Background: Unlike for eukaryote-type serine/threonine kinase of group A Streptococcus (GAS), significance of its cognate serine/threonine phosphatase (SP-STP) remains elusive.
Results: SP-STP is crucial for GAS pathophysiology.
Conclusion: SP-STP is not essential for GAS survival, but its optimal concentration is critical for cognately maintained homeostasis within GAS.
Significance: This work opens up avenues to understand the role of secretory SP-STP as an important virulence determinant in the host.
Keywords: Bacterial Protein Phosphatases, Cell Division, Gene Knockout, Metabolism, Pathogenesis, Phosphorylation, Serine/Threonine Protein Phosphatase, Virulence Factors, Streptococcus pyogenes
Abstract
Reversible phosphorylation is the key mechanism regulating several cellular events in prokaryotes and eukaryotes. In prokaryotes, signal transduction is perceived to occur primarily via the two-component signaling system involving histidine kinases and cognate response regulators. Although an alternative regulatory pathway controlled by the eukaryote-type serine/threonine kinase (Streptococcus pyogenes serine/threonine kinase; SP-STK) has been shown to modulate bacterial growth, division, adherence, invasion, and virulence in group A Streptococcus (GAS; S. pyogenes), the precise role of the co-transcribing serine/threonine phosphatase (SP-STP) has remained enigmatic. In this context, this is the first report describing the construction and characterization of non-polar SP-STP mutants in two different strains of Type M1 GAS. The STP knock-out mutants displayed increased bacterial chain lengths in conjunction with thickened cell walls, significantly reduced capsule and hemolysin production, and restoration of the phenotypes postcomplementation. The present study also reveals important contribution of cognately regulated-reversible phosphorylation by SP-STK/SP-STP on two major response regulators of two-component systems, WalRK and CovRS. We also demonstrate a distinct role of SP-STP in terms of expression of surface proteins and SpeB in a strain-specific manner. Further, the attenuation of virulence in the absence of STP and its restoration only in the complemented strains that were generated by the use of a low copy plasmid and not by a high copy one emphasize not only the essential role of STP in virulence but also highlight the tightly regulated SP-STP/SP-STK-mediated cognate functions. SP-STP thus is an important regulator of GAS virulence and plays a critical role in GAS pathogenesis.
Introduction
Streptococcus pyogenes (group A Streptococcus; GAS)3 is a Gram-positive pathogen that colonizes and invades human epithelia, leading to an array of diseases, ranging from mild pharyngitis and impetigo to severe necrotizing fasciitis and autoimmune sequelae (1). The ability of this pathogen to colonize and persist within the host and trigger infections is dependent on its wide range of virulence factors and complex regulatory networks that modulate gene expression in response to fluctuating environmental conditions (2, 3). Of these complex networks, signal transduction is one of the key regulatory processes that occur through reversible protein phosphorylation. In prokaryotes, the sensing of extracellular signal and transduction of information is primarily governed by a two-component signal transduction system, consisting of a sensor histidine kinase and its cognate response regulator (4). Parallel to this signaling pathway, prokaryotes also possess a eukaryote-type alternative signaling mechanism mediated by serine/threonine kinases (ESTKs) and co-transcribing phosphatases (ESTPs) (reviewed in Ref. 5). The latter possesses conserved I-XI motifs belonging to a distinct class of metal-dependent PPM/PP2C-type serine/threonine phosphatase (6). ESTKs found in many prokaryotic pathogens, including GAS and S. aureus, have been implicated in various cellular functions, such as stress response, biofilm formation, cell wall biosynthesis, sporulation, metabolic and developmental processes, drug resistance, and virulence (5, 7, 8). The co-transcribing nature and similar amino acid target specificity suggest that the functions of ESTKs are biochemically and physiologically coupled to ESTPs, and they are probably cognately regulated. Despite the significance of dephosphorylation events in bacterial signaling cascade(s), the precise regulatory role of ESTP still remains enigmatic (5). The STP mutants have been reported only in the organisms harboring multiple homologs of STP, such as in Myxococcus xanthus, Bacillus subtilis, Listeria monocytogenes, and Staphylococcus aureus (9, 10). Based on the phenotypic characteristics, the functional roles of STP in these organisms have been incriminated in spore formation, biofilm formation, cell wall synthesis, and virulence (11). Importantly, the ability to generate either ESTK or ESTK-ESTP double mutants but not ESTP mutants in S. pyogenes, Streptococcus agalactiae, and Bacillus anthracis (7) indicated that STP is probably essential for bacterial survival. However, recent reports on partial characterization of STP mutant in Streptococcus mutans (12) and the existing yet uncharacterized STP mutant in Streptococcus pneumoniae (13) prompted us to revisit this unsettled issue of whether STP is indeed essential for GAS survival (7). In the present investigation, we effectively addressed this by successfully deriving non-polar STP mutants in M1SF370 and M1T15448 GAS strains and studied several morphological, growth, biochemical, and biological functional parameters, including virulence. Our present study reveals that STP of S. pyogenes is not essential for bacterial survival but is essential for GAS virulence and plays an important role in GAS pathogenesis.
EXPERIMENTAL PROCEDURES
Bacterial Strain, Growth Conditions, and Cell Culture
The wild-type GAS strains M1SF370 (ATCC 700294) (14) and M1T15448 (referred to as M1T1 hereafter) (15) and the derived mutants were grown at 37 °C in Todd-Hewitt broth (Difco) supplemented with 0.5% (w/v) yeast extract (THY) with or without kanamycin (300 μg/ml). Escherichia coli strains DH5α, MC1061, and BL21(λDE3) were used (supplemental Table S1). The high copy/multicopy pDC123 (16) (chloramphenicol: 10 μg/ml for E. coli and 5 μg/ml for S. pyogenes) and low copy pREG696 (17) and pJRS9508 (18) vectors (spectinomycin: 200 μg/ml for E. coli and 100 μg/ml for S. pyogenes) were used for the complementation studies (supplemental Table S1). Detroit 562 human pharyngeal cell line (ATCC CCL138) was cultured and maintained as described (7).
Construction of STP Mutants in GAS
The M1SF370 and M1T1 strains of S. pyogenes were used for generating stp/pppL (SPy1626 and the equivalent one in M1T1) mutant employing a non-polar pVPNP-1 vector, which was derived from pFW6 plasmid (19). The pFW6 vector was modified to introduce the BstEII site (for ease in cloning of different antibiotic resistance cassettes). For this, a 240-bp fragment was PCR-amplified using specific primers (supplemental Table S2) and was digested with HindIII-PacI and cloned into pFW6, yielding pFW-HBP. The pFW-HBP plasmid was then digested with BstEII-EcoRI and ligated with a 1430-bp BstEII-EcoRI-amplified fragment encompassing the kanamycin resistance gene (kanr) from pFW13 using specific primers (supplemental Table S2). The resulting vector construct was designated as pVPNP-1 (3.877 kb). The aforementioned 1430-bp fragment (kanr) placed in this pVPNP-1 vector contains a ribosomal binding site at its 5′-end for the expression of this cassette. The stop codon and the intact ribosomal binding site contributed by the remaining 3′-end sequences (62 bp) of the stp gene allowed the independent expression of the downstream co-transcribing stk gene in the operon following homologous recombination.
To create a mutant strain lacking the full-length stp gene, the upstream 810-bp and downstream 930-bp regions were PCR-amplified with primer pairs 5/6 and 7/8 and cloned into MCS-I and MCS-II, respectively (supplemental Tables S1 and S2). The resulting plasmid pVP-SN1ΔSTP was used to transform both the wild-type GAS strains, M1SF370 and M1T1, as described previously (20) to obtain M1ΔSTP and M1T1ΔSTP mutants, respectively. The double crossover homologous recombination event was confirmed by PCR using flanking forward primer 9 in combination with kanamycin reverse primer 10, and kanamycin forward primer 11 along with flanking reverse primer 12 along with gene-specific primer pair 13/14 (supplemental Table S2). The integrity of the mutant (i.e. the replacement of stp with kanamycin cassette) was confirmed by DNA sequencing of the amplicon obtained using primer pair 13/14. The expression of STP was ascertained in the total cell lysates and extracellular fractions of the wild-type and mutant strains by immunoblotting using anti-SP-STP polyclonal antibody (7). Similarly, SP-STK in the total cell lysates was assessed by anti-SP-STK polyclonal antibody (7). The blots were probed with anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody. The signal was visualized using chemiluminescence substrate (SuperSignal West Pico, Pierce). The expression of SDH/GAPDH in the total cell lysates was taken as loading control (7, 21).
