Background: The role of ATP synthase in blood stages of malaria parasites has been unclear.
Results: Canonical subunits were targeted to the mitochondrion, could not be deleted by gene disruption, and were present in large complexes.
Conclusion: Plasmodium ATP synthase is likely essential and forms a dimeric complex.
Significance Composition, properties, structure, and drugability of the complex should be fully investigated.
Keywords: ATP Synthase, Bioenergetics, Malaria, Mitochondria, Parasite Metabolism, Plasmodium, Oxidative Phosphorylation
Abstract
The rotary nanomotor ATP synthase is a central player in the bioenergetics of most organisms. Yet the role of ATP synthase in malaria parasites has remained unclear, as blood stages of Plasmodium falciparum appear to derive ATP largely through glycolysis. Also, genes for essential subunits of the FO sector of the complex could not be detected in the parasite genomes. Here, we have used molecular genetic and immunological tools to investigate the localization, complex formation, and functional significance of predicted ATP synthase subunits in P. falciparum. We generated transgenic P. falciparum lines expressing seven epitope-tagged canonical ATP synthase subunits, revealing localization of all but one of the subunits to the mitochondrion. Blue native gel electrophoresis of P. falciparum mitochondrial membranes suggested the molecular mass of the ATP synthase complex to be greater than 1 million daltons. This size is consistent with the complex being assembled as a dimer in a manner similar to the complexes observed in other eukaryotic organisms. This observation also suggests the presence of previously unknown subunits in addition to the canonical subunits in P. falciparum ATP synthase complex. Our attempts to disrupt genes encoding β and γ subunits were unsuccessful, suggesting an essential role played by the ATP synthase complex in blood stages of P. falciparum. These studies suggest that, despite some unconventional features and its minimal contribution to ATP synthesis, P. falciparum ATP synthase is localized to the parasite mitochondrion, assembled as a large dimeric complex, and is likely essential for parasite survival.
Introduction
F1FO-ATP synthase is a nanomotor that generates ATP in mitochondria. The enzyme has two domains as follows: F1 is the hydrophilic domain, made up of subunits α, β, γ, δ, and ϵ, and FO is the membrane-associated domain and comprises subunits a, b, c, d, F6, and OSCP2 (1). The mechanism of ATP synthesis and hydrolysis has been well studied and is conserved from bacteria to mammals (2). A transmembrane proton gradient, usually generated by the mitochondrial respiratory chain enzymes, is utilized by ATP synthase to synthesize ATP. During this process, protons from the intermembrane space are transported to the matrix through aqueous half-channels in subunit a (3) that are connected by the movement of a negatively charged group on the c subunits, which form a rotational ring. Thus, the chemical potential energy of the proton gradient is converted into the mechanical energy of the oligomeric c ring rotation by this arrangement of the a subunit and c-ring in the mitochondrial membrane. The rotation of the c ring drives the rotation of the central stalk (γ, δ, and ϵ), which is surrounded by the α3β3 hexameric cap. The clockwise movement of the central stalk subunits causes a conformational change in the individual β subunits. These conformational changes are accompanied by the binding of ADP and Pi to the catalytic sites in the β subunit, followed by formation and release of ATP. The peripheral stalk (consisting of b, d, F6, and OSCP subunits) acts as a stator, holding the α3β3 hexamer in place during the movement of the central stalk.
The role of the ATP synthase in Plasmodium falciparum has remained unclear for decades now. Studies using isolated mitochondria from asexual and sexual blood stages suggested that oxidative phosphorylation is virtually absent in P. falciparum (4, 5). Studies have also shown that the major source of ATP in the parasite is the anaerobic glycolysis pathway (6–9). Furthermore, all sequenced apicomplexan parasites, including P. falciparum, seem to lack critical subunits of the enzyme. The missing subunits include subunit a, which plays an essential role in proton translocation, as well as key peripheral stalk subunits b, d, and F6, which function as a stator. In the absence of these subunits, it is inconceivable that apicomplexan ATP synthase could synthesize ATP. Intriguingly, genes encoding the same subunits also appeared to be absent from the ciliate Tetrahymena thermophila. The ciliates, along with dinoflagellates and apicomplexan parasites, form the three evolutionary lineages of the phylum Alveolata. However, despite seemingly missing the critical subunits, robust oxidative phosphorylation has clearly been demonstrated in T. thermophila mitochondria (10–14). In a recent structural and proteomic study (15), we have identified many subunits of the Tetrahymena F1FO-ATP synthase, including highly divergent candidates for the a, b, and d subunits. Our findings raised the possibility that the corresponding subunits in P. falciparum may also be highly divergent and therefore could not be identified by conventional bioinformatics tools. The presence of divergent subunits would likely mean that the malaria parasites encode all the subunits that are needed to assemble a functional ATP synthase.
In this study, we addressed some fundamental questions. (a) Is the ATP synthase essential for the growth of blood stage P. falciparum parasites? (b) Are the putative ATP synthase subunits targeted to the mitochondrion. (c) Are they assembled into a protein complex? Our results show that the enzyme is probably essential for the blood stage life cycle of the parasite and is assembled into a dimeric complex in mitochondrial membranes, as seen in other eukaryotic organisms (16–20).
EXPERIMENTAL PROCEDURES
Parasite Lines, Culture, Transfection, and Selection Methods
P. falciparum D10 strain parasites were used for knock-out (21), FKBP destabilization domain protein knockdown (22), allelic exchange (β (PFL1725w), and γ (PF13_0061) subunits) and episomal expression of OSCP (MAL13P1.47), α (PFB0795w), and c (MAL7P1.340) proteins. P. falciparum 3D7attB parasites (23) were used for localization studies of γ, δ (PF11_0485), and ϵ (MAL7P1.75) subunits. Parasites were cultured at 5% hematocrit of human O+ erythrocytes in RPMI 1640 medium supplemented with 300 mg/liter l-glutamine, 10 mg/liter hypoxanthine (Sigma), 15 mm HEPES (Hyclone), 0.225% NaHCO3 (Cellgro), 0.5% Albumax II (Invitrogen), and 50 μg/ml gentamicin (Cellgro), under a gas mixture of 5% O2, 5% CO2, and 90% N2, according to a modification of the method of Trager and Jensen (24). Parasites were transfected as described previously (25). Briefly, 0.2-cm electroporation cuvettes were loaded with 0.3 ml of 50% infected erythrocytes (5–10% parasitemia with at least 5% ring stages) and 50 μg of plasmid DNA in incomplete cytomix solution. Electroporation conditions were 0.31 kV and 960 microfarads. For knock-out studies using pCC1 (21) or pUF-1 (26) vectors, positive selection of the parasites was carried out in media containing 5 nm WR99210 (27) or 1.5 μm compound DSM1 (28), respectively. 2.5 μm fluorocytosine was used for negative selection. Three cycles of growth with no selective agent for 4 weeks followed by growth with the selective agent (“off-on cycles”) were carried out to increase the chance of integration. For knockdown experiments using FKBP destabilization domain, the parasites were selected with WR99210 and grown under the continuous presence of Shld1 (gift from Dr. D. E. Goldberg, Washington University School of Medicine) (29). For allelic replacement of the β and γ subunit genes, parasites were selected with 5 nm WR99210; two drug off-on cycles were used to obtain 3′ integrants.
