Background: Herpesviruses have evolved sophisticated strategies to escape immune surveillance.
Results: EBV BNLF2a acts as tail-anchored protein and is posttranslationally inserted into the ER membrane, where it arrests core TAP in a transport-incompetent conformation.
Conclusion: BNLF2a exploits the host tail-anchored protein insertion machinery.
Significance: This inhibition mechanism is distinct and mutually exclusive of other viral TAP inhibitors.
Keywords: ABC Transporter, Antigen Processing, Cellular Immune Response, Intracellular Trafficking, Viral Immunology, Get Pathway, Tail-anchored Proteins
Abstract
Virus-infected cells are eliminated by cytotoxic T lymphocytes, which recognize viral epitopes displayed on major histocompatibility complex class I molecules at the cell surface. Herpesviruses have evolved sophisticated strategies to escape this immune surveillance. During the lytic phase of EBV infection, the viral factor BNLF2a interferes with antigen processing by preventing peptide loading of major histocompatibility complex class I molecules. Here we reveal details of the inhibition mechanism of this EBV protein. We demonstrate that BNLF2a acts as a tail-anchored protein, exploiting the mammalian Asna-1/WRB (Get3/Get1) machinery for posttranslational insertion into the endoplasmic reticulum membrane, where it subsequently blocks antigen translocation by the transporter associated with antigen processing (TAP). BNLF2a binds directly to the core TAP complex arresting the ATP-binding cassette transporter in a transport-incompetent conformation. The inhibition mechanism of EBV BNLF2a is distinct and mutually exclusive of other viral TAP inhibitors.
Introduction
Displaying peptide epitopes at the cell surface is an essential step in the initiation of an adaptive immune response against infection and tumor progression. Cytotoxic T lymphocytes monitor peptide epitopes presented by major histocompatibility complex class I (MHC I)4 molecules at the surface of target cells (1, 2). Most epitopes displayed on MHC I are derived from proteasomal degradation in the cytosol (3, 4). The transporter associated with antigen processing (TAP), comprised of TAP1 and TAP2, fulfills the important task of translocating these peptides into the ER lumen, where they are loaded onto MHC I through the peptide-loading complex (PLC), consisting of TAP1/2, tapasin, ERp57, calreticulin, MHC I heavy chain, and β2-microglobulin (5, 6). After peptide editing and dissociation from the PLC, peptide-MHC complexes traffic to the cell surface, where they are monitored by cytotoxic T lymphocytes, which eventually eliminate the infected or malignantly transformed cell.
To circumvent presentation and cytotoxic T lymphocyte recognition of viral epitopes, viruses have evolved clever ways of immune evasion. The family of herpesviridae encodes several factors that interfere with antigen processing, in particular by affecting TAP function (7, 8). ICP47 of HSV type 1 (HSV-1) binds with high affinity to the cytosolic face of human TAP and blocks peptide binding (9–11). As a type I membrane glycoprotein, US6 of human cytomegalovirus (HCMV) inhibits ATP binding to the cytosolic nucleotide-binding domains (NBDs) of TAP via its ER-lumenal domain (12, 13). mK3 of murine γ-herpesvirus 68 (MHV-68) destabilizes MHC I, TAP, and tapasin, thus inducing rapid proteasomal degradation of the PLC (14, 15). As a type I membrane protein, UL49.5 of bovine herpesvirus 1 inhibits peptide transport and simultaneously induces proteasomal degradation of the PLC via its C-terminal tail (16, 17). Recently, BNLF2a encoded by EBV has been identified and shown to inhibit antigen presentation by preventing peptide loading of MHC I molecules (18). Nonetheless, little is known about the inhibition mechanism of BNLF2a.
EBV is a member of the γ1-herpesvirus subfamily (human herpesvirus 4) belonging to the genus of Lymphocryptovirus. EBV is known to be the causative agent for infectious mononucleosis and a subset of malignant tumors such as Hodgkin's disease and nasopharyngeal carcinoma (19). Over 90% of adult human beings worldwide have a latent EBV infection.
BNLF2a (60 amino acids, 6.5 kDa) consists of a hydrophilic N-terminal region of 40 residues followed by a predicated C-terminal transmembrane domain of 20 amino acids. Because BNLF2a lacks an N-terminal signal sequence for cotranslational membrane insertion, we anticipated that it belongs to the group of tail-anchored membrane proteins. Thus, BNLF2a and its homologues display a hitherto unique structure and the molecular inhibition mechanism needs to be captured.
Here we established both an in vitro translation and insect cell expression system to study the inhibition mechanism of this viral factor. We demonstrate that BNLF2a is a tail-anchored protein that is posttranslationally inserted into the ER membrane, where it binds directly to the core TAP complex. BNLF2a arrests the TAP heterodimer in a transport-incompetent conformation that excludes binding of the viral immune evasin US6. Thus the TAP inhibition mechanism of EBV BNLF2a is distinct and TAP binding is mutually exclusive from HCMV-US6.
