Background: General control non-derepressible (Gcn) 5 is an important histone acetyltransferase that has been implicated in diverse human diseases.
Results: Cullin4-RING E3 ubiquitin ligase (CRL4) promotes Gcn5 degradation.
Conclusion: Gcn5 protein stability is regulated via ubiquitin-mediated pathway.
Significance: Learning how Gcn5 is regulated may provide us with the mechanistic basis to develop alternative approaches to inhibit Gcn5 activity for cancer therapy.
Keywords: Cancer biology, Cell cycle, Chromatin histone modification, Ubiquitin ligase, Ubiquitin-dependent protease, And-1, Cdt2, Ctf4, Gcn5, histone acetylation
Abstract
Histone acetyltransferases play important roles in the regulation of chromatin structure and gene transcription. As one of the most important histone acetyltransferases, general control non-derepressible (Gcn) 5 has been linked to diverse cellular processes and tumorigenesis as well. We have recently identified a functional link between Gcn5 and acidic nucleoplasmic DNA-binding protein 1 (And-1) that is elevated in multiple cancer cells and is essential for Gcn5 protein stability. However, the mechanism by which And-1 regulates Gcn5 protein stability remains unknown. Here we show that the ablation of Cullin4-RING E3 ubiquitin ligase (CRL4) leads to the stabilization of Gcn5 in cells with depleted And-1, and Cdc10-dependent transcript 2 (Cdt2) serves as a substrate receptor protein of CRL4. Overexpression of Cdt2 reduces the Gcn5 protein levels, and CRLCdt2 is sufficient to ubiquitinate Gcn5 both in vivo and in vitro. And-1 stabilizes Gcn5 by impairing the interaction between Gcn5 and CRLCdt2 and thereby preventing Gcn5 ubiquitination and degradation. The degradation of Gcn5 is not dependent on proliferating cell nuclear antigen, an important player involved in CRLCdt2-mediated protein degradation. Thus, CRLCdt2 and And-1 play an essential role in the regulation of Gcn5 protein stability. This study provides us with the mechanistic basis to develop alternative approaches to inhibit Gcn5 activity for cancer therapy.
Introduction
One of the major post-transcriptional modifications of nucleosomal histones is histone acetylation, which is carried out by histone acetyltransferases (1). Histone acetylation alters higher-order chromatin structure and plays a central role in the regulation of gene expression as well as in pathogenesis of a broad set of diseases including cancers (1–3). As the first identified transcription-related histone acetyltransferase, general control non-derepressible (Gcn) 52 has been implicated in diverse cellular processes including transcription (4, 5), DNA repair (6–8), telomere maintenance (9), and nucleosome assembly (10). Significantly, the role of Gcn5 in cancer cell survival and cellular transformation by regulating oncogenic gene expression has been documented (11–13). By acting as a co-factor, Gcn5 is involved in the regulation of Myc-dependent gene transcription (12), Myc-induced global acetylation (14), and E2 transcription factor (E2F)-dependent gene transcription (15).
Due to its important roles in oncogenic expression and histone acetylation (13), Gcn5 has been exploited as an anticancer target (16, 17). Although Gcn5 histone acetyltransferase activity has been well studied (13, 18), the pathway that regulates Gcn5 protein levels remains unknown. We have recently identified a novel high-mobility group (HMG) protein acidic nucleoplasmic DNA-binding protein 1 (And-1) that is critical for the stability of Gcn5 and histone H3 acetylation (19). Intriguingly, And-1 expression is significantly increased in multiple cancer cells in a manner correlating with increased Gcn5 and acetylation of histone H3 at lysine 9 (H3K9) and lysine 56 (H3K56) (19). Thus, the functional link between Gcn5 and And-1 may provide us with a new target for cancer therapy by altering Gcn5 protein stability. For instance, we can alter histone acetylation in cancer cells by manipulating Gcn5 protein stability. To accomplish this goal, it is necessary and important to elucidate the mechanism by which And-1 stabilizes Gcn5.
As one member of the evolutionarily conserved cullin-RING E 3 family proteins, the CRL4Cdt2 E3 ligase complex3 is composed of a scaffold protein Cullin4 (Cul4), the adaptor protein DNA damage-binding protein 1 (DDB1), and a substrate receptor protein Cdc10-dependent transcript 2 (Cdt2) (20–23). Accumulating evidence shows that CRLCdt2 E3 ligase plays important roles in the regulation of genome integrity, DNA damage repair, and cell cycle progression (24). For instance, during S phase of DNA replication or in cells with genotoxic stresses, CRLCdt2 is critical to promote the degradation of Cdc10-dependent transcript 1 (Cdt1), a DNA replication “licensing” factor required for the assembly of minichromosome maintenance protein 2–7 (MCM2–7) complex to replication origins in G1 phase (22, 23, 25–27). The degradation of Cdt1 by CRLCdt2 during S phase is one of the important mechanisms to prevent DNA re-replication. CRL4Cdt2 also targets the degradation of CDK inhibitor p21 during S phase or in response to UV irradiation (28, 29). Degradation of p21 by CRL4Cdt2 in S phase regulates the nuclear export of the Cdc6 replication-licensing factor and is believed to prevent DNA re-replication (29). Recent studies showed that CRLCdt2 regulates chromatin compaction by targeting histone methyltransferase Set8 (also known as Pr-Set7) for degradation during S phase or in cells with DNA damage (30–32). Failure to degrade Set8 in S phase leads to premature chromatin compaction during DNA replication, interfering with DNA synthesis and preventing mitotic entry (30–32). Furthermore, CRLCdt2 regulates the termination of the translesion synthesis repair pathway by promoting the degradation of translesion synthesis polymerase η and the normal cell cycle progression by promoting degradation of E2F1 (33, 34). Although these CRLCdt2 targets are involved in distinct cellular processes, all of them contain a proliferating cell nuclear antigen (PCNA)-interacting protein motif (PIP box), and PCNA appears to be required for the degradation of these CRLCdt2 targets (24).
In this study, we identify a novel functional link between CRLCdt2 and And-1 that has the remarkable capability to regulate Gcn5 protein turnover. We find that the down-regulation of CRLCdt2 leads to the stabilization of Gcn5 in cells with depleted And-1. Overexpression of CRLCdt2 reduces the Gcn5 protein levels, and CRLCdt2 is sufficient to ubiquitinate Gcn5 both in vivo and in vitro. And-1 and CRLCdt2 associate with Gcn5 at the same sites, and And-1 prevents the ubiquitination of Gcn5 by impairing the interaction between Gcn5 and CRLCdt2. Surprisingly, PCNA is not required for the degradation of Gcn5 in the absence of And-1, consistent with the fact that Gcn5 does not interact with PCNA. Thus, the functional link between And-1 and CRLCdt2 is critical for Gcn5 protein stability, and this study provides us with the mechanistic basis to target Gcn5 protein stability and thereby histone acetylation for cancer therapy.