Construction of Vectors
For making the knock-out construct for the ATP synthase β subunit gene, a 5′ insert (892 bp) with EcoRI (5′) and NcoI (3′) restriction enzyme sites and a 3′ insert (800 bp) with SpeI (5′) and SacII (3′) sites were amplified from D10 genomic DNA (primers used for this and subsequent PCR amplification are listed in supplemental Table S1). The inserts were cloned in the pSC-B-amp/kan Blunt PCR Cloning Vector (StrataClone kit from Stratagene), and their sequences were verified. The inserts were then cloned into the pCC1 and the pUF-1 vectors. A similar approach was used for making the γ subunit knock-out construct, with 3′ and 5′ inserts of 578 and 553 bp, respectively. The final vectors were verified by sequencing and restriction enzyme digestion.
For the allelic replacement of the 3′ ends of β and γ subunit genes, a modified pCC1 vector3 was used. The modified vector does not contain the hsp86 5′ UTR and has an incomplete P. falciparum malate-quinone oxidoreductase-3× hemagglutinin epitope tag (MQO-3×HA) fusion gene cloned between KpnI and SalI sites, with an XhoI site between the partial MQO coding sequence and the 3×HA tag sequence. To generate 3′ integrants, 3′ partial ORF of β (986 bp) and γ (998 bp) were cloned in-frame with the HA tag, using the KpnI and XhoI sites. For FKBP knockdown studies, an 3×HA-FKBP insert was cloned into the vector described above in place of the 3×HA tag using XhoI and SalI. Before cloning into the modified pCC1 vector, the 3′ inserts and 3×HA-FKBP DNA fragment were cloned into pSC-B-amp/kan Blunt PCR cloning vector, and their sequences were verified.
For exploring alternate promoters for ectopic expression, 870 bp of the 5′ UTR immediately upstream of the mitochondrial ribosomal protein L2 (mRPL2, PF11_0337) or 948 bp of the 5′ UTR immediately upstream of the cytochrome c1 heme lyase (CC1HL, PFL0180w) gene were cloned between the ApaI and AvrII sites of the pLN plasmid (which removes the calmodulin UTR) (23). The ϵ gene was cloned between the AvrII and BsiWI sites of the pLN plasmids (with an mRPL2, CC1HL, or calmodulin promoter), which encode an in-frame GFP tag between the BsiWI and MspCI sites.
For localization studies, ATP synthase genes (α, γ, δ, ϵ, OSCP, and c) were cloned between the ApaI and AvrII sites of the pLN plasmid containing the calmodulin promoter or the mRPL2 promoter. In the pLN plasmid with mRPL2 promoter, ATP synthase genes were in frame with an 3×HA tag cloned between the BsiWI and MspCI sites. In the pLN plasmid with the calmodulin promoter, the genes were in-frame with a GFP tag between the BsiWI and MspCI sites. All inserts were initially cloned into pSC-B-amp/kan, and their sequences were verified.
Recombinant Antigens, Immunizations, and ELISA
The Plasmodium genes encoding the full-length β (1608 bp) and γ (936 bp) subunits were cloned in pSC-B-amp/kan, and their sequences were verified. The inserts were then subcloned into pET-28a(+) (Novagen). The β subunit was cloned between NheI and XhoI sites, although the γ subunit was cloned between NdeI and XhoI sites, resulting in an N-terminal His6 tag. Each plasmid was then expressed in BL21 Escherichia coli cells. Both proteins were present as inclusion bodies and were solubilized with 4 m urea (β) and 6 m guanidine hydrochloride (γ). The denatured proteins were purified using a Ni2+-nitrilotriacetic acid super flow (Qiagen) column. The β subunit was initially dialyzed against Tris buffer (20 mm Tris, 50 mm NaCl, pH 8.0) containing 4 m urea, and the urea concentration was gradually reduced to zero. The γ subunit was initially dialyzed against Tris buffer containing 6 m guanidine and 1 mm DTT, and the concentrations of guanidine and DTT were also gradually reduced to zero. Protein concentration was measured by the Bradford assay. Five mice, each 6–8 weeks old were used for immunizations. Each mouse was injected subcutaneously with 50 μg of the recombinant protein emulsified in incomplete Freund's adjuvant. Two boosters were administered at 3-weeks intervals in incomplete Freund's adjuvant. Sera were titered by ELISA. Briefly, 96-well plates were coated with 1 μg/ml of antigen in 0.1 m sodium carbonate, pH 9.6 (50 μl/well), by overnight incubation at 4 °C. After two washes with PBS, 0.1% Tween 20, the plates were blocked in the same buffer for 1–2 h at room temperature, and then washed. Diluted sera (1:1000, 1:10,000, and 1:100,000) were added (50 μl/well) and incubated for 1–2 h at room temperature and then washed. HRP-conjugated secondary goat anti-mouse HRP antibody (1:3000) was added (50 μl/well) and incubated for 1 h at room temperature. After washing, the substrate 3,3′,5,5′-tetramethylbenzidine was added to the plate and incubated at room temperature. When the color development was intense, the reaction was stopped with 50 μl of 1 m sulfuric acid, and the plate well absorbances were read at 450 nm.
Fluorescence Microscopy
Mixed stage parasitized erythrocyte cultures at 3–4% parasitemia were used. MitoTracker Red CM-H2XRos (Invitrogen) was added to a final concentration of 60 nm to 1 ml of the culture, which was then incubated at 37 °C for 30 min. These cells were washed with PBS (three times) and fixed by the method of Tonkin et al. (30). After washing with PBS (three times), the parasitized erythrocytes were permeabilized with 0.1% Triton X-100 in PBS for 10 min. After another wash with PBS (three times), the erythrocytes were treated with 0.1 mg/ml sodium borohydride in PBS for 10 min. The permeabilized parasite samples were then blocked in 3% BSA in PBS for 1 h. Anti-HA antibody (Santa Cruz Biotechnology, Santa Cruz, CA) (1:100 dilution) was added and incubated for 1–2 h in 1% BSA in PBS at room temperature. Parasites were washed with PBS (three times) to remove excess antibody. AlexaFluor 488 goat anti-mouse secondary antibody (Molecular Probes, Eugene, OR) was added at 1:350 dilution and incubated for 1–2 h in 1% BSA in PBS at room temperature. After washing twice with PBS, DAPI (Sigma) was added, and the sample was incubated for 5 min and then washed with PBS. Parasite samples were resuspended with equilibration buffer (Slowfade, Molecular Probes) for 10 min. Finally, the parasite sample pellet was resuspended with an equal volume of antifade (Slowfade, Molecular Probes) glycerol buffer, and 3 μl of the mix was placed on a glass slide, mounted with Fluoromount (Southern Biotech, Birmingham, AL), and sealed with a coverslip. After letting it mount overnight, images were taken with an Olympus BX60 epi-fluorescence microscope system.