Complementation of M1ΔSTP and M1T1ΔSTP Strains with High and Low Copy Plasmids
A DNA fragment encoding STP along with N-terminal His6 tag and a ribosomal binding site was obtained by BglII/BamHI digestion of pET-His-SP-STP plasmid (7) and cloned downstream to the tetracycline promoter (Ptet) of the pDC123 complementation vector (22) to obtain a high copy complementation construct, pDC123.stp. The resulting complementation vector pDC123.stp was used to transform M1ΔSTP and M1T1ΔSTP mutants to generate M1ΔSTP::pDC.stp and M1T1ΔSTP::pDC.stp, respectively, and M1SF370 and M1T1 wild-type GAS strains to generate STP-overexpressing strains, M1-WT::stp and M1T1-WT::stp, respectively.
To generate the low copy complementation construct, pREG696.stp, the gene encoding STP was cloned downstream of the Pspac promoter in a pIB167 plasmid (23) (supplemental Tables S1 and S2), followed by amplification and cloning of the entire promoter-gene cassette in pREG696 vector (supplemental Tables S1 and S2). To generate the low copy complementation construct, pJRS9508.stp, the stp gene was cloned downstream of the P23 promoter in pJRS9508 vector (supplemental Tables S1 and S2). The pJRS9508.stp and pREG696.stp constructs were used to transform M1ΔSTP and M1T1ΔSTP mutants, respectively, to generate M1ΔSTP::pJRS.stp and M1T1ΔSTP::pREG.stp strains. The expression of STP in both of the complemented mutant strains was analyzed in the culture supernatants and total cell lysates using anti-SP-STP antibody as described above.
Fatty Acid Composition Analysis
The fatty acid methyl esters were extracted from bacteria grown until late logarithmic phase. Total fatty acid content and the composition of individual fatty acids in the wild-type and mutant strain of M1SF370 were custom analyzed by gas chromatography (Microbial ID) as described (24). The fatty acid profiles were analyzed with the Sherlock® pattern recognition software.
Microscopic Analyses
Light and Fluorescence Microscopy
The wild-type and mutant strains of M1SF370 and M1T1 were Gram-stained and visualized under a Nikon Eclipse E600 microscope. The bacterial cultures grown until early log phase (A600 = 0.4) were stained by Fl-Van (BODIPY Fl-conjugated vancomycin (Molecular Probes); 3 μg/ml) for 2 h. Stained bacterial cells were spread on poly-l-lysine-coated microscopic slides followed by 4′,6′-diamidino-2-phenylindole (DAPI) staining (0.2 μg/ml; Molecular Probes). The cells were observed under a confocal microscope at the Ohio State University Campus Microscopy and Imaging Facility.
Transmission Electron Microscopy
The wild-type and mutant strains of M1SF370 and M1T1, grown in THY broth until late log phase (A600 = 0.8) were processed for transmission electron microscopy, employing a cryo-capable digital transmission electron microscope (Technai G2 Spirit, EFI) at the Ohio State University Campus Microscopy and Imaging Facility as described previously (7).
Expression Analysis of CdhA (CHAP Domain-containing and Chain-forming Cell Wall Hydrolase)
To assess the expression of CdhA/SibA in the GAS mutant strains devoid of STP, extracellular fractions were immunoblotted using anti-CdhA antibody (1:5000) (20), and the immunoreactive bands were visualized by nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate substrate solution.
Cloning, Expression, and Purification of Response Regulators, SP-WalR and SP-CovR, Belonging to Two Major Two-component Systems (TCSs)
The genes encoding WalR (SPy0528, 711 bp) and CovR (SPy0336, 687 bp) were cloned in pET-14b plasmid using gene-specific primers (supplemental Tables S1 and S2). The recombinant His6-tagged SP-WalR and SP-CovR were expressed and affinity-purified using Ni2+-NTA chromatography as described (7). The authenticity of the recombinant proteins was evaluated by Western blotting using custom made rabbit anti-SP-WalR and anti-SP-CovR polyclonal antibodies (supplemental Fig. S3).
In Vitro Phosphorylation and Two-dimensional Thin Layer Chromatography (TLC)
Phosphorylation of the SP-WalR and SP-CovR, 10 μg each, was carried out in the presence or absence of the recombinant SP-STKK (1 μg) in an in vitro kinase assay as described (7). Recombinant SP-STP (1 μg) (7) was included to study the reversible dephosphorylation. The SP-STKK-phosphorylated protein bands were excised from the SDS-polyacrylamide gel, followed by acid hydrolysis and identification of the phosphorylated amino acids by TLC as described previously (7).
Attributes of GAS Virulence
Bacterial Adherence and Invasion Assays
The ability of wild-type GAS, its mutant, and complemented strains to adhere to Detroit 562 cells was determined. The confluent cultures of Detroit 562 cells were infected with the indicated strains (multiplicity of infection, 100:1, bacteria/cell) for 3 h, and the adherent bacteria were enumerated as described (7). Parallel to this, internalization of the adhered population was evaluated for each strain as described previously (7). The experiments were performed twice in six independent wells. The results were statistically analyzed, and the p value significance was determined by a non-parametric t test with Welch's corrections using GraphPad Prism 4.
Phagocytosis/Bactericidal Assay
The ability of wild type, mutants, and complemented GAS strains to resist phagocytosis and survive in human blood was analyzed in bactericidal assays essentially as described previously (7). Bacterial growth (multiplication factor) was calculated as the ratio of the mean number of cfu recovered after 3 h and the initial mean number of cfu added at t = 0. The required Ohio State University Institutional Review Board approval was obtained prior to withdrawing the blood from two healthy individuals. Results were statistically analyzed by non-parametric t test as described above.
Hemolysin Assay
The hemolysis assay was performed as described previously (25). Briefly, the supernatants of wild-type, STP mutant, and complemented strains grown until late log phase (A600 = 0.8) were mixed with an equal volume of 2.5% (v/v) thoroughly washed defibrinated sheep red blood cells (shRBCs) and incubated at 37 °C for 1 h. The hemoglobin released from the lysed shRBCs was measured spectrophotometrically at 570 nm (Polarstar Galaxy). Complete lysis of shRBCs was achieved upon the addition of 1% Triton X-100 and was treated as control (100%). Samples containing only shRBCs and PBS were treated as blank or negative control. Percentage hemolysis was calculated by the equation, ((sample A − blank A)/(100% lysis A)) × 100.
Estimation of Hyaluronic Acid in the Capsule
Capsular hyaluronic acid content present in 10 ml of late log phase culture of the wild-type, mutant, and complemented GAS strains was determined as described previously using Stain-All (Sigma) (7). The amount of capsule was measured based on the standard curve generated with known concentrations of hyaluronic acid.
In Vivo Bacterial Virulence Assays
Virulence potential of the wild-type, STP mutants, the mutants complemented with high copy pDC123.stp (10 mice/group), low copy pJRS9508.stp and pREG696.stp (10 mice/group), and the wild-type GAS strains complemented with the stp gene (overexpressing STP, 8 mice/group) was assessed employing an experimental mouse infection model of peritonitis. A group of indicated mice (CD-1, 5 weeks old, 20–22 g; Charles River Laboratories) were injected intraperitoneally with 1 × 108 cfu. Morbidity and mortality in the infected animals were observed twice daily for 10 days. The data presented in the mortality/survival curve were statistically evaluated using the log rank test using GraphPad Prism4 software. All animal experiments were performed in accordance with the Ohio State University Institutional Animal Care and Use Committee-approved protocol as described previously (20).
Quantitative Real-time Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted from three independently (biological replicates) grown late log phase cultures of wild-type and mutant GAS strains using the Qiagen RNeasy kit. The purity and integrity of RNA were confirmed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). First strand cDNA was generated (Roche Applied Science), and the mRNA levels were quantitated for selected genes using SYBR Green qRT-PCR master-mix (Roche Applied Science) and specific primers (supplementary Table S3) in a LightCycler® 480 (Roche Applied Science). The copy numbers for all of the genes were normalized with the housekeeping gene, proS, using three biological and three technical replicates. The linear -fold change in the mRNA expression levels of each gene in the mutant strains with respect to their corresponding wild-type strains were analyzed using Exor4 software (Roche Applied Science). 2-fold up- or down-regulation was considered as a significant change.