EXPERIMENTAL PROCEDURES
Cells and Tissue Culture Medium
HCT116, HeLa, U2OS, and 293T cells were grown in DMEM medium supplemented with 10% FBS at 37 °C in 5% CO2 supply.
Antibodies and Immunofluorescence
The following antibodies were used: anti-H3K9Ac (9671), anti-H3K56Ac (4243), and anti-H3 (9715) were from Cell Signaling. Anti-tubulin (T6199), anti-FLAG-M2, and anti-HA were from Sigma. Anti-Gcn5 (sc-20698) (sc-130374), anti-β-actin, and anti-PCNA (SC-56) were from Santa Cruz Biotechnology. Anti-DDB1 (A300-462A), anti-Cul4A (A300-739A), and anti-Cdt2 (A300-948A) were from Bethyl Laboratories. Immunofluorescence was performed as described previously (35).
Plasmids
FLAG-And-1 (full length) and GST-And-1 plasmids were constructed as described previously (36). pEBB-GCN5 N terminus (amino acids 1–491) and pEBB-GCN5 histone acetyltransferase/bromo domain (amino acids 492–837) plasmids were gifts from Dr. Ezra Burstein. FLAG-Cdt2 plasmid was constructed by cloning Cdt2 cDNA into pEFF plasmid. Full-length FLAG-Gcn5 was constructed by cloning Gcn5 cDNA into pEFF vector. The plasmids expressing truncated Gcn5 were constructed as below; fragments of Gcn5 encoding amino acids 492–666, 667–749, 750–837, 492–706, and 707–749 were PCR-amplified using primers containing SpeI and KpnI sites, digested with SpeI and KpnI, and ligated into pEFF-vector digested with SpeI and KpnI.
Immunoprecipitation
The in vivo immunoprecipitations were performed as follows. Cells were lysed in lysis buffer (25 mm HEPES-KOH at pH 7.6, 150 mm KAc, 5 mm MgCl2, 1 mm Na2 EGTA, 10% glycerol, 0.1% Nonidet P-40, protease inhibitor, supplemented with 300 μg/ml ethidium bromide, and 15 killiunits/ml DNase I) for 20 min on ice followed by sonication. After centrifugation, the resulting supernatants were mixed with antibody or pre-bleed for immunoprecipitation overnight (2 h for FLAG-tagged protein immunoprecipitations) followed by incubation with protein G/A beads (Santa Cruz Biotechnology) for 1 h. Beads were then washed three times with lysis buffer. Associated proteins were eluted by incubating beads with SDS loading buffer for immunoblotting.
In Vivo and in Vitro Ubiquitination Assay
Details of the in vivo and in vitro ubiquitination reactions were as described (28). For the in vivo reaction, U2OS cells were transiently transfected with either FLAG-Gcn5 ± HA-ubiquitin ± FLAG-And-1-expressing or FLAG-Cdt2-expressing plasmids. Forty hours after transfection, the cells were treated with MG132 (20 μg/ml) for 2 h prior to lysis. Immunoprecipitated ubiquitinated proteins or FLAG-Gcn5 were fractionated on SDS-PAGE and immunoblotted for the indicated proteins. For the in vitro reaction, FLAG-Gcn5 and FLAG-And-1 proteins as well as FLAG-Cdt2 protein complexes were immunopurified from 293T cells expressing the above proteins from mammalian expression plasmids.
RNA Interference
siRNA oligonucleotides And-1-1 and And-1-2 were as described previously (36). The sequences for siAnd-1-1 and siAnd-1-2 oligonucleotides were as described previously (36). The sequences for siPCNA were as described before (27). The sequences for siCdt2 and siDDB1 were as described before (28). siRNA transfections were performed with 100 nm siRNA oligonucleotide duplexes using LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Western Blotting and Histone Isolation
To make total protein, cells were lysed in radioimmune precipitation buffer (150 mm NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mm Tris, pH 8.0) followed by sonication. Histone extraction was performed as described previously (37).
RESULTS
Gcn5 Is Ubiquitinated in Vivo and Degraded by the Proteasome
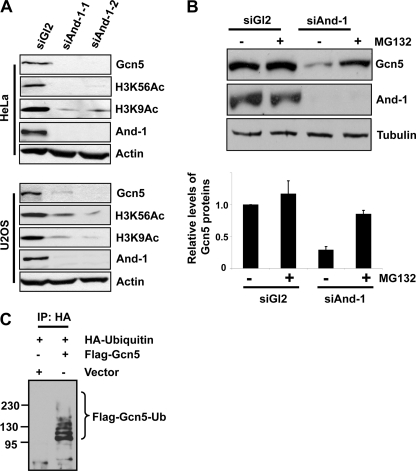
We previously observed that And-1 depletion resulted in the degradation of Gcn5 in HCT116 cells (19). To test whether this is the case in other cells, we examined Gcn5 protein levels in HeLa and U2OS cells in which And-1 was depleted by siRNA. The endogenous levels of And-1 protein were efficiently depleted after treatment by two independent siRNAs (siAnd-1-1 and siAnd-1-2) as compared with cells treated with control siRNA (siGl2) that targets firefly luciferase (Fig. 1A). Consistent with previous observations in HCT116 cells (19), Gcn5 protein levels and the acetylation of histone H3K9 and H3K56 were decreased in two cell lines treated by two independent siRNAs (Fig. 1A). Thus, the maintenance of Gcn5 protein stability by And-1 is not restricted to HCT116 cells.
FIGURE 1.