One-dimensional Blue Native-PAGE (BN-PAGE)
A mitochondrially enriched parasite extract (mitochondria) was prepared as described previously (31). Mitochondria (200 μg of protein) were resuspended in water and pelleted by centrifugation at 14,000 rpm for 10 min at 4 °C in a microcentrifuge. The pellet was resuspended in mitochondrial solubilization buffer (200 mm BisTris, 200 mm NaCl, pH 7.0) with fungal protease inhibitor mixture (Sigma). Detergent concentrations were adjusted to 5 μg of digitonin per μg of mitochondrial protein by addition of a 20% stock solution of digitonin. Samples were incubated overnight at 4 °C and were centrifuged at 14,000 rpm for 15 min at 4 °C in a microcentrifuge. Coomassie dye from a 5% G-250 stock suspension was added to the supernatant to give a dye/detergent ratio of 1:8 by weight. The sample (∼30–40 μg/lane) was loaded in a 4–16% BN-PAGE gradient gel (Invitrogen), and the gel was run for 2 h at room temperature with a constant 150 V. After 30 min, the dark blue cathode buffer (Invitrogen) was replaced by light blue cathode buffer (Invitrogen) and was run for an additional 1 h, 30 min. At the end of the run, gels were cut into strips containing one or more lanes for use in immunoblots (see under :Western Blot” below), two-dimensional PAGE (see below), or restaining with Coomassie dye to visualize protein.
Two-dimensional BN/SDS-PAGE
The two-dimensional BN/SDS-PAGE was done as described by Wittig et al. (32). The one-dimensional BN-PAGE strip was incubated in anode/running buffer (Invitrogen) with 1% SDS for 15 min. The strip was then slid into a second dimension NuPAGE® Novex 4–12% BisTris gel (Invitrogen). SDS-PAGE was carried out using NuPAGE® MES running buffer (Invitrogen) in the XCell SureLock mini-cell apparatus (Invitrogen) at 100 V for ∼2 h.
Western Blot
One-dimensional BN gels were electrophoretically transferred to a 0.45-μm PVDF membrane (GE Water & Process Technologies) using the NuPAGE® transfer buffer (Invitrogen) at 25 V overnight. The PVDF membrane was fixed with 8% acetic acid for 15 min and air-dried for 30 min. The dried membrane was washed with methanol several times to remove the Coomassie Blue dye. After rinsing with deionized water, the membrane was blocked with 5% skim milk in TBS, 0.02% Tween and was washed for 10 min (three times) in TBS, 0.02% Tween. The membrane was cut into strips and probed with 1:2000 dilution of monoclonal HA antibody (Santa Cruz Biotechnology), 1:2000 polyclonal β, or 1:1000 polyclonal γ sera for 2 h at RT. The strips were washed with TBS, 0.02% Tween (three times) and probed with 1:2000 dilution of rabbit anti-mouse-IgG HRP (Thermo Scientific (Pierce)). After washing with TBS, 0.02% Tween (three times) to remove excess antibody, signal was developed using the SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific, Rockford, IL). Two-dimensional BN/SDS-polyacrylamide gels were transferred to a 0.2-μm PVDF membrane using the Tris-glycine transfer buffer (25 mm Tris, 192 mm glycine, and 20% methanol). After transfer, the membrane was blocked, and subsequent steps were carried out as described above. For immunoblotting with parasite lysates, parasites were harvested at 5–6% parasitemia, and saponin was lysed (0.02% saponin in PBS). The saponin-lysed pellet was washed twice in PBS and resuspended in 50 μl of 1× loading buffer (62.5 mm Tris-Cl, pH 6.8, 10% glycerol, 2% SDS, 0.01% bromphenol blue and 5% 2-mercaptoethanol), and one-fourth of the sample was used for each run.
RESULTS
Plasmodium ATP Synthase Is Targeted to the Mitochondrion
All sequenced Plasmodium genomes encode putative F1 subunits α, β, γ, δ, and ϵ, similar to the corresponding subunits in mammalian and fungal ATP synthases (Table 1). In contrast, many subunits of the FO domain could not be identified in malaria species, including subunits a and b, which are essential for proton translocation and energy coupling in the ATP synthase. In fact, only two FO subunits are annotated in the Plasmodium data base as follows: subunit c and the OSCP (Table 1).
TABLE 1.
Putative F1FO subunits of P. falciparum identified at the genomic level compared with the subunits of three well characterized ATP synthases
Designations in parentheses are locus names assigned by sequencing consortia.
| Sector | Bacterial |
Mitochondrial enzymes |
||
|---|---|---|---|---|
| E. coli | Saccharomyces cerevisiae | Bos taurus | P. falciparum | |
| F1 | α | α (Atp1p) | α | α (PFB0795w) |
| β | β (Atp2p) | β | β (PFL1725w) | |
| γ | γ (Atp3p) | γ | γ (PF13_0061) | |
| ϵ | δ (Atp16p) | δ | δ (PF11_0485) | |
| ϵ (Atp15p) | ϵ | ϵ (MAL7P1.75) | ||
| FO | a | a (Atp6p) | a | |
| b | b (Atp4p) | b | ||
| c | c (Oli1p/Atp9p) | c | c (MAL7P1.340) | |
| δ | OSCP (Atp5p) | OSCP | OSCP (MAL13P1.47) | |
| Subunit 8 (Atp8p) | A6L/subunit 8 | |||
| d (Atp7p) | d | |||
| e (Tim11p) | e | |||
| f (Atp17p) | f | |||
| g (Atp20p) | g | |||
| h (Atp14p) | F6 | |||
| i/j (Atp18p) | ||||
| k (Atp19p) | ||||
| Inhibitor | Inh1p | IF1 | ||
To our knowledge, the localization of the predicted subunits of Plasmodium ATP synthase has not been investigated. The ATP synthase complex is expected to be found in the mitochondrion; however, it is important to note that the enzyme pyruvate dehydrogenase, which is targeted to the mitochondrion in most eukaryotes, where it plays a key role in linking glycolysis to the TCA cycle, is targeted solely to the apicoplast in malaria parasites (33). Furthermore, in animals, the mitochondrial ATP synthase has been shown to be ectopically targeted to the plasma membrane of different cell types, including mammalian neuronal cells (34) and hepatic cells (35), in addition to mitochondria. We chose to investigate the localization of the canonical ATP synthase subunits α, β, γ, δ, ϵ, c, and OSCP. Initially, we attempted to express the subunits with a C-terminal GFP fusion with expression driven by a constitutively strong calmodulin promoter, but parasites recovered after transfection did not express the recombinant gene. We reasoned that the strong promoter activity might be deleterious in the case of mitochondrially targeted gene products. To achieve a more balanced expression of the ATP synthase subunits, we decided to investigate the use of P. falciparum promoter regions that had been determined to drive constitutive but moderate expression of their products during the blood stage cycle of the parasite (36, 37). When the first two candidate promoters, the 5′-untranslated regions of the genes encoding mitochondrial ribosomal protein L2 (mRPL2, PF11_0337) and cytochrome c1 heme lyase (CC1HL, PFL0180w), were used to drive expression of the ϵ subunit of ATP synthase, we found that a larger number of the parasites with the 5′ UTR of mRPL2 expressed GFP when compared with parasites with the 5′ UTR of mCC1HL (data not shown).