EXPERIMENTAL PROCEDURES
Cloning and Constructs
BNLF2a was synthesized de novo (gene ID 3783720) (20) and was used as a template for PCR amplification. PCR reactions were performed under standard conditions using Phusion DNA polymerase (Finnzymes, Vantaa, Finland) and synthetic oligonucleotide primers (endonuclease cleavage sites are underlined). All constructs were verified by DNA sequencing. For expression in human cells, PCR-generated products were inserted into pIRES2-EGFP (Clontech) via the respective restriction sites upstream of the internal ribosome entry site (IRES) and enhanced GFP. The following primers were used to generate BNLF2aC8-NST: CCGGAATTCCGG ATGGTGCACGTGCTGG EcoRI forward and GCCGGATCCTCAATCCACG GTGCTGTTTTCTTCAATGCCTTCCGGGCGACCGGGCCGCGCGGCCTGCTAATCAGCAGCAGGCACAG BamHI reverse. BNLF2aHA was created with the following primers: CCGGAATTCCGGATGTACCCATACGATGTTCCGGATTACGCTGGCGGCGGCAGCATGGTGCACGTGCTGG EcoRI forward and CGCGGATCCTCAAGCGTAATCCGGAACATGTTGGGTGCTGCCGCCGCCGAGCTCAAT BamHI reverse. UL49.5 was generated using CCGGAATTCCGGATGCCAA GGTCC EcoRI forward and CGCGGATCCTCAACCTCTACC TCTACTC BamHI reverse primers and pIRES2-EGFP-UL49.5 (17) as template. Constructs for in vitro translation experiments were cloned into a modified pSP64 vector directly downstream of the 5′UTR from Xenopus globin (21). BNLF2aC8-NST was amplified with the following primers: CGATTACTCGAGTCCATGGTGCA CGTGCTGG NcoI forward and CCGGAATTC CATCATCATGGTGCTGTTTTCTTCAATGC EcoRI reverse. UL49.5 was amplified using CATGCCATGGGACCAAGGTCCCCTCTGATCG NcoI forward and CGCGGATCCACCTC TACCTCTACTC BamHI reverse primers. Ramp4-opsin was kindly provided by V. Favaloro and B. Dobberstein (Zentrum für Molekulare Biologie Heidelberg/Deutsches Krebsforschungszentrum, Heidelberg, Germany) (22). For expression in insect cells, Spodoptera frugiperda (Sf9) BNLF2aC8-NST was amplified with the NcoI forward/EcoRI reverse primers indicated above and cloned into pFastBactm1-vector (Invitrogen). TAP1 and TAP2 were cloned into the pFastBactmDual vector (Invitrogen) as described (23). Generation of the US6myc-pFastBactm was conducted with GCGGATCCATGGATCTCTTGATTCGTCTCGGTT BamHI forward and GCGAATTCTTACAAGTCTTCCTCGCTGATCAATTTCTGCTCGGAGCCACAACGTCGAATCC EcoRI reverse primer using US6 as template (13).
Antibodies
TAP1 (mAb 148.3) and TAP2 (mAb 435.3) were used for immunoblotting and immunoprecipitation (24). C8-tagged BNLF2a was detected by an anti-C8 antibody provided by D. Oprian (Brandeis University). The phycoerythrin-coupled anti-human HLA-ABC (W6/32) and the mouse IgG2a isotype control were purchased from eBioscience (San Diego, CA) and BioLegend (San Diego, CA), respectively. The monoclonal antibody directed against human β-actin and the rabbit polyclonal antibody against human calnexin were purchased from Sigma-Aldrich. US6 was detected by a monoclonal myc-tag antibody (4A6) from Millipore (Schwalbach, Germany). The monoclonal antibody against the 54-kDa subunit of the human signal recognition particle was purchased from BD Biosciences. Affinity-purified rabbit antiserum directed against Sec61α (25) was kindly provided by A. E. Johnson (Texas A&M University, College Station, TX). Affinity-purified anti-HA antibody produced in rabbit was obtained from Sigma-Aldrich. The HC10 antibody, which recognizes human MHC I heavy chain (26), was kindly provided by H. L. Ploegh (Massachusetts Institute of Technology, Cambridge, MA).
Cell Lines, Infection, and Transfection
Insect cells, S. frugiperda (Sf9) were grown in Sf900II medium (Invitrogen) following standard procedures. Sf9 insect cells were infected with recombinant baculovirus encoding for US6myc, BNLF2aC8-NST, human codon-optimized TAP1/2 (23), and human wild-type TAP1His6/TAP2 (24), respectively. 60 h after infection, cells were harvested, and membranes were prepared as described (24).
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C in a 5% CO2-humidified atmosphere. HeLa cells were transfected with BNLF2aC8-NST-pIRES2-EGFP, BNLF2aHA-pIRES2-EGFP, and UL49.5-pIRES2-EGFP, respectively. HeLa cells were seeded in 6-well plates with a density of 4 × 105 cells/well and transfected using FuGENE 6 transfection reagent (Roche) with 2 μg DNA/well following manufacturer's protocol. For coimmunoprecipitation experiments, HeLa cells were treated with 2.5 ng/ml interferon-γ (Sigma Aldrich). Start of stimulation was 18 h post-transfection and was continued for 24 h until harvesting of the cells.
Flow Cytometry
MHC I surface expression was analyzed using the phycoerythrin-coupled antibody W6/32, which recognizes peptide-MHC I complexes. 106 cells were trypsinized, washed with 1 ml of FACS buffer (2% fetal bovine serum in PBS (pH 7.3)), and centrifuged at 180 × g for 3 min at 4 °C. For blocking of nonspecific binding, the cells were incubated with 100 μl of FACS buffer containing 5% (w/v) bovine serum albumin for 10 min on ice. After two washing steps with FACS buffer, the corresponding antibody (1:5 in FACS buffer) was added to the cells and incubated for 15 min on ice in the dark. Subsequently, the cells were washed twice with FACS buffer and finally resuspended in 0.5 ml. The cells were analyzed using a FACSAria flow cytometer (BD Biosciences). For each experiment, 3 × 104 cells were evaluated.