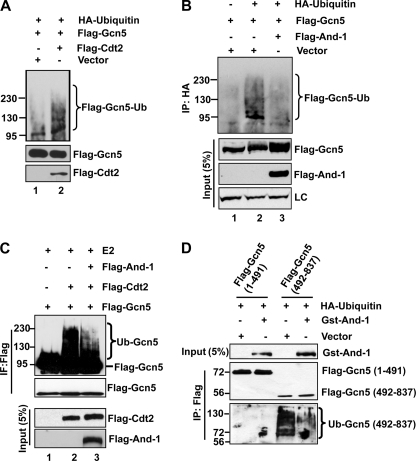
Gcn5 is ubiquitinated in vivo and degraded by proteasome. A, depletion of And-1 results in the reduction of Gcn5 proteins and acetylation of histone H3K9 and H3K56 in both HeLa and U2OS cells. Cells treated with siRNA control (siGl2) or two independent siRNAs (siAnd-1-1 or siAnd-1-2) were harvested 48 h after siRNA transfection and subjected to immunoblot using anti-Gcn5, anti-H3K56Ac, anti-H3K9Ac, anti-And-1, or anti-actin antibody. B, treatment of And-1-depleted cells with the proteasome inhibitor MG132 stabilizes Gcn5 proteins. Upper, HeLa cells transfected with the indicated siRNAs were treated with (+) or without (-) MG132 (10 μm) and lysed. Lysates were subjected to immunoblot using anti-Gcn5, anti-And-1, or anti-tubulin antibody. Lower, quantification of Gcn5 protein levels. Means ± S.D. (error bars) were obtained from three independent experiments. The signals of Gcn5 and tubulin proteins were quantified by Scion imaging software, and the background signal was subtracted. The relative levels of Gcn5 proteins were normalized by control tubulin protein signals. The Gcn5 protein levels in siGl2-treated cells alone were taken as 100%. C, Gcn5 is ubiquitinated in vivo. U2OS cells were transfected with HA-ubiquitin and FLAG-Gcn5 or vector, and the ubiquitinated species were precipitated by HA antibodies and subjected to immunoblot using anti-FLAG antibody. IP, immunoprecipitation; Ub, ubiquitin.
Given that And-1 depletion reduces Gcn5 protein levels but does not affect its mRNA levels (19), we hypothesized that Gcn5 might be degraded via ubiquitin-mediated proteolytic pathway. To test this idea, we treated siRNA transfected cells with protease inhibitor MG132. Consistently, And-1 depletion caused the degradation of Gcn5 (Fig. 1B). Strikingly, the addition of MG132 to And-1-depleted cells resulted in the stabilization of Gcn5 proteins (Fig. 1B), indicating that degradation of Gcn5 is mediated by the proteasome pathway. Degradation of proteins by the proteasome depends on conjugation of ubiquitin to the target protein. To determine whether Gcn5 is ubiquitinated in vivo, U2OS cells were co-transfected with a HA-tagged ubiquitin construct (HA-ubiquitin) and FLAG-Gcn5. Ubiquitinated proteins were isolated by immunoprecipitation using anti-HA antibody. As shown in Fig. 1C, slower migrating ubiquitin conjugates of Gcn5 were detected specifically in the HA-ubiquitin and FLAG-Gcn5 co-transfected cells. Notably, the presence of multiple slower migrating forms of Gcn5 indicates that Gcn5 is polyubiquitinated in vivo (Fig. 1C). Taken together, our data show that Gcn5 is ubiquitinated in vivo and that the degradation of Gcn5 in the absence of And-1 is mediated by the proteasome pathway.
Gcn5 Forms Complexes with CRL4Cdt2 E3 Ligase Complexes
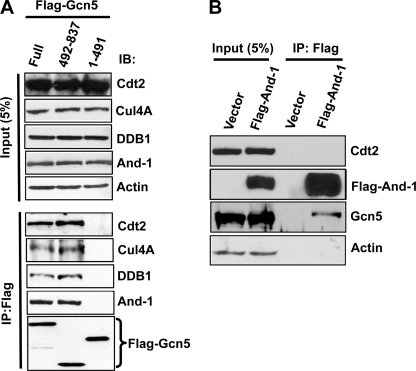
The fact that Gcn5 is ubiquitinated in vivo and is degraded via the proteasome pathway in the absence of And-1 prompted us to identify the E3 ligases that target Gcn5 for proteolysis. To this end, we searched for the E3 ligases that interact with Gcn5. Strikingly, DDB1 was previously found to interact with SPT3-TAFII31-GCN5L acetylase (STAGA) complex containing Gcn5 (38); however, the biological function of this interaction is unclear. As an adaptor protein, DDB1 forms a complex with Cul4, which serves as an E3 ubiquitin ligase targeting numerous proteins for proteolysis (39). Given that Gcn5 is degraded by the proteasome pathway (Fig. 1B), we hypothesized that Cul4-DDB1 E3 ligase might be involved in the degradation of Gcn5 in the absence of And-1. To test this idea, we examined whether Gcn5 interacts physically with DDB1. We expressed FLAG-tagged full-length Gcn5 and FLAG-Gcn5 mutants (1–491) or (492–837) in human 293T cells and monitored the presence of DDB1 in the anti-FLAG precipitates. Consistently with previous observation (38), endogenous DDB1 was detected in full-length Gcn5 immunoprecipitates (Fig. 2A), indicating that DDB1 forms a complex with Gcn5. Furthermore, we detected DDB1 in the precipitates of Gcn5 mutant (492–837) but not Gcn5 mutant (1–491), suggesting that the C terminus of Gcn5 is required for the association of Gcn5 with DDB1. This interaction is unlikely to be indirect due to DNA associations because both DNase and ethidium bromide were included in the lysis buffer for co-immunoprecipitation assays. Thus, our data indicate that Gcn5 associates with DDB1.
FIGURE 2.
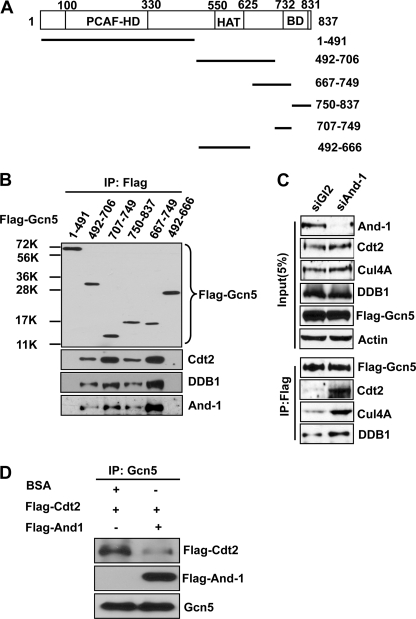
Gcn5 forms complexes with CRL4Cdt2 E3 ligase and And-1. A, Cdt2, DDB1, Cul4A, and And-1 interact with full-length (Full) Gcn5 and Gcn5 mutant (492–837). 293T cells transfected with the indicated FLAG-Gcn5 plasmids were harvested 36 h after transfection for immunoprecipitation using anti-FLAG M2-agarose beads. The FLAG immunoprecipitates (IP) were then subjected to immunoblot (IB) using anti-Cdt2, anti-Cul4A, anti-DDB1, anti-And-1, anti-actin, or anti-FLAG antibody. B, And-1 does not interact with Cdt2. 293T cells transfected with the indicated plasmids were treated as in A. Note that Gcn5 was detected in FLAG-And-1 precipitates.