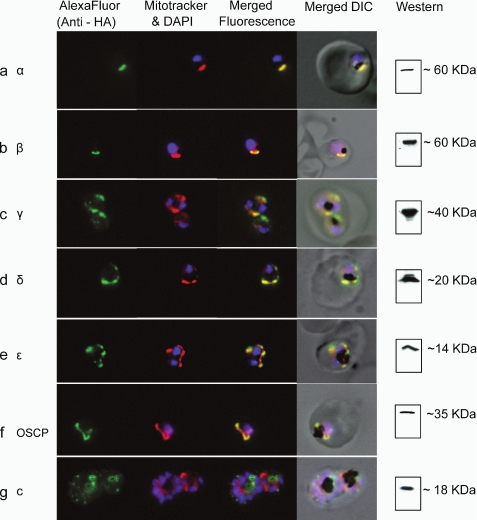
We prepared expression constructs in which the α, γ, δ, ϵ, OSCP, and c subunit genes were fused with a C-terminal 3×HA tag sequence with expression driven by the mRPL2 promoter. Upon transfection, all of these HA-tagged subunits, except the putative c subunit, localized to the mitochondrion (Fig. 1, a and c–f). The localization pattern of the HA-tagged c subunit appeared to be punctate spots throughout the parasite cytoplasm with very little mitochondrial staining (Fig. 1g); these punctate regions did not have an obvious colocation with a known organelle or structure. An alternative approach, partial allele exchange by single crossover recombination, was employed in an attempt to obtain expression of single-copy HA-tagged β and γ subunit genes under control of their own promoters. Using this method, we successfully generated parasites expressing β-3×HA, which is also targeted to the mitochondrion (Fig. 1b). In two attempts, we did not recover γ-3×HA-expressing parasites after transfection. In known ATP synthase structures, the c subunits assemble to form an oligomeric c ring and, along with the a subunit, play a key role in proton translocation. The putative Plasmodium c subunit is a small protein (18.5 kDa) and is extremely hydrophobic. It is possible that the 3×HA tag at the C terminus is interfering with the proper targeting and/or insertion of the protein into the inner mitochondrial membrane. Other possibilities are that the P. falciparum c subunit is not targeted to the mitochondrion in blood stage parasites or that it has been co-opted for another function. Additional investigation is needed to determine the localization and function of the putative c subunit.
FIGURE 1.
Localization of epitope-tagged ATP synthase subunits detected using immunofluorescence microscopy. Immunofluorescence of α (a), γ (c), δ (d), ϵ (e), OSCP (f), and c (g) subunits expressed under control of the mRPL2 promoter (see under “Experimental Procedures”). Expression of the β subunit (b) was under the control of its own promoter (see under “Experimental Procedures” and Fig. 5). Green fluorescence, anti-HA antibody (AlexaFluor 488-conjugated secondary antibody). Red fluorescence, Mitotracker Red CM-H2XRos. Blue fluorescence, DAPI (4′-6-diamidino-2-phenylindole). DIC, differential interference contrast microscopy.
Individual ATP Synthase Subunits Are Assembled into a Dimeric Complex
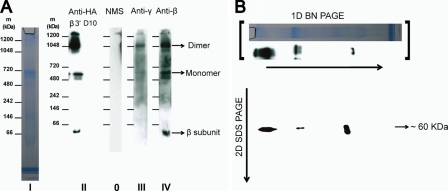
In most eukaryotes studied, the ATP synthase subunits are assembled as a dimeric complex in the mitochondrial inner membrane, which is the predominant form of the enzyme. If the key a and b subunits are in fact absent, it is unlikely that a conventional ATP synthase complex could be assembled in P. falciparum. Having established that most, if not all, of the putative subunits of the parasite ATP synthase are targeted to the mitochondrion, we analyzed mitochondria prepared from wild type P. falciparum D10 strain and from a β-3×HA transgenic clone, using BN-PAGE, for the presence of complexes containing the β and γ subunits using specific antibodies. Anti-HA sera clearly recognized two high molecular mass bands, one at ∼1.1 MDa and another at ∼550 kDa and a low molecular mass band at ∼60 kDa in immunoblots of BN-polyacrylamide gel strips (Fig. 2A, strip II). The 60-kDa band is the size of the monomeric β subunit, whereas the higher molecular mass bands of 550 kDa and 1.1 MDa are consistent with the sizes of monomeric and dimeric forms of the complex seen in other species. When we also probed BN-PAGE strips with polyclonal β and γ antisera, they too recognized the same high molecular mass bands, confirming the presence of a complex containing ATP synthase subunits (Fig. 2A, strips III and IV). To further confirm the presence of the β subunit in the high and low molecular mass bands, a mitochondrial sample from the β-3×HA transgenic clone was subjected to two-dimensional BN-PAGE/SDS-PAGE, blotted, and probed with monoclonal HA antibody. We indeed observed the expected immunoreactive spots at ∼60 kDa in the second dimension below each of the positions of the putative monomer and dimer complexes and the individual β subunit in the first dimension (Fig. 2B). It is important to note that the amount of this complex is very low (note the absence of a specific Coomassie-stained band in strip I corresponding to the high molecular mass complex) and could only be detected by using the most sensitive chemiluminescent reagents.
FIGURE 2.
BN-PAGE analysis of complex formation. A, P. falciparum mitochondria solubilized with digitonin were run on a BN-polyacrylamide gel (4–16% gradient) and stained with colloidal Coomassie Blue (strip I); a two-lane strip from a BN gel that was run with mitochondria isolated from β-3×HA transgenic parasites and with mitochondria isolated from parental D10 parasites and probed with monoclonal anti-HA antibody (strip II); BN gel strips run with mitochondria from β-3×HA parasites and probed with polyclonal γ and β antisera, respectively (strips III and IV); BN strip probed with normal mouse serum (strip 0, NMS). Digitonin was used at a concentration of 5 μg of detergent/μg of protein, and ∼30–40 μg of protein was loaded in each lane. Strips II–IV are blots from separate BN-polyacrylamide gels, but comparison of marker lanes indicated that the runs and transfers were highly reproducible, and they have the same marker protein size scales, within the uncertainty of the technique. B, strip from a one-dimensional (1D) BN gel was placed on top of a 4–12% SDS-polyacrylamide gel and was run in the second dimension. The SDS gel was then blotted onto a PVDF membrane and was probed with monoclonal anti-HA antibody. The gel lane and anti-HA blot strip shown above the two-dimensional (2D) blot are reproduced from A (strips I and II) to indicate the approximate position of protein (which is obscured by excess dye in the original unstained BN polyacrylamide gels) and of anti-HA immunoreactive protein in the first dimension.