In Vitro Translation and ER Insertion
Plasmids (pSP64-BNLF2aC8-NST containing a C-terminal C8 tag followed by an N-core glycosylation site and three extra methionines, pSP64-UL49.5 (17), pSP64-Ramp4opsin (22), and pSP64-BPL (21), 1 μg per 25-μl reaction) were transcribed and translated in vitro in rabbit reticulocytes lysate (Promega) in the presence of [35S]Met (Hartmann Analytic, Braunschweig, Germany, 10 μCi per 25-μl reaction). After incubation for 90 min at 30 °C, translation was stopped by addition of puromycin (2 mm final). For cotranslational membrane insertion, dog pancreas rough microsomes (RM, Promega) were added before the transcription/translation reaction. For posttranslational membrane insertion, translation was performed in the absence of microsomes. After puromycin treatment and translation termination, rough microsomes were added, and the samples were incubated for an additional 30 min at 30 °C.
For the generation of truncated mRNAs lacking a stop codon, BNLF2aC8-NST was amplified directly from the pSP64-BNLF2aC8-NST plasmid as described above using the GATTTAGGTGACACTATAGAATAC SP6-forward and CATCATCATGGTGCTGTTTT CTTC reverse primers. The purified PCR product was transcribed in vitro using SP6 RNA polymerase (27). In vitro translations of mRNA templates in wheat germ cell-free extract (tRNA Probes, LLC, College Station, TX) were performed for 40 min at 26 °C in the presence of [35S]Met (2 μCi per 25-μl reaction), dog pancreas rough microsomes, 40 nm canine signal recognition particle (SRP) (tRNA Probes) as indicated, and other components as described (28). WRB-67 inhibitory peptide (residue 35–101) was kindly provided by M. Mariappan and R. S. Hegde (MRC, Cambridge, UK).
Rough microsomes were collected by sedimentation through a 0.5 m sucrose cushion in HEPES buffer (10 mm HEPES (pH 7.5), 100 mm KAc, 1 mm MgAc, 1 mm DTT) at 100,000 × g for 20 min at 4 °C. Translation products were analyzed either directly or solubilized for 10 min at 100 °C in denaturation buffer (0.5% SDS, 40 mm DTT) prior to EndoH treatment (New England Biolabs, 25 units/μl for 1 h in 50 mm sodium citrate (pH 5.5), 0.25% SDS, 20 mm DTT). Proteins were then examined by Tricine/SDS-PAGE (10%) and autoradiography (PhosphoImager, GE Healthcare). Intensities were quantified using ImageJ. For carbonate extraction assays, membranes were collected by sedimentation as before. After incubation in carbonate buffer (0.1 m Na2CO3 (pH 11.5)) for 15 min on ice, the membranes were collected by centrifugation (100.000 × g for 20 min at 4 °C), washed, and centrifuged before final resuspension in carbonate buffer. The supernatants and the resuspended pellets were neutralized with glacial acetic acid and further analyzed as above.
Immunoprecipitation and Immunoblotting
Raji microsomes were prepared from human Burkitt's lymphoma cells (Raji) (29). Proteins were translated in vitro in the presence of Raji microsomes in rabbit reticulocytes lysate as described above. After completion of translation, microsomes were collected by sedimentation through a 0.5 m sucrose cushion in HEPES buffer (10 mm HEPES (pH 7.5), 100 mm KAc, 1 mm MgAc, 1 mm DTT) at 100,000 × g for 10 min at 4 °C. Membranes were solubilized in 1 ml of lysis buffer L1 (20 mm Tris/HCl (pH 7.5), 150 mm NaCl, 5 mm MgCl2, 2% (w/v) digitonin (Carl Roth, Karlsruhe, Germany)).
Membranes isolated from Sf9 insect cells (1 mg of total protein) or 2 × 106 transfected HeLa cells, respectively, were resuspended in 0.5 ml lysis buffer as described above. After solubilization for 60 min on ice, non-solubilized proteins were removed by centrifugation at 100,000 × g for 30 min at 4 °C. The supernatant was incubated with Dynabeads (M-280 sheep anti-mouse IgG, Dynal Biotech, Hamburg, Germany), which had been preloaded with antibodies. Dynabeads loaded with antibodies either directed against the unrelated ER-resident protein Sec61α or directed against other unrelated proteins (anti-myc or anti-HC10, respectively) were used as adequate negative controls. Beads were washed three times with 1 ml of washing buffer (20 mm Tris/HCl, 150 mm NaCl, 2 mm EDTA, 0.2% (w/v) digitonin (pH 7.5)). Proteins were eluted in SDS sample buffer (2% SDS, 50 mm Tris/HCl, 200 mm DTT, 10% glycerol, 0.05% bromphenol blue (pH 8.0)) for 3 min at 65 °C. Samples were denatured for 20 min at 65 °C and separated by Tricine/SDS-PAGE (10%). After electrotransfer onto polyvinylidene difluoride membranes, proteins were either detected by phosphoimaging or by incubating the membranes with specific antibodies as indicated. Horseradish peroxidase-conjugated secondary antibodies were detected with Lumi-Imager F1 (Roche).