Given that DDB1 forms a complex with Cul4 in the ubiquitin proteasome pathway (39), we next asked whether Cul4-DDB1 E3 ligase is associated with Gcn5. To test this idea, we examined the interaction between Cul4A and Gcn5. Strikingly, like DDB1, Cul4A was detected in the immunoprecipitates of full-length FLAG-Gcn5 and Gcn5 mutant (492–837) but not Gcn5 mutant (1–491) (Fig. 2A). Cdt2 is a substrate receptor protein that usually forms a complex with Cul4 and DDB1 to target proteins for degradation (24). We therefore predicted that Gcn5 might interact with Cdt2. Indeed, Cdt2 could be detected in immunoprecipitates of full-length Gcn5 (Fig. 2A). Like DDB1 and Cul4A, Cdt2 was detected in the precipitates of Gcn5 mutant (492–837) but not Gcn5 mutant (1–491) (Fig. 2A). Taken together, our data show that Gcn5 associates with CRL4Cdt2 E3 ligase complex and that the C terminus of Gcn5 is required for this association.
Our recent study showed that And-1 interacts with Gcn5 (19). We next assessed which part of Gcn5 interacts with And-1. Like CRL4Cdt2, And-1 was detected in the immunoprecipitates of full-length FLAG-Gcn5 and Gcn5 mutant (492–837) but not Gcn5 mutant (1–491) (Fig. 2A), suggesting that And-1 associates with the C terminus of Gcn5. Because Gcn5 interacts with both And-1 and CRL4Cdt2 E3 ligase, we next asked whether And-1 also interacts with Cdt2. To this end, we examined the interaction between And-1 and Cdt2 in 293T cells overexpressing vector or FLAG-And-1. Although Gcn5 was detected in FLAG-And-1 precipitates, Cdt2 was not detected in FLAG-And-1 immunoprecipitates, suggesting that And-1 does not interact with CRL4Cdt2. The fact that Gcn5 interacts with both And-1 and CRL4Cdt2, whereas And-1 does not associate with Cdt2, suggests that Gcn5 may interact with either And-1 or Cdt2 exclusively.
CRL4Cdt2 Is Required for the Degradation of Gcn5 in the Absence of And-1
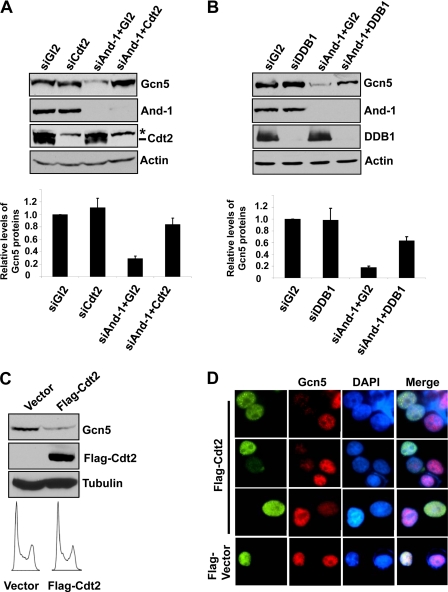
Having found the interactions between Gcn5 and CRL4Cdt2 E3 ligase complex, we hypothesized that CRL4Cdt2 E3 ligase is involved in the degradation of Gcn5 in the absence of And-1. To test this idea, we used siRNA to knock down the component of this E3 ubiquitin ligase in And-1-depleted cells. As compared with cells transfected with siGl2, cells co-transfected with siAnd-1 and siGl2 exhibited reduced Gcn5 protein levels (Fig. 3A). However, Gcn5 proteins were recovered in cells co-transfected with siAnd-1 and siCdt2 (Fig. 3A). In agreement, in cells co-transfected with siDDB1 and siAnd-1, the reduced Gcn5 protein levels were also partially recovered as compared with levels in cells co-transfected with siAnd-1 and siGl2 (Fig. 3B). Thus, CRL4Cdt2 E3 ligase is required for the degradation of Gcn5 induced by depletion of And-1.
FIGURE 3.
Down-regulation of CRL4Cdt2 E3 ligase stabilizes Gcn5 proteins in And-1-depleted cells. A, down-regulation of Cdt2 stabilizes Gcn5 proteins in cells with depleted And-1 by siRNA. Upper, U2OS cells transfected with the indicated siRNAs were harvested and subjected to immunoblot using anti-Gcn5, anti-And-1, anti-Cdt2, or anti-actin antibody. Lower, quantification of Gcn5 protein levels. Means ± S.D. (error bars) were obtained from four independent experiments. The relative levels of Gcn5 proteins were measured as in Fig. 1B. *, nonspecific bands recognized by anti-Cdt2 antibody. B, down-regulation of DDB1 stabilizes Gcn5 in cells with depleted And-1 by siRNA. Upper, U2OS cells transfected with the indicated siRNAs were harvested and subjected to immunoblot using anti-Gcn5, anti-And-1, anti-DDB1, or anti-actin antibody. Lower, quantification of Gcn5 protein levels. Means ± S.D. (error bars) were obtained from four independent experiments. The relative levels of Gcn5 proteins were measured as in Fig. 1B. C, overexpression of Cdt2 results in the reduction of Gcn5 protein levels. U2OS cells transfected with vector or FLAG-Cdt2 plasmids were harvested 36 h after transfection for immunoblot using anti-Gcn5, anti-FLAG, or anti-tubulin antibody. Lower, cells treated as above were harvested for FACS analysis. D, cells treated as in C were harvested for immunofluorescence using anti-FLAG and Gcn5 antibody. FLAG-vector is a plasmid expressing FLAG-And-1 (HMG domain). Note that cells with expression of FLAG-Cdt2 displayed reduced Gcn5 fluorescence signals as compared with cells without expression of FLAG-Cdt2.
To further explore the role of CRL4Cdt2 in the degradation of Gcn5, we next explored whether CRL4Cdt2 is sufficient to down-regulate Gcn5 in vivo. To this end, we overexpressed Cdt2 in U2OS cells and examined Gcn5 proteins. Notably, overexpression of Cdt2 specifically decreased the Gcn5 protein levels by immunoblot analyses (Fig. 3C). To confirm this observation, we performed indirect immunofluorescence in U2OS cells with expressed FLAG-Cdt2. As shown in Fig. 3D, cells expressing FLAG-Cdt2 exhibited lower levels of Gcn5 as indicated by reduced Gcn5 fluorescent signal as compared with that in cells without overexpression of FLAG-Cdt2. Taken together, our data suggest that CRL4Cdt2 is sufficient to induce the degradation of Gcn5 in vivo.