β and γ Subunit Genes of ATP Synthase Appear to Be Essential
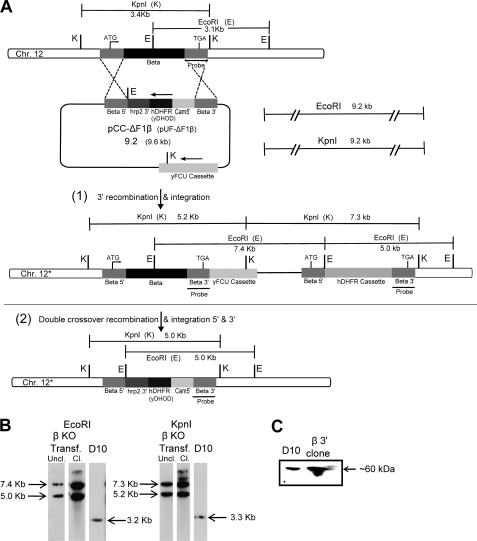
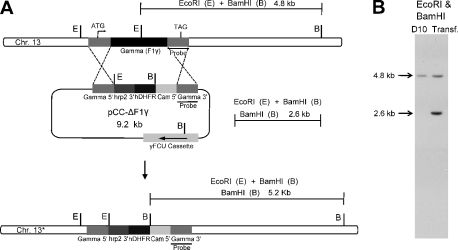
Because the major source of ATP in parasites is glycolysis, we investigated the essentiality of ATP synthase in blood stages. We selected the β and γ subunits for our knock-out experiments as these are very critical subunits of the enzyme, and their high degree of sequence similarity to the corresponding F1 subunits from other species leaves little doubt that they are true orthologues. The β subunit contains the catalytic sites where ATP synthesis/hydrolysis takes place, whereas the γ subunit is the main part of the central stalk whose rotation leads to conformational changes in the β subunit. We used the classical gene disruption approach, which involves the attempted replacement of all or part of the gene by double crossover recombination. To decrease the possibility that our experimental results could be due to an unknown interaction of the target gene product with the pathway blocked by the selective agent or other artifact, we transfected parasites with two different disruption plasmid constructs for each gene, one selected using the antifolate WR99210 and one selected by DSM1 (26, 28), which blocks the pyrimidine biosynthesis pathway (Fig. 3A). Despite three selective agent off-on cycles, we did not observe any double crossover integrants for either the β or the γ subunit gene with either of the two types of transfection constructs. After cycling, we did obtain parasites with a 3′ integration in the β subunit gene using the WR99210-selected plasmid, pCC1 (Fig. 3A-1), as indicated by the Southern analysis (Fig. 3B). In these parasites, the 3′ UTR following the β subunit gene has been truncated by the integration; therefore, we wanted to see if the shorter 3′ UTR affects protein expression. A Western blot of a lysate from a clone isolated from the 3′ integrant parasites demonstrates that this β subunit gene is transcribed and translated (Fig. 3C). We did not see any evidence of 3′ integration in the disruption experiments for the γ subunit gene (Fig. 4, A and B). These experiments suggest that both these genes are essential in P. falciparum blood stages. To rule out the possibility that the genes encoding the subunits were inaccessible to genetic recombination, we carried out allelic replacement experiments using partial 3′ ORFs of β and γ genes. The allelic replacement constructs were designed to introduce a 3×HA tag at the C terminus of the β (Fig. 5A) and γ gene products, allowing detection of expression using immunological techniques. After two selective off-on cycles, integrants were obtained in the β gene and were cloned (Fig. 5B). Protein expression was confirmed by immunofluorescence assay and Western blot (Fig. 1b). However, in the case of the γ gene, the level of 3′ integration detected by PCR in the culture appeared to be a very minor fraction, and clones of integrants could not be obtained.
FIGURE 3.
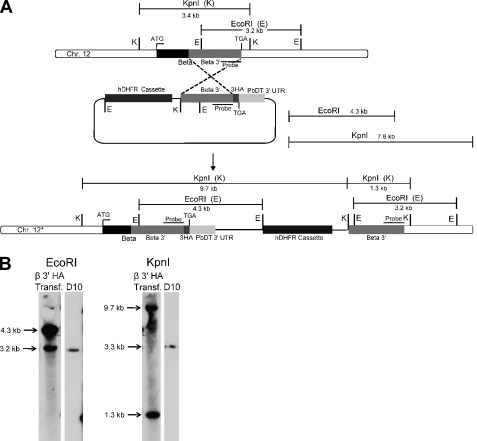
ATP synthase β subunit gene could not be disrupted. A, diagram of the β gene locus and the vectors used for gene disruption by double crossover recombination at the β gene. In the disruption constructs, the hDHFR/yDHOD cassette was flanked by 5′ and 3′ inserts of the β gene (see under “Experimental Procedures”); the plasmids also contain the negative selectable marker yFCU. With pCC-ΔF1β, 3′ integration at the β gene (diagrammed in A(1)) was observed (B); however, no double crossover replacement (A(2)) was observed with either knock-out construct. B, Southern blot analysis of genomic DNA from transfected parasites digested separately with EcoRI and KpnI and using the probe indicated in A shows 3′ integration in uncloned (Uncl.) and cloned (CL) β KO transgenic parasites (EcoRI, 7.4, 5.0 kb; KpnI, 7.3, and 5.2 kb). Southern blot analysis of D10 genomic DNA cut with EcoRI and KpnI results in 3.2- and 3.3-kb bands, respectively. White spaces between the lanes in B indicate the cropping of irrelevant and unused lanes for clarity. C, Western blot of parasite lysates of D10 and a β KO 3′ integrant clone probed with β polyclonal antibody confirms expression of the β subunit protein.
FIGURE 4.
ATP synthase γ subunit gene could not be disrupted. A, schematic of the knock-out strategy utilized for the attempted disruption of the γ gene. B, Southern blot of DNA recovered from transfected parasites after three cycles of selection (see under “Experimental Procedures”) and digested with EcoRI and BamHI shows a 4.8-kb fragment, as in the DNA from wild type D10 parasites, and a 2.6-kb band indicative of the continued presence of the episomal γ construct.
FIGURE 5.
Integration of an epitope tag sequence at the end of the β gene, indicating locus accessibility. A, schematic of the β gene locus and the vector used for the integration of a 3×HA tag sequence at the 3′ end of the β gene by allelic exchange. B, Southern blot analysis of genomic DNA from transfected parasites digested with EcoRI/KpnI shows 3′ integration in β 3′ HA transgenic parasites (EcoRI, 4.3 and 3.2 kb; KpnI, 9.7 and 1.3 kb). A Southern blot of D10 wild type genomic DNA with EcoRI and KpnI results in 3.2- and 3.3-kb bands, respectively. Expression of the tagged protein is shown in Fig. 1b.