RESULTS
Construction, Integrity, and Growth Characteristics of the Non-polar Serine/Threonine Phosphatase (STP) Mutant of S. pyogenes
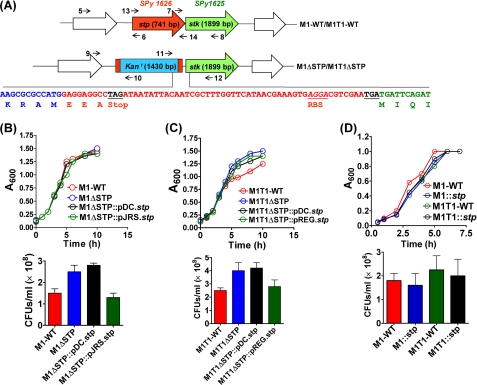
Because stp/pppL and stk are co-transcribed (7), with stp transcription preceding the stk (Fig. 1A), to prevent the downstream polar effects following stp gene deletion, the pFW6 with spectinomycin-resistant gene marker (aad9) and a transcription terminator was modified to generate a non-polar vector, pVPNP-1, which resulted in the generation of STP mutants in M1SF370 and M1T1 GAS strains. The genotype of the stp mutants, M1ΔSTP and M1T1ΔSTP, was confirmed by PCR using primers 9/10 (supplemental Fig. S1A, lanes 3 and 7), 11/12 (supplemental Fig. S1A, lanes 4 and 8), and stp gene-specific primers 13/14, showing the replacement of the stp gene in the wild type (supplemental Fig. S1A, 741 bp, lanes 1 and 5) with kanamycin resistance cassette in both of the mutants (supplemental Fig. S1A, 1430 bp, lanes 2 and 6). The identity and integrity of the M1ΔSTP and M1T1ΔSTP mutants were further confirmed by DNA sequencing of the amplicon generated by the 13/14 primer pair (Fig. 1A).
FIGURE 1.
Construction of non-polar stp mutants in M1SF370 and M1T1 strains. A, schematic representation of the genome organizations of wild-type and mutant strains of S. pyogenes stp/stk operon. The numbers and arrows depict the primers used for confirming gene deletion in the mutants. The indicated nucleotide sequence shows the replacement of the stp gene with the sequence encoding the kanamycin resistance cassette and uninterrupted stk gene sequence. The transcription terminator after kanr and the ribosomal binding site for the stk are also highlighted. Shown are growth characteristics of the wild-type M1SF370 (B) and M1T1 (C) in comparison with their respectively derived isogenic mutants and complemented strains, grown in THY. D, the growth profile of stp-overexpressing M1SF370 and M1T1 wild-type strains. Each data point of the spectrophotometrically (A600) monitored growth curve represents an average of three independent readings. The panels below each graph represent the cfu of the indicated strains grown until A600 = 0.8. Error bars, S.D.
Deletion of the stp gene in the mutant strains at the transcriptional level was assessed by qRT-PCR analysis. Western blot analysis was performed to assess the level of STP protein in the cell lysates of the wild type and the mutant strains. The blots probed with anti-SP-STP antibody show the absence of STP in the cell lysates of M1ΔSTP and M1T1ΔSTP mutant strains (supplemental Fig. S1B, lanes 2 and 4) unlike the wild-type strains (supplemental Fig. S1B, lanes 1 and 3). Because SP-STP is a secretory protein (7), the extracellular fractions were also analyzed for the STP protein to obviate the possibility of its presence in the extracellular medium of the mutant strains. We did not observe STP in the extracellular secretion of the mutants (supplemental Fig. S1C, lanes 2 and 4) when compared with the wild-type strains (supplemental Fig. S1C, lanes 1 and 3), confirming the integrity of the mutants in both of the GAS strains.
To restore the expression and function of STP in the mutants and also to determine the suitability of complementation plasmids, high and low copy complementation plasmids were used. Complementation of the M1ΔSTP and M1T1ΔSTP mutants with the wild-type stp using multicopy plasmid pDC123 yielded an expected high level of STP expression (supplemental Fig. S2A). However, when these mutants were complemented with a low copy plasmid pJRS9508 (P23 promoter, in M1ΔSTP) and pREG696 (Pspac promoter in M1T1ΔSTP), the wild-type levels of SP-STP were restored (supplemental Fig. S2B), indicating the strain-specific efficiency of transcription from a distinct promoter in driving the STP expression.
Along with the absence of STP described above, the following findings confirmed the non-polarity of these mutants: (i) Western blot analysis of the cell lysates obtained from the mutant strains showing an intense anti-STK-reacting protein band (supplemental Fig. S1D, lanes 2 and 4) comparable with their corresponding wild types (supplemental Fig. S1D, lanes 1 and 3); (ii) the maintained mRNA expression levels of stk (SPy1625) (see Fig. 6, C and D; see also supplemental Table S4); and (iii) the DNA sequence showing the presence of a strong Rho-independent transcription terminator (UUCUUUUUAACUACU, ΔG = −15.9 kcal/mol) (26) after the stk gene.
FIGURE 6.
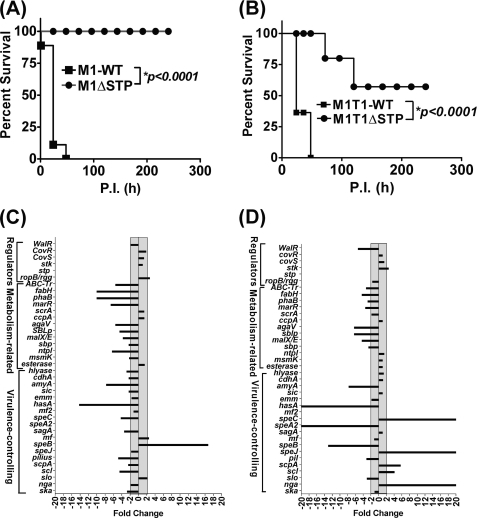
Deletion of STP attenuates GAS virulence in an experimental mouse intraperitoneal infection model. Effects of the deletion of the stp gene on GAS virulence of CD-1 mice (n = 10) as observed for the wild-type and mutant strains of M1SF370 (A) and M1T1 (B). Survival/mortality for the strains was monitored for 10 days, and the data were statistically evaluated and analyzed by the log rank test using GraphPad Prism 4 software. For real-time (qRT-PCR) analysis, the expression profiles of 19 virulence-related genes, 13 metabolism-related genes, and the other six one-/two-component transcription regulators in the stp-deleted mutant strains derived from M1SF370 (C) and M1T1 (D) were compared with respect to their corresponding wild-type strains. The transcripts were quantified by a SYBR Green-based real-time PCR assay as described under “Experimental Procedures.” The gray zone across the y axis depicts the threshold for determining significant (≥2-fold) change.
The growth patterns of both of the STP mutants (Fig. 1, B and C), monitored over a period of 10 h in THY broth, were comparable to those of their corresponding wild-type GAS strains. Further analysis of the growth patterns of the complemented strains, M1ΔSTP::pDC.stp, M1T1ΔSTP::pDC.stp, M1ΔSTP::pJRS.stp, M1T1ΔSTP::pREG.stp, and STP-overexpressing wild-type GAS strains M1::stp and M1T1::stp (Figs. 1, B–D), in comparison with their respective wild types, revealed no discernible differences in their doubling time, thus confirming no aberrant effect on the growth patterns as a result of complementation.
Enumeration of cfu after 8 h of growth revealed that the STP mutant, stp-complemented, and stp-overexpressing wild-type strains grew equally well as their respective wild-type strains (Fig. 1, B–D, bottom).
Role of Serine/Threonine Phosphatase in Cell Division/Septa and Chain Formation
The phenotypic characteristics like colony size and morphology of the GAS mutants were comparable with those of their corresponding wild types (data not shown). The Gram staining showed formation of long chains in the M1ΔSTP and M1T1ΔSTP mutant strains compared with their respective wild-type strains, indicating a plausible role of STP in cell division (Fig. 2A).
FIGURE 2.
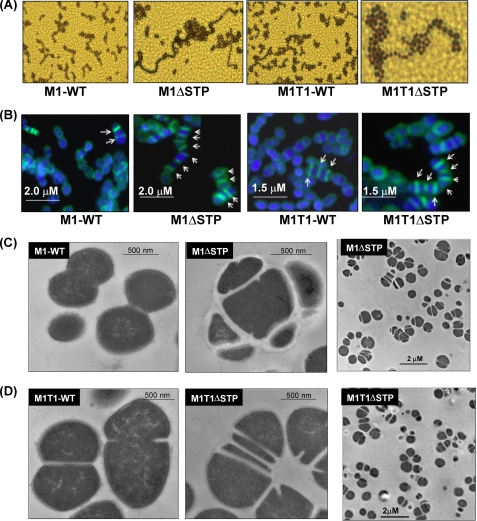
STP is involved in cell division and septa and chain formation. Shown are Gram-stained (A) and FL-Van-stained (B) images of the wild-type and mutant GAS strains. The arrows shown in the overlay images of green FL-van and blue DAPI point to the distinct septum in each case with the scale bars shown. Transmission electron micrographs of the wild type (M1SF370 (C) and M1T1 (D)) and their respectively derived stp-deleted mutant strains are shown at the indicated magnification.