Cross-linking of TAP
Membranes isolated from Sf9 insect cells (0.5 mg of total protein) were resuspended in 100 μl of ice-cold PBS buffer. The homo-bifunctional cross-linker ethylene glycol bis(succinimidyl succinate) (Thermo Scientific, Rockford, IL) was added to a final concentration of 0.5 mm. After incubation for 30 min at 4 °C, the reaction was stopped by adding Tris/HCl buffer (pH 7.5) (50 mm final concentration). Membranes were collected by centrifugation (20,000 × g for 8 min at 4 °C) and analyzed by SDS-PAGE (6%) and immunoblotting using TAP1- or TAP2-specific antibodies. As specifically indicated, membranes were preincubated with 10 μm peptide RRYQKSTEL or ICP47 at 4 °C for 1 h prior to cross-linking. ICP47 was expressed and purified as described (9).
Peptide Transport
Membranes isolated from Sf9 cells (100 μg of protein) were resuspended in 50 μl of AP buffer (5 mm MgCl2 in PBS (pH 7.4)) in the presence of 3 mm ATP. The transport reaction was started by adding 1 μm of the peptide RRYQNSTØL (Ø, fluorescein-labeled cysteine) for 3 min at 32 °C and terminated with 1 ml of ice-cold stop buffer (10 mm EDTA in PBS (pH 7.0)). After centrifugation (20,000 × g for 8 min), the pellet was solubilized in 0.5 ml of lysis buffer L2 (50 mm Tris/HCl (pH 7.5), 150 mm NaCl, 5 mm KCl, 1 mm CaCl2, 1 mm MnCl2, 1% Nonidet P-40) for 30 min at 4 °C. Non-solubilized proteins were removed by centrifugation, and the supernatant was incubated with 60 μl of ConA-Sepharose (50% w/v, Sigma-Aldrich) for 1 h at 4 °C. After three washing steps with 0.5 ml of lysis buffer each, ConA-bound peptides were specifically eluted with methyl-α-d-mannopyranoside (200 mm) and quantified by a fluorescence plate reader (λex/em 485/520 nm, Polarstar Galaxy, BMG Labtech, Offenburg, Germany). Background transport was determined in the presence of apyrase (1 units/sample). All measurements were performed in triplicate.
Peptide Binding
Membranes (100 μg of total protein) prepared from Sf9 cells were incubated with 0.5 μm of the peptide R9LFlu (RRYØKSTEL; Ø, fluorescein-labeled cysteine) in 50 μl of AP buffer for 15 min at 4 °C. Free peptides were removed by washing the membranes twice with 100 μl of ice-cold AP buffer using a vacuum manifold with 96-well filter plates (0.65 μm of polyvinylidene difluoride membranes, MultiScreen, Millipore). After elution with AP buffer containing 1% SDS, peptides were quantified by a fluorescence plate reader. All measurements were performed in triplicate. Background binding was determined in 100-fold excess of unlabeled peptide R9L (RRYQKSTEL). The soluble domain of BNLF2a (amino acids 2–42, BNLF2a2–42) was prepared by solid-phase synthesis using the Fmoc/tBu-strategy. BNLF2a2–42 was purified by reverse-phase C18 HPLC and labeled with 5(6)-carboxyfluorescein succinimidyl ester (Invitrogen) at the N terminus. The identity of the unlabeled and labeled BNLF2a2–42 was confirmed by mass spectrometry.
RESULTS
EBV BNLF2a Has a Tail-anchored Topology
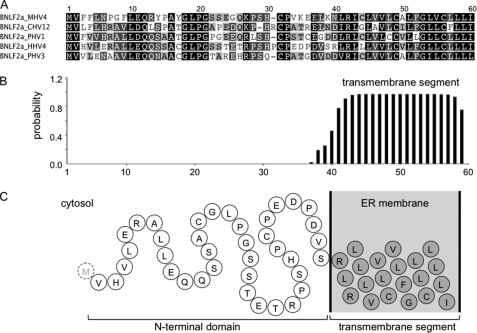
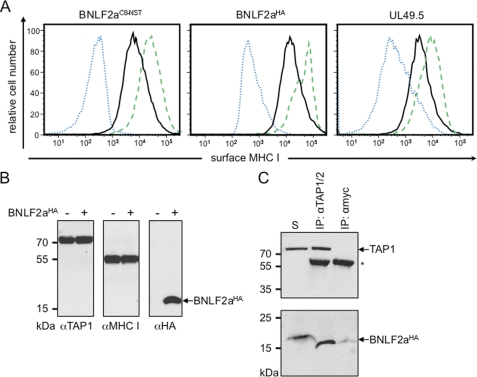
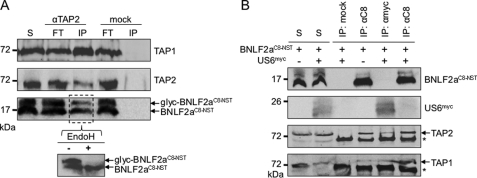
BNLF2a is composed of a hydrophilic N-terminal region (40 amino acids) followed by a hydrophobic segment (20 amino acids) with a predicted α-helical structure (Fig. 1). Notably, BNLF2a does not harbor an N-terminal signal sequence. These characteristic traits are indicative for tail-anchored (TA) proteins, which are targeted to subcellular membranes by their C-terminal transmembrane segment, hereby exposing their N-terminal domain toward the cytosol (30). TA proteins are released from free ribosomes and are posttranslationally inserted into membranes (31). To examine whether BNLF2a has indeed a tail-anchored topology, we extended its C terminus by an N-glycosylation site (NST) placed sufficiently far from the transmembrane segment by an intervening C8 epitope tag (BNLF2aC8-NST). To ensure that these extra residues do not affect BNLF2a activity, we expressed BNLF2aC8-NST