In addition, we attempted knockdown experiments with an FKBP destabilization domain (22). The constructs were designed to introduce a sequence encoding a dual 3×HA -FKBP tag at the 3′ end of the β and γ genes (supplemental Fig. S1, A and C). After two selection cycles, the fraction of 3′ integrants was undetectable for the β and γ genes (supplemental Fig. S1, B and D). We were thus unable to observe a knockdown phenotype. In an assembled ATP synthase, γ forms the central stalk, and both the N and C termini of the protein are surrounded by the α3β3 hexameric cap. It is possible that introducing a 3×HA/3×HA-FKBP tag at the C-terminal end of the γ gene interferes with the rotation of the central stalk and hence affects the function of ATP synthase. This could be a reason why the parasites are refractory to introduction of a domain at the C terminus of the γ gene.
DISCUSSION
The mitochondrion in malaria parasites is highly unusual in many respects; it has the smallest genome encoding just three proteins and rRNA genes in pieces (38–41); it possesses a unique branched tricarboxylic acid metabolism with no contributions from a pyruvate dehydrogenase (33, 42); and it does not seem to be a significant source of ATP production in blood stages of the parasite (6–9). Yet the Plasmodium mitochondrion clearly has a functional electron transport chain that is a validated target for antimalarial drugs (43–45). We have previously shown that the main function of the mitochondrial electron transport chain in blood stages of P. falciparum is to service the essential pyrimidine biosynthesis enzyme dihydroorotate dehydrogenase (46). In contrast, the main function of mitochondrial electron transport chain in most other eukaryotic organisms is to generate an electrochemical proton gradient across the inner membrane of the mitochondrion that is utilized by the ATP synthase to drive the synthesis of ATP (a process called oxidative phosphorylation). However, because oxidative phosphorylation is not a significant source of ATP in P. falciparum blood stages, the functional significance of ATP synthase has remained intriguing. Given the apparent absence of genes encoding key subunits (a, b, d, and F6) of the enzyme at the genomic level, we earlier proposed that the absence of these subunits may correlate with the dearth of oxidative phosphorylation in Plasmodium mitochondria (44, 47). Of the missing subunits, subunit a plays an important role in proton translocation, and subunit b functions as the backbone of the peripheral stator and anchors it to subunit a and the inner membrane. If these subunits are indeed missing, a conventional ATP synthase could not be assembled, let alone be fully functional as a proton-coupled ATP synthase. In any event, in the apparent absence of its expected primary function, it seemed possible that the enzyme might not be fulfilling a vital role, and the genes for the individual subunits might be dispensable for the growth of intraerythrocytic parasites. This view was further advanced recently by the demonstration that the gene encoding the mitochondrial NADH dehydrogenase could be deleted in blood stages of a rodent malaria parasite (48). Despite several attempts, however, we could not disrupt either of the genes encoding the catalytic β subunit or the rotary γ subunit of the enzyme. Thus, it appears that these subunits are serving an important function in the blood stage parasites. We attempted to corroborate the gene knock-out results by generating parasites expressing conditional protein knockdowns for the β and γ subunits, using fusions to an FKBP destabilization domain, but we were unable to detect any integration into the β or the γ subunit genes after two cycles. It is possible that addition of a domain the size of FKBP is deleterious to the β and γ subunits. We note that we were able to add a smaller 3×HA tag to the β subunit but not to the γ subunit. In a conventional ATP synthase, γ subunit forms the principal part of the central stalk, and both the N and C termini of the protein are surrounded by the α3β3 hexameric F1 cap. It thus appears plausible that the introduction of a 3×HA/3×HA-FKBP tag at the C-terminal end of the γ subunit interferes with the function of ATP synthase.
The localization studies demonstrate that most of the known subunits of the enzyme are targeted to the mitochondrion. More importantly, we show by BN-polyacrylamide gels that the β and γ subunits are associated with large complexes in the mitochondrial membrane. These correspond to the monomeric and dimeric ATP synthase complexes found in other eukaryotic organisms (16, 20), with the dimeric form in predominance. The assembly of such a large, dimeric complex suggests that the parasite is probably not missing key subunits. The apparently missing genes encoding subunits of the FO sector are likely to be present in the genome but cannot be detected using current bioinformatic tools because of a high degree of divergence. A recent article (49) did suggest candidates for the P. falciparum a and b subunits; however, no proteomic results, trafficking data or predictions, or sequence conservation among other apicomplexan parasites were presented to support these choices, and we consider these unlikely candidates. However, the current release of the Pfam data base (50) contains a candidate for the Plasmodium b subunit (PF11_0262) with statistically significant similarity to the revised Pfam model for the mitochondrial ATP synthase B chain precursor family. PF11_0262 has apparent homologues throughout the Apicomplexa and several predicted attributes expected in a b subunit (N-terminal mitochondrial targeting sequence, two transmembrane helical segments near the N terminus, and a C-terminal domain with an extended region of α-helical secondary structure). We are presently investigating whether this b-like protein is also associated with the complexes containing ATP synthase subunits that we identified in P. falciparum mitochondria.
Role of ATP Synthase in P. falciparum
In most organisms, ATP synthase taps the energy stored in the proton gradient generated by the respiratory complexes to synthesize ATP from ADP and phosphate (Pi). However, the enzyme can also operate in reverse, as an ATPase hydrolyzing ATP into ADP and Pi and pumping protons into the intermembrane space, thereby establishing a proton gradient. This reverse reaction is important in some tissues to maintain the membrane potential during anoxia, as well as in organisms that lack or have down-regulated the mitochondrial respiratory enzymes, such as blood-stage trypanosomes (51–54). The membrane potential generated by ATPase might be needed in addition to the transmembrane potential generated by the mitochondrial electron transport chain, because of the very low oxygen consumption rate of P. falciparum parasites. A previous study from our laboratory provided evidence of an alternate source that generates membrane potential, in addition to the mitochondrial electron transport chain, in the parasite mitochondrion (46). Thus, in blood stages, the ATP synthase could be operating in reverse to generate the membrane potential needed for the optimum functioning of the mitochondrion. Although low, mitochondrial ATPase activity could be measured in Plasmodium mitochondrial preparations (31). This ATPase activity was highly resistant to classical inhibitors like oligomycin and azide, suggesting the possibility of novel FO structural domains, as seen in the Tetrahymena ATP synthase (15).
Dimeric ATP synthases are the predominant form of the enzyme in most eukaryotic organisms (16–19). The cristae morphology of yeast and mammalian mitochondria is suggested to be primarily due to the angle formed by the dimeric enzyme (19, 55). The curvature of the cristae tip in yeast mitochondria has been shown to be caused by rows of oligomeric dimeric ATP synthase complexes (56). In the ciliates T. thermophila and Paramecium, the structure and arrangement of dimeric ATP synthase have been suggested to determine the tubular cristae morphology seen in the mitochondria of these organisms (15, 57, 58).