PCNA Is Dispensable for the Degradation of Gcn5 in the Absence of And-1
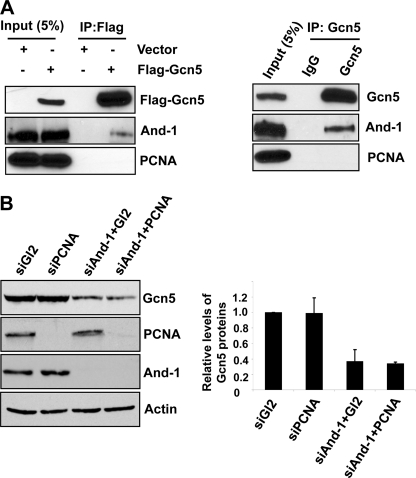
PCNA has been shown to be required for CRL4Cdt2-mediated degradation of several target proteins such as Cdt1, p21, Set8, and polymerase η (24). All these proteins bind to PCNA through a PIP box and are degraded on chromatin in a PCNA- and CRL4Cdt2-dependent manner (24). Interestingly, Gcn5 does not contain a PIP box. We therefore assumed that Gcn5 is unable to bind to PCNA. To test this possibility, we expressed FLAG-Gcn5 in 293T cells and monitored for the presence of PCNA in FLAG-Gcn5 precipitates. Consistent with our previous observation (19), we could detect And-1 from FLAG-Gcn5 precipitates (Fig. 4A). However, PCNA was not detected from FLAG-Gcn5 immunoprecipitates (Fig. 4A). We also failed to detect PCNA from endogenous Gcn5 precipitates (Fig. 4A). Taken together, Gcn5 does not exist in a complex with PCNA.
FIGURE 4.
PCNA is dispensable for the degradation of Gcn5 by CRL4Cdt2 E3 ligase. A, Gcn5 does not interact with PCNA. Left, 293T cells transfected with the indicated plasmids were harvested for immunoprecipitation (IP) using anti-FLAG M2-agarose beads. FLAG precipitates were subjected for immunoblot using anti-FLAG, anti-And-1, or anti-PCNA antibody. Note that And-1 was detected in FLAG-Gcn5 precipitates. Right, 293T cells were harvested for immunoprecipitation using anti-Gcn5 antibody or IgG. The Gcn5 precipitates were subjected for immunoblot using anti-And-1, anti-Gcn5, or anti-PCNA antibody. B, down-regulation of PCNA does not stabilize Gcn5 proteins in cells with depleted And-1 by siRNA. Left, U2OS cells transfected with the indicated siRNAs were harvested and subjected to immunoblot using anti-Gcn5, anti-PCNA, anti-And-1, or anti-actin. Right, quantification of Gcn5 protein levels. Means ± S.D. (error bars) were obtained from four independent experiments. The relative levels of Gcn5 proteins were measured as in Fig. 1B.
Given that Gcn5 is not associated with PCNA, we hypothesized that PCNA might not be involved in the degradation of Gcn5 in the absence of And-1. To test this idea, we co-transfected siAnd-1 and siPCNA and compared the Gcn5 protein levels in co-transfected cells with levels in And-1-depleted cells. Although PCNA protein is depleted in cells co-transfected with siAnd-1 and siPCNA, PCNA depletion had little effect on Gcn5 protein levels in co-transfected cells as compared with And-1-depleted cells (Fig. 4B). Thus, our data indicate that Gcn5 does not associate with PCNA and that PCNA is dispensable for the CRL4Cdt2-dependent degradation of Gcn5 in the absence of And-1.
And-1 Prevents the Ubiquitination of Gcn5
To assess whether Gcn5 is a bona fide substrate of CRL4Cdt2 E3 ligase, we overexpressed Cdt2 along with the FLAG-Gcn5 and HA-ubiquitin in U2OS cells. To prevent the degradation of Gcn5 by Cdt2 overexpression, MG132 was added to cells before harvest. Gcn5 ubiquitination was detected in cells co-expressed with HA-ubiquitin and FLAG-Gcn5 (Fig. 5A, lane 1). Notably, Cdt2 overexpression enhanced Gcn5 polyubiquitination (Fig. 5A, lane 2), indicating that CRL4Cdt2 is able to ubiquitinate Gcn5 in vivo. We next asked whether the CRL4Cdt2 E3 ligase is capable of ubiquitinating Gcn5 in vitro. To this end, we purified FLAG-Cdt2 complexes and FLAG-And-1 from 293T cells, respectively. The incubation of FLAG-Cdt2 complexes with E2 and Gcn5 produced high-molecular weight ubiquitinated Gcn5 species (Fig. 5C, lanes 1 and 2). Thus, Gcn5 is a bona fide direct substrate of CRL4Cdt2 ubiquitin ligase.
FIGURE 5.
And-1 prevents the ubiquitination of Gcn5 by CRL4Cdt2 E3 ligase. A, overexpression of Cdt2 enhances the ubiquitination of Gcn5. U2OS cells transfected with the indicated plasmids were harvested 48 h after transfection. Cells were treated with MG132 for 6 h before harvest. The ubiquitinated species were precipitated by HA antibodies and subjected to immunoblot for ubiquitinated FLAG-Gcn5 using anti-FLAG antibody. Ub, ubiquitin. B, overexpression of And-1 suppresses the ubiquitination of Gcn5. U2OS cells transfected with the indicated plasmids were harvested for 48 h and treated as in A. Note that And-1 overexpression increased the FLAG-Gcn5 protein levels and reduced the ubiquitination of Gcn5. IP, immunoprecipitation; LC, loading control. C, And-1 prevents Gcn5 ubiquitination by CRL4Cdt2 in vitro. Incubation of immunopurified CRL4Cdt2 with Gcn5 in an in vitro ubiquitin ligase assay increased the formation of Gcn5 ubiquitinated species. The addition of immunopurified And-1 suppressed the Gcn5 ubiquitination by CRL4Cdt2. IF, immunofluorescence. D, And-1 overexpression suppresses the ubiquitination of the C terminus of Gcn5. U2OS cells transfected with the indicated plasmids were harvested. FLAG-And-1(1–491) or FLAG-And-1(492–837) was precipitated using anti-FLAG M2-agarose beads. FLAG precipitates and inputs were subjected to immunoblot using anti-ubiquitin, anti-FLAG, or anti-And-1 antibody.