Fl-Van staining of the M1ΔSTP and M1T1ΔSTP mutants and their respective wild-types strains with comparable vancomycin susceptibilities (MIC 0.25 μg/ml) revealed unusual staining patterns with multiple asymmetric and parallel septa formation (Fig. 2B). Similar defects observed by transmission electron microscopy (Fig. 2, C and D) in conjunction with relatively thicker cell walls indicated that SP-STP plays a role in the regulation of septa formation and in turn cell division in GAS.
CdhA-regulated Cell Division in GAS Is Cognately Controlled by SP-STK/STP-mediated Reversible Phosphorylation of WalR
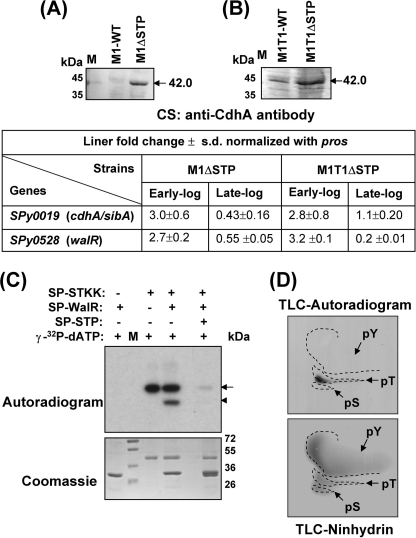
Our previous results demonstrating the correlation between long chain formation with the overexpression of CdhA (20), the STP mutants displaying long chain-forming phenotypes, loss of plane recognition, and multiple parallel septa formation led us to hypothesize that the loss of STP directly or indirectly affects CdhA in a temporal fashion. We validated this hypothesis by demonstrating an increase in the level of CdhA protein in the culture supernatants of early log phase-grown mutant strains, M1ΔSTP (Fig. 3A) and M1T1ΔSTP (Fig. 3B). In accordance with the increased CdhA levels, we also observed a corresponding up-regulation in the mRNA transcript encoding CdhA (2.8–3-fold) during the early growth phase in both of the mutant strains (Fig. 3, tabulated below A and B).
FIGURE 3.
The chain formation in STP mutants is attributed to CdhA controlled by SP-WalR. Shown is Western blotting of the extracellular fractions obtained from the wild-type M1SF370 and M1ΔSTP (A) and the wild-type M1T1 and M1T1ΔSTP (B) using anti-CdhA (SPy0019) antibody. The arrows indicate detection of an immunoreactive band at ∼45 kDa corresponding to CdhA. C, in vitro phosphorylation of SP-WalR by SP-STKK and dephosphorylation by SP-STP are shown in the autoradiogram. The arrows and arrowheads point toward the phosphorylated SP-STKK and SP-WalR, respectively. The parallel Coomassie-stained gel depicting the integrity of the proteins in each case is shown. M, migration of molecular mass standards. D, two-dimensional TLC of phosphorylated SP-WalR revealing phosphorylation at threonine in the autoradiogram and the migration of phosphoamino acid standards (tyrosine (pY), serine (pS), and threonine (pT)) by ninhydrin.
Because STK has been shown previously to positively regulate cdhA (20) and its pneumococcal homolog is controlled by the WalRK regulon in S. pneumoniae (27, 28), we attempted to explore the possibility of reversible phosphorylation of SP-WalR by SP-STK/STP couple. The in vitro phosphorylation assay revealed that SP-STK was able to phosphorylate SP-WalR (Fig. 3C) specifically at threonine residues, as is evident by TLC (Fig. 3D). The SP-STP was able to dephosphorylate the SP-STK-phosphorylated SP-WalR (Fig. 3C), demonstrating that this couple functions as a cognate kinase-phosphatase pair. In support of this, we also observed a simultaneous up-regulation of walR (2.7–3.2-fold) transcript at an early time point in both of the strains (Fig. 3, table below A and B).
Deletion of stp Adversely Affects Capsule Biosynthesis and SagA-mediated Hemolysin Production in GAS
Our previous study on the M1ΔSTK mutant of GAS demonstrated that upon deletion of SP-STK, capsule biosynthesis (encoded by the has operon) and hemolysin expression (encoded by sagA) (7) are up-regulated. In both of the STP mutants, we observed a 3–4-fold decrease in the capsule production (Fig. 4A) and 14–100-fold down-regulation of the corresponding has transcript as revealed by qRT-PCR (supplemental Table S4; see also Fig. 6, C and D).
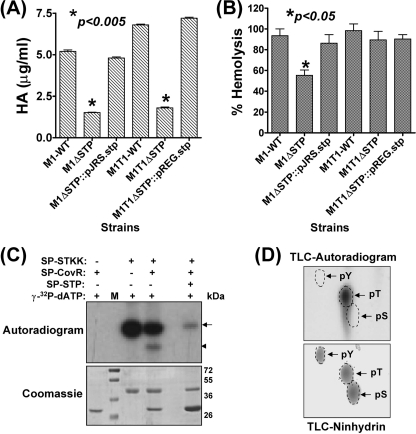
FIGURE 4.
Capsule and hemolysin production is cognately regulated by SP-STK/STP-mediated reversible phosphorylation of CovR. A, the comparison of capsule content based on the biochemical estimation of the hyaluronic acid for the STP mutants and complemented GAS strains with respect to their corresponding wild-type strains. Error bars, S.D. of two independent experiments performed in six wells. B, hemolysin production in the wild-type M1SF370 and M1T1 and their corresponding mutant and complemented strains. Complete lysis of shRBCs achieved upon the addition of 1% Triton X-100 was taken as control (100%). Error bars, S.D. of two independent experiments performed in six wells. C, autoradiogram showing phosphorylation of SP-CovR by SP-STKK and its dephosphorylation by SP-STP in an in vitro kinase assay. Arrows and arrowheads point toward the phosphorylated SP-STKK and SP-CovR, respectively. The parallel Coomassie-stained gel is shown to assess the integrity of the proteins in each case. M, migration of molecular mass standards. D, two-dimensional TLC of phosphorylated SP-CovR revealed phosphorylation at threonine residues in the autoradiogram and the migration of phosphoamino acid standards (tyrosine (pY), serine (pS), and threonine (pT)) by ninhydrin.
The significant decrease in the hemolytic activity of the M1ΔSTP (30%, p < 0.05) versus only 10% decrease in the M1T1ΔSTP (Fig. 4B) strain concurred with the down-regulation of sagA-specific mRNA transcript (3.4-fold) in the M1ΔSTP mutant strain (Fig. 6C; see also supplemental Table S4) and the unaltered mRNA transcript in the M1T1ΔSTP mutant (Fig. 6D; see also supplemental Table S4). These results thus indicated that the STP-mediated sagA regulation is strain-specific.
CovRS TCS has been shown to govern capsule and hemolysin production in GAS (29). Based on the phenotypes with contrasting characteristics observed for the SP-STK mutant in our previous study (7) and SP-STP mutants in the present study, we hypothesized that CovR could be a possible substrate for SP-STK-mediated reversible phosphorylation. We demonstrated that the SP-STK was able to catalyze the phosphorylation of CovR in vitro (Fig. 4C). The specificity of SP-STK mediated CovR phosphorylation was confirmed by its subsequent dephosphorylation by the cognate SP-STP (Fig. 4C). Also, the TLC revealed that SP-STK mediates phosphorylation of CovR specifically at its threonine residues (Fig. 4D).
Strain-specific Role of STP in Bacterial Adherence and Invasion of Human Pharyngeal Cells
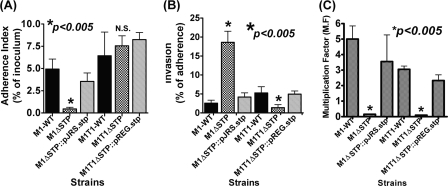
Upon close examination of the electron micrographs of the STP mutant strains (Fig. 2, C and D), we observed a substantial loss of the outermost electron dense fuzzy layer, which is contributed primarily by several surface proteins. Notably, these changes were more obvious in the M1ΔSTP mutant strain (Fig. 2C) as compared with the M1T1ΔSTP mutant (Fig. 2D). This prompted us to evaluate whether the ability of the mutant strains to adhere to the pharyngeal cells is strain-specific. The results showed a significantly reduced adherence pattern for the M1ΔSTP strain and unaltered adherence for the M1T1ΔSTP mutant in comparison with their corresponding wild-type strains (Fig. 5A). Further, the M1ΔSTP mutant was found to be more invasive than the M1T1ΔSTP mutant, reiterating the strain-specific STP-mediated regulation in GAS adherence and invasion (Fig. 5B). Upon complementation with stp, the altered phenotypes regained characteristics comparable with those of the wild-type strains (Fig. 5, A and B).
