. Mol. Evol. 72, 296–305 [DOI] [PubMed] [Google Scholar]
- 30. Kojima K. K., Fujiwara H. (2004) Mol. Biol. Evol. 21, 207–217 [DOI] [PubMed] [Google Scholar]
- 31. Burke W. D., Malik H. S., Lathe W. C., 3rd, Eickbush T. H. (1998) Nature 392, 141–142 [DOI] [PubMed] [Google Scholar]
- 32. Rezende-Teixeira P., Siviero F., da Costa Rosa M., Machado-Santelli G. M. (2009) Chromosome Res. 17, 455–467 [DOI] [PubMed] [Google Scholar]
- 33. Kawaoka S., Hayashi N., Katsuma S., Kishino H., Kohara Y., Mita K., Shimada T. (2008) Insect Biochem. Mol. Biol. 38, 1058–1065 [DOI] [PubMed] [Google Scholar]
- 34. De la Peña M., Gago S., Flores R. (2003) EMBO J. 22, 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khvorova A., Lescoute A., Westhof E., Jayasena S. D. (2003) Nat. Struct. Biol. 10, 708–712 [DOI] [PubMed] [Google Scholar]
- 36. Jimenez R. M., Delwart E., Lupták A. (2011) J. Biol. Chem. 286, 7737–7743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perreault J., Weinberg Z., Roth A., Popescu O., Chartrand P., Ferbeyre G., Breaker R. R. (2011) PLoS Comput. Biol. 7, e1002031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Been M. D., Perrotta A. T., Rosenstein S. P. (1992) Biochemistry 31, 11843–11852 [DOI] [PubMed] [Google Scholar]
- 39. Nishikawa F., Roy M., Fauzi H., Nishikawa S. (1999) Nucleic Acids Res. 27, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chadalavada D. M., Knudsen S. M., Nakano S., Bevilacqua P. C. (2000) J. Mol. Biol. 301, 349–367 [DOI] [PubMed] [Google Scholar]
- 41. Jeong S., Sefcikova J., Tinsley R. A., Rueda D., Walter N. G. (2003) Biochemistry 42, 7727–7740 [DOI] [PubMed] [Google Scholar]
- 42. Hoskins R. A., Carlson J. W., Kennedy C., Acevedo D., Evans-Holm M., Frise E., Wan K. H., Park S., Mendez-Lago M., Rossi F., Villasante A., Dimitri P., Karpen G. H., Celniker S. E. (2007) Science 316, 1625–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tay W. T., Behere G. T., Batterham P., Heckel D. G. (2010) BMC Evol. Biol. 10, 144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ferbeyre G., Smith J. M., Cedergren R. (1998) Mol. Cell. Biol. 18, 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de la Pena M., Garcia-Robles I. (2010) EMBO Rep. 11, 711–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de la Peña M., García-Robles I. (2010) RNA 16, 1943–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McFarlane C. T., Docker M. F. (2009) Conservation Genet. Resour. 1, 377–380 [Google Scholar]
- 48. Bryan M. B., Zalinski D., Filcek K. B., Libants S., Li W., Scribner K. T. (2005) Mol. Ecol. 14, 3757–3773 [DOI] [PubMed] [Google Scholar]
- 49. Kidd S. J., Glover D. M. (1981) J. Mol. Biol. 151, 645–662 [DOI] [PubMed] [Google Scholar]
- 50. Long E. O., Collins M., Kiefer B. I., Dawid I. B. (1981) Mol. Gen. Genet. 182, 377–384 [DOI] [PubMed] [Google Scholar]
- 51. Kamoshita N., Nomoto A., RajBhandary U. L. (2009) Mol. Cell 35, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berry K. E., Waghray S., Doudna J. A. (2010) RNA 16, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cech T. R. (1993) in The RNA World (Atkins J. F. ed) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Winkler W. C., Nahvi A., Roth A., Collins J. A., Breaker R. R. (2004) Nature 428, 281–286 [DOI] [PubMed] [Google Scholar]
- 55. Fedor M. J., Williamson J. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 399–412 [DOI] [PubMed] [Google Scholar]
- 56. Pfingsten J. S., Costantino D. A., Kieft J. S. (2006) Science 314, 1450–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhu J., Korostelev A., Costantino D. A., Donohue J. P., Noller H. F., Kieft J. S. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 1839–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bibiłło A., Eickbush T. H. (2002) J. Mol. Biol. 316, 459–473 [DOI] [PubMed] [Google Scholar]
- 59. Bibillo A., Eickbush T. H. (2004) J. Biol. Chem. 279, 14945–14953 [DOI] [PubMed] [Google Scholar]
- 60. Liu F., Gu J. (2011) Protein Cell 2, 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Duhamel J., Liu D. M., Evilia C., Fleysh N., Dinter-Gottlieb G., Lu P. (1996) Nucleic Acids Res. 24, 3911–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tang J., Breaker R. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5784–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Salehi-Ashtiani K., Szostak J. W. (2001) Nature 414, 82–84 [DOI] [PubMed] [Google Scholar]
- 64. £egiewicz M., Wichłacz A., Brzezicha B., Ciesiołka J. (2006) Nucleic Acids Res. 34, 1270–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nehdi A., Perreault J. P. (2006) Nucleic Acids Res. 34, 584–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Malik H. S., Burke W. D., Eickbush T. H. (1999) Mol. Biol. Evol. 16, 793–805 [DOI] [PubMed] [Google Scholar]
- 67. Blesa D., Gandía M., Martínez-Sebastián M. J. (2001) Mol. Biol. Evol. 18, 585–592 [DOI] [PubMed] [Google Scholar]
- 68. Burke W. D., Malik H. S., Rich S. M., Eickbush T. H. (2002) Mol. Biol. Evol. 19, 619–630 [DOI] [PubMed] [Google Scholar]
- 69. Epstein L. M., Gall J. G. (1987) Cell 48, 535–543 [DOI] [PubMed] [Google Scholar]
- 70. Saville B. J., Collins R. A. (1990) Cell 61, 685–696 [DOI] [PubMed] [Google Scholar]
- 71. Green B., Pabón-Peña L. M., Graham T. A., Peach S. E., Coats S. R., Epstein L. M. (1993) Mol. Biol. Evol. 10, 732–750 [DOI] [PubMed] [Google Scholar]
- 72. Rojas A. A., Vazquez-Tello A., Ferbeyre G., Venanzetti F., Bachmann L., Paquin B., Sbordoni V., Cedergren R. (2000) Nucleic Acids Res. 28, 4037–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Khan H., Smit A., Boissinot S. (2006) Genome Res. 16, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Belfort M., Derbyshire V., Parker M. M., Cousineau B., Lambowitz A. M. (2002) in Mobile DNA II (Craig N. L., Craigie R., Gellert M., Lambowitz A. M. eds) pp. 761–783, American Society for Microbiology, Washington, D. C [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.