in HeLa cells and analyzed the effect on MHC I surface expression by flow cytometry (Fig. 2A). Similar to UL49.5 (17, 18), expression of BNLF2aC8-NST causes a down-regulation of MHC I surface expression, thereby confirming the activity of C-terminally extended constructs. Similar results were obtained for wild-type and differently tagged versions, e.g. BNLF2aHA. It is worth mentioning that the total amount of MHC I was not altered in BNLF2a transiently transfected HeLa cells (Fig. 2B). BNLF2aHA coimmunoprecipitated with TAP1 and TAP2 in IFN-γ stimulated HeLa cells (Fig. 2C, supplemental Fig. S1A), demonstrating that C-terminally tagged BNLF2a binds to the PLC (18, 32). Notably, the slight difference in migration behavior of directly solubilized or coimmunoprecipitated BNLF2a is explained by different lipid-to-detergent ratios associated with the small hydrophobic peptide. BNLF2aC8-NST-transfected cells were analyzed by immunoblotting (supplemental Fig. S1B). Apart from non-glycosylated BNLF2a, the glycosylated form was detected by a 10-kDa upshift (supplemental Fig. S1B). Removal of N-linked oligosaccharides with endoglycosidase H (EndoH) resulted in the disappearance of the 16-kDa protein and an increase in non-glycosylated BNLF2a, indicating that the viral factor was inserted into the ER with tail-anchored topology.
FIGURE 1.
Primary structure and membrane topology of BNLF2a homologs. A, sequence alignment of BNLF2a from macacine herpesvirus 4 (MHV4), cercopithecine herpesvirus 12 (CHV12), pongine herpesvirus 1/3 (PHV1/3), and human herpesvirus 4 (HHV4). Identical amino acids and conserved residues are boxed in black and gray, respectively. B, transmembrane prediction of EBV BNLF2a. The TMHMM algorithm was applied. Similar results were obtained by SOSUI, HMMTOP, PredictProtein, and TMpred. A signal sequence or a corresponding cleavage site was not predicted by iPSORT, Phobius, PSORT II, SIG-Pred, SignalP 3.0, SOSUIsignal, or TargetP1.1. C, schematic presentation of EBV BNLF2a (N-terminal methionine is removed in the mature protein).
FIGURE 2.
BNLF2a inhibits the MHC I antigen presentation by targeting the PLC. A, down-regulation of MHC I surface expression. HeLa cells were transiently transfected with BNLF2aC8-NST-pIRES2-EGFP, BNLF2aHA-pIRES2-EGFP, or UL49.5-pIRES2-EGFP, respectively (black solid histogram). Peptide-loaded MHC I molecules at the cell surface were stained with PE-coupled W6/32 antibodies. During flow cytometry, only transfected cells (EGFP-positive) were analyzed. As controls, cells were transfected with empty pIRES2-EGFP vector and stained either with W6/32 (green dashed histogram) or the isotype control (blue dotted histogram). B, transfection of BNLF2a does not affect the expression level of the MHC I and TAP. Equal amounts of MHC I and TAP1 were confirmed by SDS-PAGE (10%) and immunoblotting with the corresponding antibodies. C, BNLF2aHA associates with the PLC. γ-IFN-stimulated HeLa cells were solubilized with 2% digitonin. Solubilized proteins were immunoprecipitated (IP) using TAP1/2 specific antibodies. As negative controls, either myc-antibody (mock) or an antibody specific for the ER translocon protein Sec61α (shown in supplemental Fig. S1A) were used. Samples were analyzed by Tricine/SDS-PAGE (10%) and immunoblotting with the corresponding antibodies. An aliquot (1/20) of the solubilized input (S) is shown. Asterisk, immunoglobulin heavy chain.
BNLF2a Inserts Posttranslationally into the ER Membrane
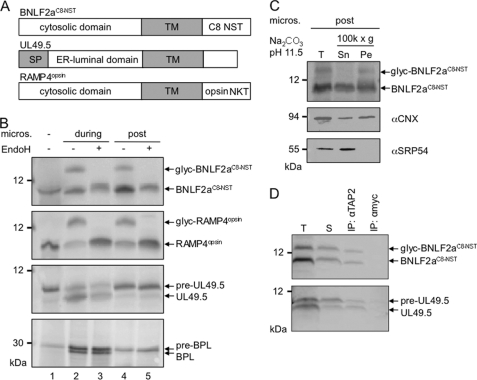
We next established an in vitro translation system for BNFL2aC8-NST (Fig. 3). As the single N-terminal methionine of BNLF2a is posttranslationally removed, we added three methionines at the C terminus for [35S]Met labeling. After in vitro translation in the presence of dog pancreas microsomes, two BNLF2a species were detected, corresponding to the non-glycosylated and glycosylated species (Fig. 3B, lanes 2 and 4). Glycosylated BNLF2a was EndoH-sensitive.