FIGURE 5.
Lack of STP affects bacterial adherence to human pharyngeal cells and abrogates GAS anti-phagocytic function. The ability of the mutants and stp-complemented strains to adhere to and invade Detroit 562 human pharyngeal cells in comparison with that of the wild-type strains was determined by calculating adherence (percentage of initial inoculum) (A) and invasion (percentage of adherence) (B) indices. The multiplicity of infection was 100:1 (bacteria/cell). Error bars, S.D. of three independent experiments, each performed in six wells. C, phagocytic index of the wild-type M1SF370 and M1T1 and their corresponding mutant and complemented strains. The phagocytic index was measured as multiplication factor (M.F.), demonstrating the ability of the GAS strain to multiply in the whole human defibrinated blood as described. Error bars, S.D. of the two independent experiments performed in triplicates using blood drawn from two healthy donors.
STP Deletion Abrogates Anti-phagocytic Activity of GAS
Capsule, which was found to be severely down-regulated in both of the mutant strains, is an important determinant in imparting anti-phagocytic activity to GAS. In bactericidal assays, both of the M1ΔSTP and M1T1ΔSTP mutants were effectively phagocytosed and were rapidly cleared from the whole blood (Fig. 5C). The complementation of the mutants with STP led to the restoration of both capsule expression (Fig. 4A) and concomitant anti-phagocytic activity (Fig. 5C), indicating that SP-STP, being a cognate phosphatase to SP-STK, participates in the regulation of “has” transcription via CovR and thus impacts capsule production in GAS.
Role of STP in Fatty Acid Biosynthesis of GAS
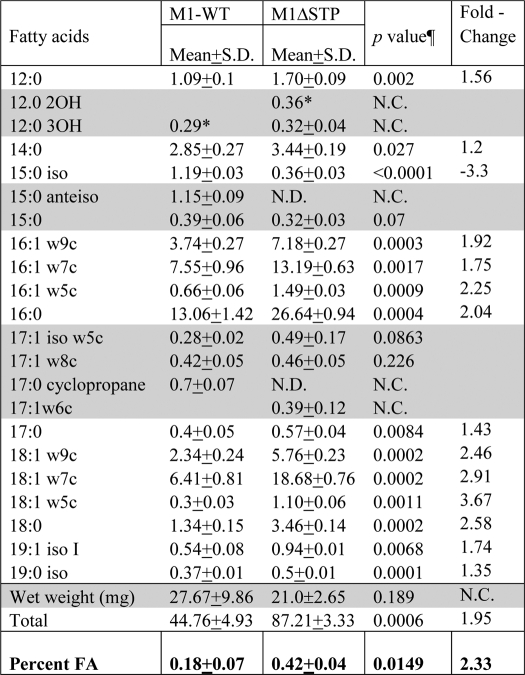
Unlike the wild-type GAS strains, the overnight liquid cultures of M1ΔSTP and M1T1ΔSTP mutant strains remained in suspension and did not settle. The increase in the capsule production had conferred increased buoyancy to the M1ΔSTK mutant (7). Because we observed significant down-regulation of capsular polysaccharides in the STP mutants, we speculated that this increased buoyancy could be due to the increased total lipid content. The total lipid content analysis (fatty acid methyl ester) of the wild type and its corresponding STP mutant revealed that both saturated fatty acid (SFA) and unsaturated fatty acid (UFA) content in the mutant were 2–3-fold up-regulated (Table 1). This was reflected in the overall 3-fold increase in total lipid content. Interestingly, in the wild-type M1SF370 strain, distribution of SFA and UFA was relatively equal (UFA/SFA = 1.03), whereas in the M1ΔSTP, this ratio was found to be 1.37, indicating a significant increase (34%) in the unsaturated fatty acid synthesis. The total lipid composition analysis also revealed the absence of C15:0 anteiso and C17:0 cyclopropane fatty acids in the M1ΔSTP mutant strain (Table 1).
TABLE 1.
Total fatty acid analysis in the wild-type M1SF370 and its isogenic M1ΔSTP mutant strain
Unless otherwise indicated, values indicate weight in μg. The ratio of UFA/SFA in M1-WT and M1ΔSTP is 1.02 and 1.33, respectively. -Fold change in SFA and UFA in M1ΔSTP versus M1-WT is 1.74 and 2.27, respectively. N.D., not detected; N.C., not calculated. Shaded rows, no significant difference.
* Amount detected only in one sample.
¶ Significance p values were statistically calculated by non-parametric t test using one-tailed parameter and applying Welch correction.
STP Is Essential for GAS Virulence
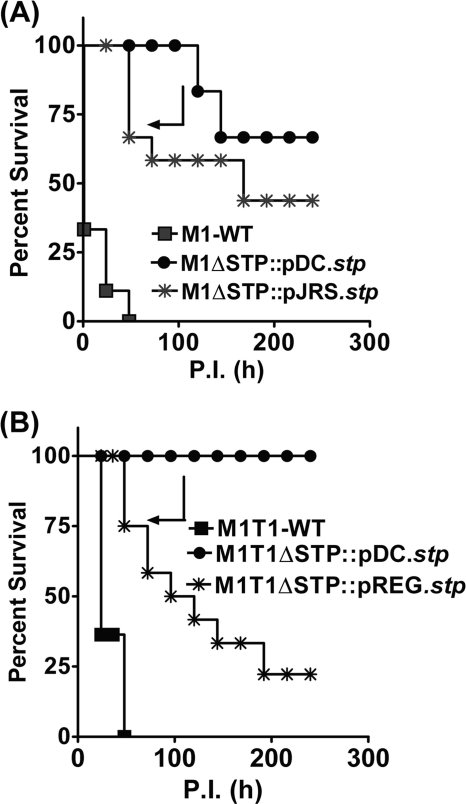
The defective cell division pattern in the STP mutants, their reduced ability to adhere to the host, and their increased susceptibility to phagocytosis indicated that SP-STP positively regulates GAS virulence and plays an important role in GAS pathogenesis. To confirm this, we employed the mouse infection model of peritonitis. Mice intraperitoneally infected with the wild-type M1SF370 strain showed 90% mortality in 2–3 days, whereas the M1ΔSTP mutant was completely attenuated for virulence because no morbidity or mortality was observed in the infected mice for the entire observation period (10 days) (Fig. 6A). On the other hand, whereas wild-type M1T1 caused 100% mortality in mice within 3 days postinfection, the virulence of M1T1ΔSTP mutant was significantly reduced (p < 0.0001) and caused mortality only in 40% of the infected mice (Fig. 6B).
The real-time PCR analyses of 38 genes (including 19 virulence-associated genes, 13 metabolism-related genes, and six one- or two-component regulators) for both of the mutant strains revealed different expression patterns (Fig. 6, C and D; see also supplemental Table S4). Except for the up-regulation of SpeB (16-fold) in M1ΔSTP, in essence, the majority of the genes (10 of 19) (SPy0128-pilin, SPy0711-SpeC, SPy0738-sagA, SPy1302-amyA, SPy1600-hyaluronidase, SPy1979-ska, SPy1983-scl, Spy2010-scpA, SPy2016-sic, and SPy2200-hasA) in M1ΔSTP were significantly (2–14-fold) down-regulated. Although in the M1T1ΔSTP strain, only 6 of 19 virulence genes (slo, pilin (SPy0128 equivalent), speB, speA2, hasA, and amyA) were down-regulated (3–500-fold), 5 of 19 genes (nga, scl, scpA, speJ, and speC) were significantly (4 to >5000-fold) up-regulated. Additionally, in both the mutants, the majority of the metabolism-related genes were also found to be down-regulated (Fig. 6, C and D). Together, these results (Fig. 6, A–D) clearly demonstrated that STP is essential for GAS virulence, and this regulation is strain-specific with more pronounced effects on the expression profile of the natively hypervirulent M1T1 strain (30).