We have previously shown that the interaction between And-1 and Gcn5 is critical for And-1 to maintain the stability of Gcn5 (19). Given that Gcn5 is degraded via the ubiquitin-mediated proteasome degradation pathway, we hypothesized that And-1 might stabilize Gcn5 proteins by preventing Gcn5 ubiquitination. To test this idea, we co-expressed HA-ubiquitin together with Gcn5 or with Gcn5 and And-1, respectively, in U2OS cells. Consistent with our previous observation (19), And-1 overexpression elevated the FLAG-Gcn5 protein levels in the whole cell extracts (Fig. 5B). Gcn5 was polyubiquitinated in cells co-expressing FLAG-Gcn5 and HA-ubiquitin (Fig. 5B, lane 2). Strikingly, co-expression of And-1 with Gcn5 significantly reduced ubiquitination of Gcn5 (Fig. 5B, lane 3), suggesting that And-1 interferes with the Gcn5 ubiquitination. To further confirm this observation, we next performed in vitro ubiquitination assays. As shown in Fig. 5C, FLAG-Cdt2 complexes were able to ubiquitinate Gcn5 (Fig. 5C, lane 2); however, the addition of purified FLAG-And-1 substantially reduced the ubiquitination of Gcn5 (Fig. 5C, lane 3). Taken together, our data show that And-1 prevents Gcn5 ubiquitination by CRL4Cdt2 both in vivo and in vitro.
Gcn5 interacts with CRL4Cdt2 via its C terminus (492–837) (Fig. 2A). We predicted that CRL4Cdt2 may ubiquitinate Gcn5 at its C terminus (492–837). To test this idea, we co-expressed FLAG-Gcn5(1–491) or FLAG-Gcn5(492–837) with HA-ubiquitin or vector. As expected, FLAG-Gcn5(492–837) but not FLAG-Gcn5(1–491) was detected with ubiquitination (Fig. 5D), suggesting that the C terminus of Gcn5 contains ubiquitin sites. Because And-1 also interacts with FLAG-Gcn5(492–837), it is possible that overexpression of And-1 may impair the ubiquitination of FLAG-Gcn5(492–837). Indeed, overexpression of And-1 substantially reduced the ubiquitination of FLAG-Gcn5(492–837) (Fig. 5D). Thus, Gcn5 ubiquitin sites are located within the C terminus of Gcn5 that is the binding site for both And-1 and CRL4Cdt2 E3 ligase.
Gcn5 Exclusively Interacts with And-1 and CRL4Cdt2
We have previously reported that the interaction between Gcn5 and And-1 is critical for And-1 to maintain the stability of Gcn5 (19). Given that C terminus of Gcn5 interacts with both And-1 and CRL4Cdt2, whereas And-1 does not interact with Cdt2 (Fig. 2), we hypothesized that And-1 might compete with Cdt2 for the interaction with Gcn5 at the same site or adjacent sites at the C terminus of Gcn5. To explore this possibility, we constructed a series of truncated Gcn5 mutants to delineate the And-1 and CRL4Cdt2 binding regions within Gcn5 (Fig. 6A). Because both And-1 and CRL4Cdt2 interact with the C terminus of Gcn5 (Fig. 2A), all of the Gcn5 truncation mutants excluded the N-terminal regions of Gcn5 (Fig. 6A). We expressed these Gcn5 mutants in 293T cells and examined their interactions with And-1 or Cdt2 by co-immunoprecipitation assays. Consistent with previous observations (Fig. 2A), Cdt2 was not detected in FLAG-Gcn5(1–491) precipitates. We failed to detect Cdt2 from precipitates of Gcn5 mutant (492–666), suggesting that Gcn5 histone acetyltransferase domain is not required for the association of Gcn5 with Cdt2. Notably, Cdt2 was readily detected in precipitates of Gcn5 mutants (Gcn5(492–706), Gcn5(707–749), Gcn5(750–837), and Gcn5(667–749)) (Fig. 6B), suggesting that the region between 667 and 837 of Gcn5 was the binding site for Cdt2. The stronger association between Cdt2 and Gcn5(667–749) or Gcn5(707–749) indicates that the major site on Gcn5 for the association with Cdt2 is located within the region between amino acids 667–749. In agreement, similar interaction patterns were observed between DDB1 and Gcn5 truncation mutants (Fig. 6B). Strikingly, And-1 was also detected in the precipitates of Gcn5 mutants (Gcn5(492–706), Gcn5(707–749), Gcn5(750–837), or Gcn5(667–749)). Like Cdt2 and DDB1, And-1 also exhibited a stronger interaction with Gcn5(667–749) or Gcn5(707–749). Taken together, our data show that both And-1 and CRL4Cdt2 complexes associate with Gcn5 at the same site within the region of 667–749.
FIGURE 6.
And-1 competes with CRL4Cdt2 for the association with Gcn5. A, schematic of Gcn5 truncation mutants used for protein-protein interactions as in B. The P300/CBP-associated factor (PCAF) homology domain (PCAF-HD), histone acetyltransferase (HAT) domain, and bromo domain (BD) are indicated. B, And-1 and CRL4Cdt2 interact with Gcn5 at the same site within the C terminus. 293T cells transfected with the indicated plasmids were harvested and immunoprecipitated using anti-FLAG M2-agarose beads. FLAG-Gcn5 immunoprecipitates (IP) were subjected to immunoblot using anti-FLAG, anti-Cdt2, anti-DDB1, or anti-And-1 antibody. C, And-1 depletion increased the interaction between Gcn5 and CRL4Cdt2. U2OS cells transfected with the indicated siRNAs and FLAG-Gcn5 plasmid were treated with MG132 to stabilize Gcn5 before harvest. Cells were harvested 55 h after siRNA transfection for immunoprecipitation using anti-FLAG M2-agarose beads. FLAG-Gcn5 precipitates were subjected to immunoblot using anti-And-1, anti-Cdt2, anti-Cul4A, anti-actin, or anti-FLAG antibody. D, And-1 blocks the interaction between Cdt2 and Gcn5 in vitro. Immunopurified Gcn5 proteins on agarose beads were mixed with immunopurified FLAG-Cdt2 in the presence BSA or immunopurified FLAG-And-1 for 3 h at 4 °C. The beads were then washed three times using lysis buffer for immunoprecipitation. Gcn5 precipitates were subjected to immunoblot using anti-FLAG or anti-Gcn5 antibody.
Given that And-1 and CRL4Cdt2 complexes interact with Gcn5 at the same site and that And-1 does not interact with Cdt2, it is possible that And-1 and Cdt2 may compete with each other for the interaction with Gcn5. To explore this possibility, we examined the interaction between Gcn5 and Cdt2 in And-1-depleted cells. If our hypothesis is correct, And-1 depletion should result in an increased interaction between Gcn5 and Cdt2. Indeed, the interaction between Gcn5 and Cdt2 was substantially increased in the absence of And-1 (Fig. 6C). Consistently, And-1 blocked the interaction between Cdt2 and Gcn5 in an in vitro protein-protein interaction assay (Fig. 6D). Taken together, our data suggest that And-1 prevents the interaction between CRL4Cdt2 and Gcn5.