FIGURE 3.
BNLF2a inserts posttranslationally into ER membranes where it binds to TAP. A, schematic illustration of the in vitro-translated proteins BNLF2aC8-NST, UL49.5, and RAMP4opsin. Predicted transmembrane segments (TM) are shown as gray boxes. An N-glycosylation site was added to epitope-tagged BNLF2a and RAMP4. UL49.5 has a signal sequence (SP) that is cleaved by the signal peptidase. B, BNLF2a inserts posttranslationally into the ER membrane. In vitro translation reactions were performed in rabbit reticulocyte lysate in the presence of [35S]Met using templates encoding BNLF2aC8-NST, RAMP4opsin, UL49.5, or bovine prolactin (BPL), respectively. Microsomal membranes were added either at the start of translation (during, lanes 2 and 3) or after termination of translation by puromycin (post, lanes 4, 5). Translation products were analyzed either directly (lane 1, without microsomes) or solubilized (lanes 2-5) and treated with EndoH (lanes 3 and 5). Proteins were separated by Tricine/SDS-PAGE (10%) and visualized by phosphoimaging. glyc, glycosylated protein; pre, preprotein with an uncleaved signal sequence. C, glycosylated BNLF2a is an integral membrane protein. BNLF2aC8-NST was posttranslationally inserted into ER membranes and then subjected to extraction at pH 11.5. Supernatant (Sn), membrane pellet (Pe), and an aliquot before centrifugation (T) were analyzed by SDS-PAGE and visualized by phosphoimaging (BNLF2aC8-NST) or immunoblotting for SRP54 (peripheral membrane protein) and calnexin (CNX; integral membrane protein) as controls. D, posttranslationally inserted BNLF2a interacts with TAP. BNLF2aC8-NST and UL49.5 were in vitro-translated and inserted into TAP-containing Raji microsomes post- or cotranslationally, respectively. After translation, microsomes were collected by sedimentation through a sucrose cushion, solubilized with 2% digitonin, and subjected to immunoprecipitations using antibodies specific for TAP2 or anti-myc (mock). Immune complexes (IP) and 1/20 aliquot of the translation reaction (T) and of the solubilizate (S) were separated by Tricine/SDS-PAGE (10%) and visualized by phosphoimaging. pre-UL49.5, preprotein with signal sequence.
To determine whether BNLF2a is posttranslationally inserted into the ER membrane, the translation reaction was performed in the absence of microsomes. After translation termination by puromycin and addition of microsomes, BNLF2a still inserted into ER membranes as demonstrated by N-glycosylation (Fig. 3B, lanes 4 and 5). In contrast, the viral TAP inhibitor UL49.5, as a type I membrane protein and a model secretory protein, bovine prolactin, both with cleavable leader sequences, were only processed and translocated into the ER membrane cotranslationally.
In contrast, the tail-anchored model protein Ramp4 (22), harboring a C-terminal opsin tag (including an N-glycosylation site), inserts posttranslationally into microsomes (Fig. 3B). In contrast to the signal recognition particle protein SRP54, both glycosylated and unglycosylated BNLF2a are resistant to carbonate extraction to a similar extent as calnexin, an ER-resident integral membrane protein (Fig. 3C). Protease K treatment results in a slight downshift of both glycosylated and non-glycosylated BNLF2a, demonstrating that the C terminus remained intact (supplemental Fig. S2). Taken together, these results demonstrate the ER insertion and the tail-anchored topology of BNLF2a.
Because tail-anchored insertion requires that the hydrophobic substrate and any associated factors must be kept soluble until ER membranes are added, membrane targeting in vitro is typically less efficient than in vivo, which may explain why not all BNLF2a was glycosylated posttranslationally. The important finding here is that BNLF2aC8-NST does undergo posttranslational targeting, glycosylation, and membrane integration to a significant extent.
Another strategy to uncouple translation and membrane insertion is to translate, in vitro, BNLF2aC8-NST in wheat germ extract using truncated mRNAs lacking a stop codon. During translation, BNLF2a nascent chains are therefore not released and remain tethered to the ribosome. Such BNLF2a-containing ribosome-nascent chain complexes were then collected by centrifugation and subsequently incubated with mammalian microsomes and cell lysate. After release from the ribosomes, BNLF2aC8-NST was still inserted into the ER membranes as demonstrated by N-glycosylation and carbonate extraction (supplemental Fig. S3). In the ribosome-nascent chain complexes, the hydrophobic transmembrane segment of BNLF2a remains hidden within the ribosomal exit tunnel until ribosomal release. Therefore, less BNLF2a aggregates during translation, as shown by increased carbonate resistance and, hence, membrane insertion (compare Fig. 3C and supplemental Fig. S3). However, a fraction of membrane-inserted BNLF2aC8-NST molecules remained non-glycosylated. These unmodified viral proteins are either inaccessible for the oligosaccharyltransferase or insufficiently processed by the glycosylation machinery. However, carbonate extraction indicates clearly that BNFL2a, even if translated in a plant cell-free system, efficiently inserts posttranslationally into ER membranes if mammalian cytosolic factors are present (supplemental Fig. S3).