STP Complementation with High and Low Copy Plasmids Differentially Affects the in Vivo Virulence
Because pDC123 is the most commonly used multicopy plasmid, we initially complemented M1ΔSTP and M1T1ΔSTP mutants using pDC123.stp plasmid. Despite the increased expression of STP in M1ΔSTP::pDC.stp and M1T1ΔSTP::pDC.stp (supplemental Fig. S1D), it could minimally restore the virulence in M1ΔSTP strain (i.e. only 20% mortality was observed) (Fig. 7A). On the other hand, complementation of the M1T1ΔSTP strain using the same construct could not restore the virulence, and 100% survival was obtained (Fig. 7B). We therefore hypothesized that this observed attenuation in the virulence despite complementation could be either due to the instability of the plasmid in the host, as reported earlier (16), or due to the overexpression of STP from a multicopy plasmid, resulting in the aberrant dephosphorylation.
FIGURE 7.
In vivo virulence of the STP mutants complemented with high copy (pDC123) and low copy (pREG696 and pJRS9508) plasmids. Effects of the stp complementation on GAS virulence in CD-1 mice (n = 10) for the wild-type and complemented strains of M1SF370 (A) and M1T1 (B) are shown. Survival/mortality for the strains was monitored for 10 days, and the data were statistically analyzed as described above in the legend to Fig. 6.
To obviate the problems associated with the plasmid instability and copy number, we harnessed the potential of two low copy number plasmids expressing SP-STP driven by two different promoters (M1ΔSTP::pJRS.stp and M1T1ΔSTP::pREG.stp) (supplemental Fig. S1E). Both the pREG696 and pJRS9508 plasmids with the stp gene placed under the control of Pspac and P23 promoters, respectively, also harbor the toxin-antitoxin (axe-txe) locus, which imparts substantial segregational stability when propagated in the host (17). The virulence potential of these aforementioned complemented strains revealed 60% mortality in mice infected with M1ΔSTP::pJRS.stp and 90% mortality in M1T1ΔSTP::pREG.stp (Fig. 7, A and B). These results thus indicated that the STP expression levels are crucial to restore the virulence.
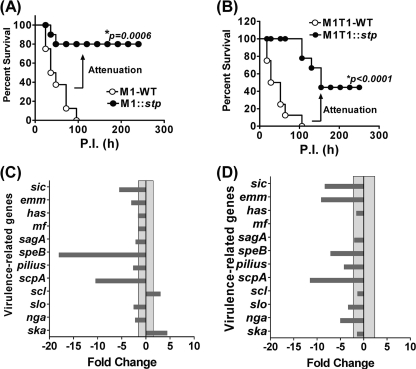
Wild-type GAS Strains Overexpressing SP-STP Are Substantially Attenuated for Virulence in Mice
To precisely address the hypothesis pertaining to the adverse effect of STP overexpression on GAS virulence, we constructed two wild-type GAS strains overexpressing STP by complementing them with pDC123.stp. The mice infected with both of the STP overexpressing strains, M1::stp and M1T1::stp, displayed a substantial decrease in the mortality (80% survival in M1::stp (p < 0.0006) and 40% survival in M1T1::stp (p < 0.0001)) as compared with their respective wild-type strains (Fig. 8, A and B). These results thus provide a supporting explanation in favor of the observed inability of the complemented strains to restore the wild-type virulence using a high copy plasmid, pDC123, which seemed to remain stable throughout the observation period.
FIGURE 8.
Effect of overexpression of STP on GAS virulence. CD-1 mice (n = 8) infected with STP-overexpressing M1SF370 (A) and M1T1 (B) strains were compared with their respective wild types using an experimental mouse model of peritonitis. Survival/mortality curves for all of the strains were monitored and evaluated as described in the legend to Fig. 6. Real-time PCR analysis was used to evaluate the changes in the expression profile of major virulence genes in the M1SF370-derived (C) and M1T1-derived (D) STP-overexpressing strains in comparison with their corresponding wild-type strains. The transcript abundance was quantified and analyzed as described in the legend to Fig. 6.
In accordance with the in vivo experimental mouse infection studies, the qRT-PCR analysis for the 12 major virulence-associated genes revealed substantial down-regulation of the majority of the genes analyzed in both of the STP-overexpressing wild-type GAS strains (Fig. 8, C and D, and supplemental Table S5). Thus, these results clearly indicated that the overexpression of a regulator beyond its physiological concentration can have adverse effects on the bacterial physiology and virulence.
DISCUSSION
Reversible phosphorylation is the key mechanism regulating cell growth, division, differentiation, pathogenicity, and secondary metabolism in both prokaryotes and eukaryotes. Although ESTKs are recognized as important one-component regulators controlling a variety of cellular functions in GAS (7) and other Gram-positive pathogens (5), the mechanism underlying their regulatory functions is not well understood. This is in part due to the paucity in the knowledge of the role of co-transcribing ESTP-mediated physiologically relevant functions. In the present investigation, by creating and characterizing the non-polar, viable STP mutants in two different Type M1 GAS strains, we have bridged this important knowledge gap, which existed due to the inability to generate an STP-specific knock-out mutant in GAS.
Because ESTKs and ESTPs are co-transcribing proteins and belong to a one-component system (31), several affected functions in our SP-STP mutants were found to be cognately regulated by the SP-STK/SP-STP-mediated reversible phosphorylation (7). In this context, the long chain formation in M1/M1T1ΔSTP mutants is reminiscent of our recent report on the implications of the overexpression of the CdhA protein (SPy0019) in GAS chain formation and cell division-plane recognition (20). Because STK positively regulates CdhA expression (20), we speculated that this regulation is mediated via WalR/VicR/YycF, a two-component response regulator (32). Because STK and STP mediated reversible phosphorylation of WalR in vitro, we believe that in the absence of STP, the constitutively phosphorylated STK will phosphorylate WalR, thereby facilitating its binding to the cdhA promoter, resulting in its transactivation and increased CdhA expression, leading to the chain-forming phenotype. The cell division defects observed at the level of septa formation could be a consequence of decreased cdhA transcript (2.3-fold) at the later stages of growth.
The bacterial cell division and cell wall synthesis are closely related and interdependent phenomena (33), wherein penicillin-binding proteins (PBPs) play an important role (34). However, unaltered expression profiles of several pbp genes in the M1ΔSTP and M1T1ΔSTP mutants (supplemental Table S4) displaying thick cell walls indicate that in the absence of SP-STP, the SP-STK with its kinase activity enhances cell wall synthesis and modulation. As a result of this, the STP mutants display relatively thicker cell walls and unaltered antibiotic susceptibility patterns for cell wall-acting antibiotics (supplemental Table S6), a phenotype similar to the STP mutant derived from S. aureus N315 (8). The defective cell wall synthesis and extreme susceptibility to the cell wall-acting antibiotics in the STK mutants derived from S. aureus and GAS (8, 20) further signify the cognate regulation by SP-STK and SP-STP in modulating bacterial cell division and cell wall synthesis.
The GAS mutants lacking SP-STK display increased capsule as well as hemolysin production (7), both of which have been shown to be regulated by CovRS two-component regulatory system (29). Similar evidence of cognate regulation was also observed for hemolysin production in the STP mutants of S. agalactiae (35) and S. aureus (10). The significantly decreased expression of the hasA and sagA genes with a corresponding decrease in the hyaluronic acid content and hemolysin production in the M1ΔSTP and M1T1ΔSTP mutants underscores the cognate regulation and the importance of reversible phosphorylation carried out by SP-STK and SP-STP. Because the expression levels of covR and covS transcripts remained unaltered in these mutants (supplemental Table S4 and Fig. 6, C and D), it is rational to propose that the capsule and hemolysin production in GAS is also regulated by the reversible phosphorylation of CovR by SP-STK and SP-STP in addition to CovS, with one compensating for the function of the other in GAS. In fact, the present study also demonstrates the in vitro phosphorylation of CovR by STK, indicating that this one component signal transduction pair (SP-STK/STP) integrates with the known TCSs to coordinately regulate GAS physiology and pathogenesis.
The significantly decreased adherence pattern of the M1SF370-derived STP mutant can be attributed to the severalfold down-regulation of transcripts encoding surface proteins, such as pilin (SPy0128) (5-fold), scpA (2.4-fold), sic (2.3-fold), and scl (4.5-fold), which is also responsible for the evident loss of the surface electron-dense layer (Fig. 2C). However, in contrast to this, we did not observe a significant reduction in the outermost fibrillar electron-dense layer in the M1T1-derived STP mutant strain (Fig. 2D), which corroborated with the observed up-regulation of the transcripts encoding two surface proteins, scl (4.0-fold) and scpA (5.6-fold), which is consequently reflected in the unaltered adherence pattern of this mutant strain.