DISCUSSION
CRL4Cdt2-And-1, a Novel Functional Link Regulating Gcn5 Protein Stability
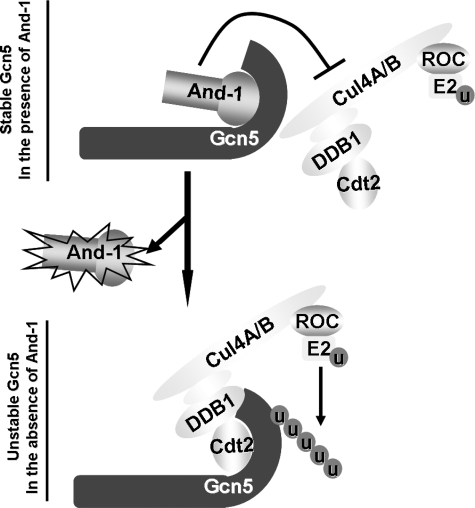
We have previously shown that And-1 has remarkable capability to regulate the stability of Gcn5 proteins and thereby histone H3 acetylation (19). To gain further insight into the mechanism by which And-1 maintains the stability of Gcn5, we identified a novel functional link between CRL4Cdt2 and And-1 that is critical to regulate Gcn5 protein turnover. We find that CRL4Cdt2 E3 ligase complexes target Gcn5 for degradation in the absence of And-1. Both And-1 and Cdt2 bind to the C terminus of Gcn5 (Fig. 7). Although Gcn5 interacts with And-1 and CRL4Cdt2, And-1 and CRL4Cdt2 do not exist in the same complex, and And-1 prevents the interaction between CRL4Cdt2 and Gcn5. CRL4Cdt2 is sufficient to ubiquitinate Gcn5 both in vivo and in vitro. Thus, we conclude that And-1 stabilizes the Gcn5 protein by preventing the association of Gcn5 with CRL4Cdt2 and subsequently the ubiquitination and degradation of Gcn5 (Fig. 7). This discovery explains our previous observation that the interaction between And-1 and Gcn5 is required for And-1 to maintain Gcn5 protein stability (19).
FIGURE 7.
A proposed model for the role of And-1 and CRL4Cdt2 in the regulation of Gcn5 protein turnover. Both And-1 and CRL4Cdt2 associate with Gcn5 at the same site within its C terminus. In the presence of And-1, the And-1 module associates with Gcn5 and stabilizes Gcn5 protein levels by preventing the interaction between Gcn5 and CRL4Cdt2 and thereby the ubiquitination of Gcn5. In the absence of And-1, CRL4Cdt2 interacts with the C terminus of Gcn5 and ubiquitinates Gcn5, resulting in the degradation of Gcn5. Note that the C-terminal domain of Cul4 associates with a small RING protein regulator of cullin (ROC), which recruits and activates E2 enzyme to transfer ubiquitin (u) to Gcn5.
CRL4Cdt2 Targets Gcn5 for Degradation in a PCNA-independent Manner
CRL4Cdt2 regulates the degradation of multiple cell cycle-regulating proteins (24). Our study indicates that Gcn5 is a new bona fide target of CRL4Cdt2 E3 ligase. This conclusion is based on three solid lines of evidence. First, CRL4Cdt2 interacts with Gcn5 (Fig. 2). Second, CRL4Cdt2 promotes the ubiquitination of Gcn5 both in vivo and in vitro (Fig. 2). Finally, silencing Cdt2 or DDB1 rescues the degradation of Gcn5 in And-1-depleted cells (Fig. 3). Intriguingly, CRL4Cdt2 recently was shown to be involved in chromatin modification by promoting the destruction of histone methyltransferase Set8 in S phase, a process that prevents aberrant chromatin compaction during DNA synthesis (30–32). Thus, CRL4Cdt2 appears to be an important player that regulates chromatin structure and function via the regulation of proteins involved in histone modification. It remains unclear how CRL4Cdt2 regulates cell cycle progression by maintaining the balance between the histone acetyltransferase and histone methyltransferase.
PCNA plays a critical role in the CRL4Cdt2-mediated degradation pathway. CRL4Cdt2 targets such as Cdt1, p21, Set8, and polymerase η contain a conserved PIP box, which is required for CRL4Cdt2-mediated degradation (24). Gcn5 does not contain a PIP box and does not interact with PCNA. In support of this, silencing of PCNA fails to rescue degradation of Gcn5 in And-1-depleted cells (Fig. 4). Thus, PCNA is dispensable for Gcn5 degradation in the absence of And-1. Why is PCNA dispensable in CRL4Cdt2-mediated degradation of Gcn5 in the absence of And-1? One possible explanation is that Gcn5 physically associates with CRL4Cdt2 E3 ligase complexes and that the ubiquitination of Gcn5 by CRL4Cdt2 E3 ligase does not need PCNA as a platform. Indeed, we could detect an interaction between Gcn5 and CRL4Cdt2 complexes, and this interaction is substantially increased in the absence of And-1 (Figs. 2 and 6). Thus, Gcn5 is ubiquitinated by CRL4Cdt2 E3 ligase immediately upon loss of its interacting protector And-1. Further experiments with careful examination of chromatin structure in the absence of And-1 will be needed to determine how Gcn5 is targeted by CRL4Cdt2 without PCNA as a docking site.
The Functional Link between CRL4Cdt2 and And-1, a Potential Target for Cancer Therapy
We have reported that And-1 protein is overexpressed in multiple cancer cells in a manner correlating with increased Gcn5 and acetylation of histone H3 at lysine 9 and 56 (19). Interestingly, Cdt2 expression is also elevated in aggressive hepatocellular carcinomas and breast and gastric cancers (40–42). How the elevated functional link between And-1 and Cdt2 contributes to the tumorigenesis is unclear. In fact, our study indicates that the disruption of the balance between And-1 and Cdt2 significantly alters Gcn5 protein levels. One possibility is that And-1 and Cdt2 maintain a physiological level of Gcn5 that is critical for cancer cell proliferation or survival. Given that Gcn5 plays an important role in the regulation of oncogenic pathways (13), the future work should address whether and how And-1 and Cdt2 regulate Gcn5-mediated oncogenic pathways during tumorigenesis. Although our observation underscores the significance of CRL4Cdt2 in the regulation of Gcn5-mediated oncogenic pathways, we could not rule out the possibility that other targets of CRL4Cdt2 may play an important role in CRL4Cdt2-mediated tumorigenesis. Gcn5 has been exploited as a new target for cancer therapy largely because of its role in the regulation of oncogenic pathways and histone acetylation (16, 17). The current approach is to identify compounds that inhibit Gcn5 histone acetyltransferase activity. Our study identified a novel pathway governing Gcn5 protein stability. Thus, targeting the And-1-CRL4Cdt2 link may be an alternative approach to alter histone acetylation by manipulating Gcn5 protein stability for cancer therapy.