Our inability to genetically disrupt ATP synthase subunits in P. falciparum suggests an essential role of the complex in blood stages. There is significant evidence to suggest that most, if not all, ATP is derived from glycolysis in blood stages of P. falciparum with little contribution from oxidative phosphorylation. Thus, the question arises as to what critical role the ATP synthase complex could be playing in these parasites. We suggest three mutually nonexclusive possibilities. (i) ATP synthase may provide an adjunct mechanism for the maintenance of electropotential across the mitochondrial inner membrane. We have previously shown that although mtETC functions are dispensable in transgenic P. falciparum expressing a cytosolic dihydroorotate dehydrogenase, maintenance of mitochondrial membrane potential is essential for other critical functions of the organelle (46). ATP hydrolysis, with or without concomitant proton pumping, by the ATP synthase complex could serve this function as has been shown in other systems (54, 59). (ii) Production of ATP is required for the local consumption in the mitochondrion. Various physiological processes within the parasite mitochondrion demand energetic contributions from ATP. The parasite mitochondrion may rely on locally produced ATP without making significant contributions to the cytosol. The relatively low rate of respiration by the parasite mitochondrion may be sufficient for the low level of locally critical oxidative phosphorylation. There is some evidence that the expression of the ATP synthase subunits and some other mitochondrially targeted proteins are up-regulated in other stages of the parasite life cycle (60, 61), so that the ATP synthase could be making its main contribution in another part of the life cycle. (iii) ATP synthase complex may participate in mitochondrial morphogenesis. Studies in other systems (57, 62–65) have shown that formation of respiratory supercomplexes is required for functional morphogenesis of mitochondria. Dimerization of the ATP synthase complex has been shown to be critical for the curvature of mitochondrial cristae (17, 19, 66). Minimal cristae structures seen in P. falciparum mitochondria are still likely to be critical for normal mitochondrial functions, a process in which dimers of the ATP synthase complex may participate as seen in other organisms (67, 68). Although further studies would be necessary to distinguish between these possible functions, it is clear that malaria parasites possess an ATP synthase complex that is highly divergent from that of their hosts. In light of this divergence, it may prove fruitful to investigate the potential of the Plasmodium complex as a therapeutic target.
Acknowledgments
We thank the Malaria Research and Reference Reagent Resource of the NIAID, National Institutes of Health, for the provision of the 3D7attB-integrated parasite line.
This work was supported, in whole or in part, by National Institutes of Health Grant AI28398 (to A. B. V.). This work was also supported by Drexel University College of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
S. M. Ganesan, unpublished data.
- OSCP
- oligomycin sensitivity conferring protein (subunit)
- BN
- blue native
- FKBP
- FK506-binding protein (domain)
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Walker J. E., Dickson V. K. (2006) Biochim. Biophys. Acta 1757, 286–296 [DOI] [PubMed] [Google Scholar]
- 2. Devenish R. J., Prescott M., Rodgers A. J. (2008) Int. Rev. Cell Mol. Biol. 267, 1–58 [DOI] [PubMed] [Google Scholar]
- 3. Angevine C. M., Herold K. A., Vincent O. D., Fillingame R. H. (2007) J. Biol. Chem. 282, 9001–9007 [DOI] [PubMed] [Google Scholar]
- 4. Fry M., Beesley J. E. (1991) Parasitology 102, 17–26 [DOI] [PubMed] [Google Scholar]
- 5. Krungkrai J., Burat D., Kudan S., Krungkrai S., Prapunwattana P. (1999) Southeast Asian J. Trop. Med. Public Health 30, 636–642 [PubMed] [Google Scholar]
- 6. Bryant C., Voller A., Smith M. J. (1964) Am J. Trop. Med. Hyg. 13, 515–519 [DOI] [PubMed] [Google Scholar]
- 7. Saliba K. J., Kirk K. (1999) J. Biol. Chem. 274, 33213–33219 [DOI] [PubMed] [Google Scholar]
- 8. Saliba K. J., Krishna S., Kirk K. (2004) FEBS Lett. 570, 93–96 [DOI] [PubMed] [Google Scholar]
- 9. Sherman I. W. (1998) in Malaria: Parasite Biology, Pathogenesis and Protection (Sherman I. W. ed) pp. 135–144, American Society for Microbiology, Washington, D. C [Google Scholar]
- 10. Nishi A., Scherbaum O. H. (1962) Biochim. Biophys. Acta 65, 419–424 [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi S. (1965) J. Biochem. 58, 444–457 [DOI] [PubMed] [Google Scholar]
- 12. Turner G., Lloyd D., Chance B. (1971) J. Gen. Microbiol. 65, 359–374 [DOI] [PubMed] [Google Scholar]
- 13. Conklin K. A., Chou S. C. (1972) Comp. Biochem. Physiol. B 41, 45–54 [DOI] [PubMed] [Google Scholar]
- 14. Kilpatrick L., Erecińska M. (1977) Biochim. Biophys. Acta 462, 515–530 [DOI] [PubMed] [Google Scholar]
- 15. Balabaskaran Nina P., Dudkina N. V., Kane L. A., van Eyk J. E., Boekema E. J., Mather M. W., Vaidya A. B. (2010) PLoS Biol. 8, e1000418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arnold I., Pfeiffer K., Neupert W., Stuart R. A., Schägger H. (1998) EMBO J. 17, 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudkina N. V., Sunderhaus S., Braun H. P., Boekema E. J. (2006) FEBS Lett. 580, 3427–3432 [DOI] [PubMed] [Google Scholar]
- 18. Vázquez-Acevedo M., Cardol P., Cano-Estrada A., Lapaille M., Remacle C., González-Halphen D. (2006) J. Bioenerg. Biomembr. 38, 271–282 [DOI] [PubMed] [Google Scholar]
- 19. Minauro-Sanmiguel F., Wilkens S., García J. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12356–12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krause F., Reifschneider N. H., Goto S., Dencher N. A. (2005) Biochem. Biophys. Res. Commun. 329, 583–590 [DOI] [PubMed] [Google Scholar]
- 21. Maier A. G., Braks J. A., Waters A. P., Cowman A. F. (2006) Mol. Biochem. Parasitol. 150, 118–121 [DOI] [PubMed] [Google Scholar]
- 22. Armstrong C. M., Goldberg D. E. (2007) Nat. Methods 4, 1007–1009 [DOI] [PubMed] [Google Scholar]