One of the intriguing phenomena to be noted in the present study, as also we observed previously for the M1ΔSTK mutant (7), is that although the M1ΔSTP mutant lost the ability to adhere to pharyngeal cells, the adherent mutant strain was 6-fold more invasive. We believe that this is in part due to ∼14-fold up-regulation of the speB gene in M1ΔSTP and several fold down-regulation of this gene in M1T1ΔSTP mutant strain. Although the role of SpeB in GAS invasion is debatable, the recent reports have clearly demonstrated that the direct cysteine protease activity may confer an advantage to GAS in tissue invasion, and its presence in systemic infection may remain disadvantageous for GAS because its secreted proteolytic activity degrades GAS anti-phagocytic surface proteins, rendering them vulnerable to host innate immunity (36). On the other hand, GAS with down-regulated SpeB status is shown to be more invasive and able to cause metastatic lesion through the bloodstream due to unaffected intact anti-phagocytic molecules on its surface (37). Therefore, in support of the former hypothesis, the M1ΔSTP mutant strain producing elevated levels of SpeB, although it remains highly invasive in vitro, was attenuated for in vivo virulence in mice.
The role of lipid biosynthesis in virulence regulation has been studied for S. pneumoniae (38), S. aureus (39), and L. monocytogenes (40) but not for GAS. Like S. pneumoniae, the FAS-II operon in the M1SF370 GAS strain is constituted by SPy1743 to SPy1755. Fatty acid biosynthesis (FAS-II) in Streptococcus species is regulated by the fabT transcription regulator (41), which negatively regulates the expression of FAS-II operon genes (41). As in the case of S. pneumoniae (38), we also observed down-regulation of the fabT homolog of GAS, marR/SPy1755 (3–6-fold), in both of the mutant strains (supplemental Table S4; see also Fig. 6, C and D) and a concomitant increase in the total fatty acid content in M1ΔSTP. Further, 3–10-fold down-regulation of enolyl-CoA hydratase encoding SPy1758/phaB in both of the mutants (supplemental Table S4; see also Fig. 6, C and D) responsible for fatty acid catabolism correlates with the 4-fold increased total lipid content in the M1ΔSTP mutant (Table 1). Our study showing increased fatty acid biosynthesis in the M1ΔSTP mutant is in agreement with the recently demonstrated role of SP-STK in positively regulating fatty acid biosynthesis (42). This highlights another level of cognate regulation mediated by this SP-STK/STP couple in controlling metabolic processes within GAS and is speculated to be manifested by SP-STK-mediated phosphorylation of the fabT repressor. The deletion of the FAS-II operon in S. mutans resulted in decreased fatty acid biosynthesis with attenuation for virulence (43), which is contrary to the M1ΔSTP mutant showing complete virulence attenuation despite the increased fatty acid biosynthesis, which calls into question the precise role of FAS-II genes in regulating GAS virulence. Interestingly, the absence of C15:0 anteiso branched chain fatty acid in the M1ΔSTP mutant (Table 1) is attributed to the 4–10-fold down-regulation of the fabH gene (SPy1754), which is involved in the branched chain fatty acid synthesis (44). The down-regulation of the fabH/SPy1754 in M1ΔSTP and corresponding absence of the C15:0 anteiso branched chain fatty acid (44) corroborate with the attenuation of virulence, as was also observed in S. aureus (45) and L. monocytogenes (40). Further, the undetected C17:0 cyclopropane fatty acid, which is responsible for conferring acid tolerance to bacteria by altering the membrane rigidity and fluidity (46), may play an indirect role in reduced survival of the M1ΔSTP mutant within the host, especially while encountering an unfavorable intracellular environment.
Based on the transmission electron microscopy data, qRT-PCR analyses, and animal experiments, the observed phenotypes of stp mutants in two evolutionarily distinct M strains of the same type, allow us to conclude that SP-STP and its dephosphorylation activity seem to interface directly or indirectly with several regulatory networks, including those governed by two-component regulatory systems and stand-alone regulators that regulate expression of several genes associated with virulence, carbohydrate metabolism, lipid metabolism, and cell division (3, 47). In essence, the effect on the expression levels of many genes, especially those associated with virulence, was more pronounced in the M1T1ΔSTP strain than in the M1ΔSTP strain, possibly due to the fact that M1T1 strain inherently produces more nga-encoded NADase, slo-encoded streptolysin-O, and amyA-encoded cyclomaltodextrin glucanotransferase (30).
In the present study, STP mutants showing observed strain-specific variation in the expression of SpeB and other virulence factors and resulting variation in the virulence phenotypes is not unprecedented (48). The observed differences in the SpeB expression could likely be due to the strain-specific differences in the rgg expression (supplemental Table S4) because intact expression of Rgg is essential for SpeB expression (36). The strain-associated differences in the regulon are probably due to the ability of STP and STK to interface with various other regulatory systems, including Rgg. The latter in turn interacts with other subregulons, which are found to be species-specific (48). Therefore, it can be inferred that the deletion of STP will not only affect the genes that are directly under its control but will also have multifarious effects on the genes that are indirectly controlled by other regulators, as is also highlighted clearly in Fig. 9.
FIGURE 9.
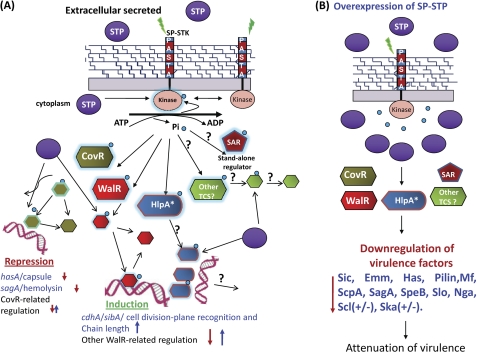
Schematic representation of the regulatory role of the SP-STK/SP-STP couple in modulating GAS physiology and virulence. A, the external cues sensed by the wall-associated PASTA domain of SP-STK results in the activation of its cytoplasmically located kinase domain (STKK). The autophosphorylated (activated) SP-STK interfaces with certain well established TCSs (e.g. CovR and WalR) and possibly other stand-alone regulators (SAR). In that response, regulators of TCS get Thr-phosphorylated by SP-STK. The latter also recognizes other substrates, such as histone-like protein HlpA (7). The Thr-phosphorylated TCS regulators bind to specific promoters and regulate the transcription of several genes encoding for virulence factors such as capsule (Has), hemolysin (Sag), and CdhA. The schematic diagram summarizes the findings of the present study by showing that these function are cognately regulated by co-transcribing SP-STP, whose optimal balance in cytoplasm and its phosphatase activity at various levels may be required to allow or prevent the binding of regulators to its promoters and thus help maintain homeostasis within GAS (i.e. expression levels of various genes responsible for physiological/metabolic processes, growth, cell wall structure, cell division, and virulence). The absence of SP-STP thus allows uncontrolled SP-STK-mediated phosphorylation, which is manifested in the form of unique phenotypes. B, schematic diagram showing the implications of overexpression of SP-STP in GAS. Overexpression of SP-STP may dephosphorylate STK, and the Thr phosphorylation is abrogated even in the presence of appropriate external cues. This may result in down-regulation of virulence genes and attenuation and strain-specific differential expression of genes (M1SF370/M1T1). Normally secreted and overexpressed SP-STP may affect host cell signaling. + and ↑, up-regulation; − and ↓, down-regulation.
Further, the results of the complementation experiments provide an important message with regard to the SP-STK/SP-STP-mediated regulation. They emphasize that the signal transduction events regulated by the reversible phosphorylation process demand a crucial balance of phosphorylation and dephosphorylation. Thus, on one hand, the inability to restore the lost function by the use of commonly employed multicopy plasmid pDC123 (22) clearly indicated that the increased expression of STP is not only deleterious to the STP mutants (Fig. 7) but also to the wild-type GAS strains (Fig. 8). On the other hand, the ability to restore the wild-type functions by the use of a low copy plasmid emphasizes the fact that the optimal concentration of any multifunctional regulatory protein is a prerequisite to maintain the cellular homeostasis.
Thus, GAS pathogenesis is multifactorial and is regulated by the interplay of highly dynamic and temporally regulated processes. In summary, because the phosphatase activity plays a key role in signaling events both in eukaryotes and prokaryotes, SP-STP-mediated GAS virulence is invariably an outcome of the cumulative effects of (i) the expressed virulence factors under their direct or indirect regulation, (ii) the nature of the host response to those expressed virulence factors, (iii) GAS strain specificity, and (iv) the host that they are infecting. Considering these factors in perspective, the activity of SP-STP and its secretory nature (7) could have important implications in host signaling events. Based on the present study, we believe that the overexpression and hence accumulation of STP within GAS might be detrimental to its pathogenicity. Thus, despite not having a classical signal sequence, GAS has strategically devised a hitherto unknown mechanism in favor of its secretion during the late log phase of its growth. We are presently investigating this hypothesis. In conclusion, our study is a first significant step in understanding the role of SP-STP in regulating strain-specific GAS virulence and host-specific GAS pathogenesis.