Acknowledgments
We thank Anindya Dutta for And-1 plasmids and antibodies, Ezra Burstein for Gcn5 plasmids, and Zhiguo Zhang and Lee Zou for reading or suggestions during manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant 4R00CA136555 (to W. Z.).
The CRL4Cdt2 E3 ligase complex is a complex of CRL4 with Cdt2.
- Gcn5
- general control non-derepressible 5
- And-1
- acidic nucleoplasmic DNA-binding protein 1
- Cdt2
- Cdc10-dependent transcript 2
- CRL4
- Cullin4-RING E3 ubiquitin ligase
- Cul4
- Cullin4
- PCNA
- proliferating cell nuclear antigen
- PIP
- PCNA-interacting peptide
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1. Shahbazian M. D., Grunstein M. (2007) Annu. Rev. Biochem. 76, 75–100 [DOI] [PubMed] [Google Scholar]
- 2. Jones P. A., Baylin S. B. (2002) Nat. Rev. Genet. 3, 415–428 [DOI] [PubMed] [Google Scholar]
- 3. Toyota M., Issa J. P. (2005) Semin. Oncol. 32, 521–530 [DOI] [PubMed] [Google Scholar]
- 4. Brownell J. E., Zhou J., Ranalli T., Kobayashi R., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Cell 84, 843–851 [DOI] [PubMed] [Google Scholar]
- 5. Kuo M. H., Brownell J. E., Sobel R. E., Ranalli T. A., Cook R. G., Edmondson D. G., Roth S. Y., Allis C. D. (1996) Nature 383, 269–272 [DOI] [PubMed] [Google Scholar]
- 6. Tamburini B. A., Tyler J. K. (2005) Mol. Cell. Biol. 25, 4903–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brand M., Moggs J. G., Oulad-Abdelghani M., Lejeune F., Dilworth F. J., Stevenin J., Almouzni G., Tora L. (2001) EMBO J. 20, 3187–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robert T., Vanoli F., Chiolo I., Shubassi G., Bernstein K. A., Rothstein R., Botrugno O. A., Parazzoli D., Oldani A., Minucci S., Foiani M. (2011) Nature 471, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Atanassov B. S., Evrard Y. A., Multani A. S., Zhang Z., Tora L., Devys D., Chang S., Dent S. Y. (2009) Mol. Cell 35, 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgess R. J., Zhou H., Han J., Zhang Z. (2010) Mol. Cell 37, 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kikuchi H., Kuribayashi F., Takami Y., Imajoh-Ohmi S., Nakayama T. (2011) Biochem. Biophys. Res. Commun. 405, 657–661 [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Tesfai J., Evrard Y. A., Dent S. Y., Martinez E. (2003) J. Biol. Chem. 278, 20405–20412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagy Z., Tora L. (2007) Oncogene 26, 5341–5357 [DOI] [PubMed] [Google Scholar]
- 14. Knoepfler P. S., Zhang X. Y., Cheng P. F., Gafken P. R., McMahon S. B., Eisenman R. N. (2006) EMBO J. 25, 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lang S. E., McMahon S. B., Cole M. D., Hearing P. (2001) J. Biol. Chem 276, 32627–32634 [DOI] [PubMed] [Google Scholar]
- 16. Dekker F. J., Haisma H. J. (2009) Drug Discov. Today 14, 942–948 [DOI] [PubMed] [Google Scholar]
- 17. Rekowski M. W., Giannis A. (2010) Biochim. Biophys. Acta 1799, 760–767 [DOI] [PubMed] [Google Scholar]
- 18. Burgess R. J., Zhang Z. (2010) Cell Cycle 9, 2979–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y., Jaramillo-Lambert A., Yang Y., Williams R., Lee N., Zhu W. (July 4, 2011) Oncogene 10.1038/onc.2011.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 21. Higa L. A., Yang X., Zheng J., Banks D., Wu M., Ghosh P., Sun H., Zhang H. (2006) Cell Cycle 5, 71–77 [DOI] [PubMed] [Google Scholar]
- 22. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 23. Sansam C. L., Shepard J. L., Lai K., Ianari A., Danielian P. S., Amsterdam A., Hopkins N., Lees J. A. (2006) Genes Dev. 20, 3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abbas T., Dutta A. (2011) Cell Cycle 10, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arias E. E., Walter J. C. (2005) Genes Dev. 19, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arias E. E., Walter J. C. (2006) Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 27. Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006) J. Biol. Chem 281, 6246–6252 [DOI] [PubMed] [Google Scholar]
- 28. Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim Y., Starostina N. G., Kipreos E. T. (2008) Genes Dev. 22, 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abbas T., Shibata E., Park J., Jha S., Karnani N., Dutta A. (2010) Mol. Cell 40, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Centore R. C., Havens C. G., Manning A. L., Li J. M., Flynn R. L., Tse A., Jin J., Dyson N. J., Walter J. C., Zou L. (2010) Mol. Cell 40, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oda H., Hübner M. R., Beck D. B., Vermeulen M., Hurwitz J., Spector D. L., Reinberg D. (2010) Mol. Cell 40, 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim S. H., Michael W. M. (2008) Mol. Cell 32, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shibutani S. T., de la Cruz A. F., Tran V., Turbyfill W. J., 3rd, Reis T., Edgar B. A., Duronio R. J. (2008) Dev. Cell 15, 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu W., Chen Y., Dutta A. (2004) Mol. Cell. Biol. 24, 7140–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 38. Martinez E., Palhan V. B., Tjernberg A., Lymar E. S., Gamper A. M., Kundu T. K., Chait B. T., Roeder R. G. (2001) Mol. Cell. Biol. 21, 6782–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson S., Xiong Y. (2009) Trends Biochem. Sci. 34, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan H. W., Chou H. Y., Liu S. H., Peng S. Y., Liu C. L., Hsu H. C. (2006) Cell Cycle 5, 2676–2687 [DOI] [PubMed] [Google Scholar]
- 41. Ueki T., Nishidate T., Park J. H., Lin M. L., Shimo A., Hirata K., Nakamura Y., Katagiri T. (2008) Oncogene 27, 5672–5683 [DOI] [PubMed] [Google Scholar]
- 42. Li J., Ng E. K., Ng Y. P., Wong C. Y., Yu J., Jin H., Cheng V. Y., Go M. Y., Cheung P. K., Ebert M. P., Tong J., To K. F., Chan F. K., Sung J. J., Ip N. Y., Leung W. K. (2009) Br. J. Cancer 101, 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]