- 23. Nkrumah L. J., Muhle R. A., Moura P. A., Ghosh P., Hatfull G. F., Jacobs W. R., Jr., Fidock D. A. (2006) Nat. Methods 3, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trager W., Jensen J. B. (1976) Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 25. Fidock D. A., Wellems T. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ganesan S. M., Morrisey J. M., Ke H., Painter H. J., Laroiya K., Phillips M. A., Rathod P. K., Mather M. W., Vaidya A. B. (2011) Mol. Biochem. Parasitol. 177, 29–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Koning-Ward T. F., Fidock D. A., Thathy V., Menard R., van Spaendonk R. M., Waters A. P., Janse C. J. (2000) Mol. Biochem. Parasitol. 106, 199–212 [DOI] [PubMed] [Google Scholar]
- 28. Phillips M. A., Gujjar R., Malmquist N. A., White J., El Mazouni F., Baldwin J., Rathod P. K. (2008) J. Med. Chem. 51, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., Wandless T. J. (2006) Cell 126, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. (2004) Mol. Biochem. Parasitol. 137, 13–21 [DOI] [PubMed] [Google Scholar]
- 31. Mather M. W., Morrisey J. M., Vaidya A. B. (2010) Mol. Biochem. Parasitol. 174, 150–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418-428 [DOI] [PubMed] [Google Scholar]
- 33. Foth B. J., Stimmler L. M., Handman E., Crabb B. S., Hodder A. N., McFadden G. I. (2005) Mol. Microbiol. 55, 39–53 [DOI] [PubMed] [Google Scholar]
- 34. Xing S. L., Yan J., Yu Z. H., Zhu C. Q. (2011) Cell Biol. Int. 35, 81–86 [DOI] [PubMed] [Google Scholar]
- 35. Mangiullo R., Gnoni A., Leone A., Gnoni G. V., Papa S., Zanotti F. (2008) Biochim. Biophys. Acta 1777, 1326–1335 [DOI] [PubMed] [Google Scholar]
- 36. Bozdech Z., Llinás M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. (2003) PLoS Biol. 1, E5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Llinás M., Bozdech Z., Wong E. D., Adai A. T., DeRisi J. L. (2006) Nucleic Acids Res. 34, 1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vaidya A. B., Akella R., Suplick K. (1989) Mol. Biochem. Parasitol. 35, 97–107 [DOI] [PubMed] [Google Scholar]
- 39. Suplick K., Morrisey J., Vaidya A. B. (1990) Mol. Cell. Biol. 10, 6381–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feagin J. E., Werner E., Gardner M. J., Williamson D. H., Wilson R. J. (1992) Nucleic Acids Res. 20, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vaidya A. B., Lashgari M. S., Pologe L. G., Morrisey J. (1993) Mol. Biochem. Parasitol. 58, 33–42 [DOI] [PubMed] [Google Scholar]
- 42. Olszewski K. L., Mather M. W., Morrisey J. M., Garcia B. A., Vaidya A. B., Rabinowitz J. D., Llinás M. (2010) Nature 466, 774–778 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Vaidya A. B. (2004) Curr. Drug Targets Infect. Disord. 4, 11–23 [DOI] [PubMed] [Google Scholar]
- 44. Mather M. W., Henry K. W., Vaidya A. B. (2007) Curr. Drug Targets 8, 49–60 [DOI] [PubMed] [Google Scholar]
- 45. Vaidya A. B., Mather M. W. (2009) Annu. Rev. Microbiol. 63, 249–267 [DOI] [PubMed] [Google Scholar]
- 46. Painter H. J., Morrisey J. M., Mather M. W., Vaidya A. B. (2007) Nature 446, 88–91 [DOI] [PubMed] [Google Scholar]
- 47. Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., Paulsen I. T., James K., Eisen J. A., Rutherford K., Salzberg S. L., Craig A., Kyes S., Chan M. S., Nene V., Shallom S. J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M. W., Vaidya A. B., Martin D. M., Fairlamb A. H., Fraunholz M. J., Roos D. S., Ralph S. A., McFadden G. I., Cummings L. M., Subramanian G. M., Mungall C., Venter J. C., Carucci D. J., Hoffman S. L., Newbold C., Davis R. W., Fraser C. M., Barrell B. (2002) Nature 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boysen K. E., Matuschewski K. (2011) J. Biol. Chem. 286, 32661–32671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mogi T., Kita K. (2009) Mitochondrion 9, 443–453 [DOI] [PubMed] [Google Scholar]
- 50. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Di Lisa F., Blank P. S., Colonna R., Gambassi G., Silverman H. S., Stern M. D., Hansford R. G. (1995) J. Physiol. 486, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. St-Pierre J., Brand M. D., Boutilier R. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8670–8674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Traba J., Froschauer E. M., Wiesenberger G., Satrústegui J., Del Arco A. (2008) Mol. Microbiol. 69, 570–585 [DOI] [PubMed] [Google Scholar]
- 54. Brown S. V., Hosking P., Li J., Williams N. (2006) Eukaryot. Cell 5, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dudkina N. V., Heinemeyer J., Keegstra W., Boekema E. J., Braun H. P. (2005) FEBS Lett. 579, 5769–5772 [DOI] [PubMed] [Google Scholar]
- 56. Rabl R., Soubannier V., Scholz R., Vogel F., Mendl N., Vasiljev-Neumeyer A., Körner C., Jagasia R., Keil T., Baumeister W., Cyrklaff M., Neupert W., Reichert A. S. (2009) J. Cell Biol. 185, 1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Allen R. D. (1995) Protoplasma 189, 1–8 [Google Scholar]
- 58. Allen R. D., Schroeder C. C., Fok A. K. (1989) J. Cell Biol. 108, 2233–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Giraud M. F., Velours J. (1997) Eur. J. Biochem. 245, 813–818 [DOI] [PubMed] [Google Scholar]
- 60. Young J. A., Fivelman Q. L., Blair P. L., de la Vega P., Le Roch K. G., Zhou Y., Carucci D. J., Baker D. A., Winzeler E. A. (2005) Mol. Biochem. Parasitol. 143, 67–79 [DOI] [PubMed] [Google Scholar]
- 61. Zhou Y., Ramachandran V., Kumar K. A., Westenberger S., Refour P., Zhou B., Li F., Young J. A., Chen K., Plouffe D., Henson K., Nussenzweig V., Carlton J., Vinetz J. M., Duraisingh M. T., Winzeler E. A. (2008) PLoS One 3, e1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schägger H., Pfeiffer K. (2000) EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Acín-Pérez R., Bayona-Bafaluy M. P., Fernández-Silva P., Moreno-Loshuertos R., Pérez-Martos A., Bruno C., Moraes C. T., Enríquez J. A. (2004) Mol. Cell 13, 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wittig I., Carrozzo R., Santorelli F. M., Schägger H. (2006) Biochim. Biophys. Acta 1757, 1066–1072 [DOI] [PubMed] [Google Scholar]
- 65. Dudkina N. V., Kouril R., Peters K., Braun H. P., Boekema E. J. (2010) Biochim. Biophys. Acta 1797, 664–670 [DOI] [PubMed] [Google Scholar]
- 66. Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D. M., Brèthes D., di Rago J. P., Velours J. (2002) EMBO J. 21, 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bornhövd C., Vogel F., Neupert W., Reichert A. S. (2006) J. Biol. Chem. 281, 13990–13998 [DOI] [PubMed] [Google Scholar]
- 68. Strauss M., Hofhaus G., Schröder R. R., Kühlbrandt W. (2008) EMBO J. 27, 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]