Acknowledgments
We sincerely thank June R. Scott (Emory University, Atlanta, GA) for the low copy number plasmids (pREG696 and pJRS9508), Indranil Biswas (University of Kansas Medical Center) for pIB167 plasmid, Rita Kansal and Malak Kotb for M1T1 strain, and Daniel Nelson and Vincent Fischetti (Rockefeller University, New York) for recombinant phage lysin (PlyC). We also thank Rajamohan Govindan for the modification of the pWF6 vector and Vijaya Bharathi Srinivasan and Hong Jin for two-dimensional thin layer chromatography.
This work was supported, in whole or in part, by National Institutes of Health Grants AI-64912 and AI-76889 (to V. P.). This work was also supported by Ohio State University departmental funds (to P. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S6 and Figs. S1–S3.
- GAS
- group A Streptococcus
- STK
- serine/threonine kinase
- STP
- serine/threonine phosphatase
- ESTK
- eukaryote-like serine/threonine kinase
- ESTP
- eukaryote-like serine/threonine phosphatase
- SP-STP
- S. pyogenes serine/threonine phosphatase
- SP-STK
- S. pyogenes serine/threonine kinase
- SP-WalR
- SP-CovR, and STKK, S. pyogenes WalR, CovR, and kinase domain of serine/threonine kinase, respectively
- shRBC
- sheep red blood cell
- qRT-PCR
- quantitative RT-PCR
- SFA
- saturated fatty acid
- UFA
- unsaturated fatty acid
- TCS
- two-component system.
REFERENCES
- 1. Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005) Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 2. Musser J. M., Shelburne S. A., 3rd (2009) J. Clin. Invest. 119, 2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kreikemeyer B., McIver K. S., Podbielski A. (2003) Trends Microbiol. 11, 224–232 [DOI] [PubMed] [Google Scholar]
- 4. Stock A. M., Robinson V. L., Goudreau P. N. (2000) Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 5. Pereira S. F., Goss L., Dworkin J. (2011) Microbiol. Mol. Biol. Rev. 75, 192–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Y. (2009) Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 7. Jin H., Pancholi V. (2006) J. Mol. Biol. 357, 1351–1372 [DOI] [PubMed] [Google Scholar]
- 8. Beltramini A. M., Mukhopadhyay C. D., Pancholi V. (2009) Infect. Immun. 77, 1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Treuner-Lange A., Ward M. J., Zusman D. R. (2001) Mol. Microbiol. 40, 126–140 [DOI] [PubMed] [Google Scholar]
- 10. Burnside K., Lembo A., de Los Reyes M., Iliuk A., Binhtran N. T., Connelly J. E., Lin W. J., Schmidt B. Z., Richardson A. R., Fang F. C., Tao W. A., Rajagopal L. (2010) PLoS One 5, e11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madec E., Laszkiewicz A., Iwanicki A., Obuchowski M., Séror S. (2002) Mol. Microbiol. 46, 571–586 [DOI] [PubMed] [Google Scholar]
- 12. Banu L. D., Conrads G., Rehrauer H., Hussain H., Allan E., van der Ploeg J. R. (2010) Infect. Immun. 78, 2209–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ulijasz A. T., Falk S. P., Weisblum B. (2009) Mol. Microbiol. 71, 382–390 [DOI] [PubMed] [Google Scholar]
- 14. Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chatellier S., Ihendyane N., Kansal R. G., Khambaty F., Basma H., Norrby-Teglund A., Low D. E., McGeer A., Kotb M. (2000) Infect. Immun. 68, 3523–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rajagopal L., Clancy A., Rubens C. E. (2003) J. Biol. Chem. 278, 14429–14441 [DOI] [PubMed] [Google Scholar]
- 17. Grady R., Hayes F. (2003) Mol. Microbiol. 47, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 18. Barnett T. C., Bugrysheva J. V., Scott J. R. (2007) J. Bacteriol. 189, 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Podbielski A., Spellerberg B., Woischnik M., Pohl B., Lütticken R. (1996) Gene 177, 137–147 [DOI] [PubMed] [Google Scholar]
- 20. Pancholi V., Boël G., Jin H. (2010) J. Biol. Chem. 285, 30861–30874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pancholi V., Fischetti V. A. (1992) J. Exp. Med. 176, 415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chaffin D. O., Rubens C. E. (1998) Gene 219, 91–99 [DOI] [PubMed] [Google Scholar]
- 23. Biswas I., Jha J. K., Fromm N. (2008) Microbiology 154, 2275–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin H., Agarwal S., Agarwal S., Pancholi V. (2011) MBio 2, e00068–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ravins M., Jaffe J., Hanski E., Shetzigovski I., Natanson-Yaron S., Moses A. E. (2000) J. Infect. Dis. 182, 1702–1711 [DOI] [PubMed] [Google Scholar]
- 26. de Hoon M. J., Makita Y., Nakai K., Miyano S. (2005) PLoS Comput. Biol. 1, e25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ng W. L., Robertson G. T., Kazmierczak K. M., Zhao J., Gilmour R., Winkler M. E. (2003) Mol. Microbiol. 50, 1647–1663 [DOI] [PubMed] [Google Scholar]
- 28. Barendt S. M., Land A. D., Sham L. T., Ng W. L., Tsui H. C., Arnold R. J., Winkler M. E. (2009) J. Bacteriol. 191, 3024–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Churchward G. (2007) Mol. Microbiol. 64, 34–41 [DOI] [PubMed] [Google Scholar]
- 30. Sumby P., Porcella S. F., Madrigal A. G., Barbian K. D., Virtaneva K., Ricklefs S. M., Sturdevant D. E., Graham M. R., Vuopio-Varkila J., Hoe N. P., Musser J. M. (2005) J. Infect. Dis. 192, 771–782 [DOI] [PubMed] [Google Scholar]
- 31. Ulrich L. E., Zhulin I. B. (2007) Nucleic Acids Res. 35, D386–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winkler M. E., Hoch J. A. (2008) J. Bacteriol. 190, 2645–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabeen M. T., Jacobs-Wagner C. (2005) Nat. Rev. Microbiol. 3, 601–610 [DOI] [PubMed] [Google Scholar]
- 34. Zapun A., Contreras-Martel C., Vernet T. (2008) FEMS Microbiol. Rev. 32, 361–385 [DOI] [PubMed] [Google Scholar]
- 35. Rajagopal L., Vo A., Silvestroni A., Rubens C. E. (2006) Mol. Microbiol. 62, 941–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hollands A., Aziz R. K., Kansal R., Kotb M., Nizet V., Walker M. J. (2008) PLoS One 3, e4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aziz R. K., Kansal R., Aronow B. J., Taylor W. L., Rowe S. L., Kubal M., Chhatwal G. S., Walker M. J., Kotb M. (2010) PLoS One 5, e9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu Y. J., Rock C. O. (2006) Mol. Microbiol. 59, 551–566 [DOI] [PubMed] [Google Scholar]
- 39. Balemans W., Lounis N., Gilissen R., Guillemont J., Simmen K., Andries K., Koul A. (2010) Nature 463, E3–E5 [DOI] [PubMed] [Google Scholar]
- 40. Sun Y., O'Riordan M. X. (2010) Infect. Immun. 78, 4667–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujita Y., Matsuoka H., Hirooka K. (2007) Mol. Microbiol. 66, 829–839 [DOI] [PubMed] [Google Scholar]
- 42. Bugrysheva J., Froehlich B. J., Freiberg J. A., Scott J. R. (2011) Infect. Immun. 79, 4201–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fozo E. M., Scott-Anne K., Koo H., Quivey R. G., Jr. (2007) Infect. Immun. 75, 1537–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Choi K. H., Heath R. J., Rock C. O. (2000) J. Bacteriol. 182, 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh V. K., Hattangady D. S., Giotis E. S., Singh A. K., Chamberlain N. R., Stuart M. K., Wilkinson B. J. (2008) Appl. Environ. Microbiol. 74, 5882–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fozo E. M., Quivey R. G., Jr. (2004) Appl. Environ. Microbiol. 70, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McIver K. S. (2009) Contrib. Microbiol. 16, 103–119 [DOI] [PubMed] [Google Scholar]
- 48. Wehenkel A., Bellinzoni M., Schaeffer F., Villarino A., Alzari P. M. (2007) J. Mol. Biol. 374, 890–898 [DOI] [PubMed] [Google Scholar]