In Vitro-translated BNLF2a Binds to TAP
Because translated proteins typically comprise less than 0.1% of the microsomal protein in cell-free translation (33), we determined whether in vitro-translated BNLF2a would still target to TAP. After in vitro translation of BNLF2aC8-NST in the presence of Raji isolated from B lymphoblastic Raji cells, the TAP complex was solubilized and coimmunoprecipitated using a TAP2-specific antibody. As control, the type I membrane protein UL49.5 was translated cotranslationally. Our results show that both viral factors coimmunoprecipitate with TAP (Fig. 3D). Notably, glycosylated and non-glycosylated BNLF2a interact with TAP. We demonstrate that in vitro-translated BNLF2a and UL49.5 are not only inserted properly into the ER membrane but also bind to their physiological target.
BNLF2a Exploits the Asna-1/WRB Machinery for ER Insertion
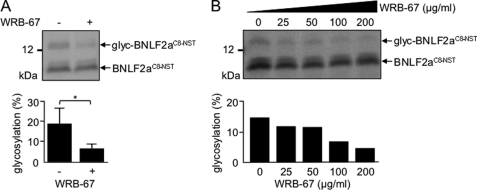
Recently, the structural basis and mechanism for membrane-associated steps in tail-anchored protein insertion by the yeast Get3/receptor complex composed of the ER membrane proteins Get1 and Get2 (34) have been deciphered (35–37). The large cytosolic loop of Get1 (amino acids 21–104, including a coiled-coil region) inhibited tail-anchored protein insertion into yeast microsomes (35). The mammalian tryptophan-rich basic protein (WRB) shows sequence similarity to yeast Get1 and acts as the ER membrane receptor for TRC40/Asna-1, the mammalian homologue of yeast Get3 (38). In analogy to Get1, the coiled-coil domain of WRB interfered with Asna-1 mediated insertion of TA proteins (38), and a similar recombinant inhibitory peptide (WRB-67) reduced glycosylation and, hence, membrane insertion of in vitro-translated Ramp4opsin from 65 ± 8% to 40 ± 5% in the presence of 200 μg/ml WRB-67 (data not shown). Strikingly, WRB-67 reduced the tail-anchored membrane insertion of BNLF2aC8-NST in a dose-dependent manner (Fig. 4). In conclusion, BNLF2a inserts into the ER membrane posttranslationally, exploiting the WRB/Asna-1 (Get3/Get1) machinery, and thus represents the first viral TAP inhibitor with a tail-anchored targeting mechanism.
FIGURE 4.
Asna-1 and WRB are involved in the posttranslational ER membrane insertion of BNLF2a. In vitro translation reactions were performed in wheat germ extract in the presence of [35S]Met using truncated BNLF2aC8-NST mRNA templates lacking a stop codon. After translation, BNLF2aC8-NST containing ribosome-nascent chain complexes were collected by centrifugation through a sucrose cushion and resuspended in microsomal membrane containing rabbit reticulocyte lysate in the presence of either 200 μg/ml R9L peptide as mock control (-) or WRB-67 inhibitory peptide (A, 200 μg/ml; B, concentrations as indicated). After ribosomal release by puromycin and RNase treatment, samples were further incubated for 30 min at 32 °C. Translation products were analyzed by Tricine/SDS-PAGE (10%) and visualized by phosphoimaging. Histograms show the average amount of glycosylated BNLF2a protein. The error bars indicate the mean ± S.D. of three independent experiments. *, p = 0.05 (Student's t test).
Posttranslational ER Targeting of BNLF2a Is Independent of TAP
It still remains unclear whether BNLF2a binds directly or indirectly to TAP and whether BNLF2a requires or not the assistance of TAP for a proper ER insertion. We therefore expressed BNLF2a in insect cells, which achieve high-level expression of membrane proteins, including core TAP subunits, but lack any other factors of the adaptive immune system (39). Despite the fact that these cells do not contain endogenous TAP, BNLF2a was properly inserted into ER membranes and N-core glycosylated (supplemental Fig. S4). Thus, targeting of BNLF2a to the ER membrane is independent of TAP.
Interaction of BNLF2a and TAP
We next examined whether BNLF2a binds to TAP1/2 in the absence of components of the adaptive immune system and peptide-loading complex. Insect cells were coinfected with a baculovirus encoding both TAP subunits and with an additional virus encoding BNLF2aC8-NST. As shown by coimmunoprecipitation using TAP2-specific antibodies, both non-glycosylated and glycosylated BNLF2a are associated with the TAP complex (Fig. 5A). EndoH treatment further indicated that the higher molecular band corresponds to N-core glycosylated and ER-resident BNLF2a. These results demonstrate that the interaction between BNLF2a and TAP1/2 does not require other components of the peptide-loading complex. Because BNLF2a consists of a rather small hydrophilic N-terminal region of approximately 40 residues, it might be possible that this region is capable to bind to TAP in analogy to a TAP-substrate peptide. We therefore examined whether a BNLF2a-soluble domain (amino acids 2–42, BNLF22–42), if provided in 100-fold molar excess, is able to outcompete the binding of a fluorescein-labeled peptide (R9LFlu) to TAP. However, neither did BNLF22–42 compete with R9LFlu for binding to TAP1/2, nor did fluorescein-labeled BNLF22–42 bind directly to TAP with detectable affinity (supplemental Fig. S5). Therefore, these results suggest that the interaction between TAP1/2 and BNLF2a requires the full-length protein.
FIGURE 5.
Binding of BNLF2a to TAP is mutually exclusive from HCMV-US6. A, interaction of BNLF2a with TAP. Membranes isolated from Sf9 insect cells coexpressing TAP1/2 and BNLF2aC8-NST were solubilized with 2% digitonin. Solubilized proteins were immunoprecipitated using TAP2-specific antibodies (IP). As a negative control, HC10-antibody was used (mock). Aliquots of the anti-TAP2 immunoprecipitate were treated with EndoH. Samples were analyzed by SDS-PAGE (10%) and subsequent immunoblotting with the corresponding antibodies. Aliquots (1/20) of solubilized input (S) and unbound proteins after immunoprecipitation (FT) are shown. B, BNLF2a and US6 are mutually exclusive in their interaction with TAP. Membranes isolated from Sf9 cells coexpressing TAP1/2, BNLF2aC8-NST and US6myc were subjected to coimmunoprecipitation experiments (IP) as described above using anti-C8 and anti-myc antibodies, respectively. As a negative control, HC10-antibody was used (mock). Input (S, 1/20 aliquot) and immune complexes (IP) were analyzed by immunoblotting. Asterisk, immunoglobulin heavy chain.
BNLF2a and US6 Are Mutually Exclusive in TAP Inhibition
We next examined how two viral proteins compete for TAP inhibition. The ER-luminal domain of US6 of HCMV specifically blocks ATP binding to and peptide translocation by TAP (12, 13). After coexpression of TAP1/2, BNLF2aC8-NST and US6myc in Sf9 cells, coimmunoprecipitations were performed using antibodies against C8-tagged BNLF2a (anti-C8) or myc-tagged US6 (anti-myc), respectively. TAP1 and TAP2 coprecipitated with either BNLF2aC8-NST or US6myc, confirming the specific interaction of each viral inhibitor with the TAP complex (Fig. 5B). Strikingly, US6 was not coprecipitated with BNLF2a-TAP complexes. Vice versa, BNLF2a was not detected in coprecipitated US6-TAP complexes. Because both BNLF2a and US6 are expressed in high excess, as demonstrated by almost complete inhibition of peptide transport by TAP (see below), the total amount of both viral factors is sufficient to occupy all TAP complexes. In conclusion, TAP complexes cannot interact with US6 and BNLF2a at the same time.
BNLF2a Prevents a Peptide-induced Conformational Change of TAP
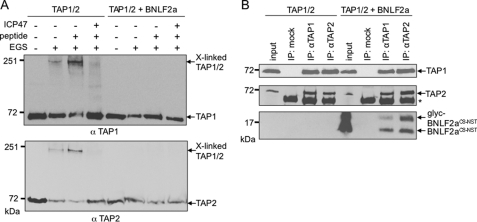
Interestingly, the ER-lumenal domain of US6 arrests TAP in a conformation that inhibits ATP binding to the cytosolic NBDs (13). Because both factors exclude each other from binding to TAP, BNLF2a and US6 may arrest distinct conformations of the TAP complex. Moreover, it has been reported that viral inhibitors have a direct influence on TAP conformation. The presence of either ICP47 or US6 prevents peptide-induced conformational rearrangements of the transporter (12, 13, 40). To investigate the influence of BNLF2a on the conformational change following peptide binding, Sf9 membranes containing TAP1/2 were incubated with the lysine-specific bivalent chemical cross-linker ethylene glycol bis(succinimidyl succinate). In the presence of peptides, cross-linking of the TAP1/2 heterodimer, most likely via the NBDs (12, 40), was detected at 200 kDa (Fig. 6A). ICP47 abolished the chemical cross-linking of the TAP heterodimer. The weak cross-linking of TAP, even without addition of peptides, was explained by the presence of endogenous peptides. Strikingly, when BNLF2a and TAP1/2 were coexpressed, we could not detect a cross-linked TAP heterodimer, even after the addition of peptides. It is worth mentioning that BNLF2a did not interfere with TAP heterodimerization per se, as BNLF2a was coprecipitated with fully assembled TAP1/2 complexes (Fig. 6B). The total amount of BNLF2a was sufficient to occupy all TAP1/2-heterodimers, as shown by almost complete inhibition of TAP-mediated peptide transport (see below). In the absence and presence of BNLF2a, comparable amounts of TAP1 were coimmunoprecipitated with TAP2 and vice versa (Fig. 6B). Thus, BNLF2a prevents a conformational change of TAP induced by peptide binding.
FIGURE 6.
BNLF2a inhibits peptide-induced conformational change of TAP. Membranes isolated from insect cells either expressing TAP1/2 with or without BNLF2aC8-NST were incubated in the presence or absence of the peptide RRYQKSTEL (10 μm) or ICP47 (10 μm) before cross-linking with ethylene glycol bis(succinimidyl succinate) (EGS). Subsequently, samples were analyzed by SDS-PAGE (6%) and immunoblotting with TAP1- and TAP2-specific antibodies. B, BNLF2aC8-NST does not interfere with TAP1/2 heterodimerization. Membranes isolated from Sf9 insect cells expressing either TAP1/2 or BNLF2aC8-NST together with TAP1/2 were subjected to coimmunoprecipitation experiments (IP) using TAP1- or TAP2-specific antibodies. As a negative control, HC10-antibody was used (mock). Input (1/10 aliquot) and immune complexes (IP) were analyzed by immunoblotting. glyc, glycosylated BNLF2a; asterisk, immunoglobulin heavy chain.
BNLF2a Targets the Core TAP and Inhibits Peptide Binding and Translocation
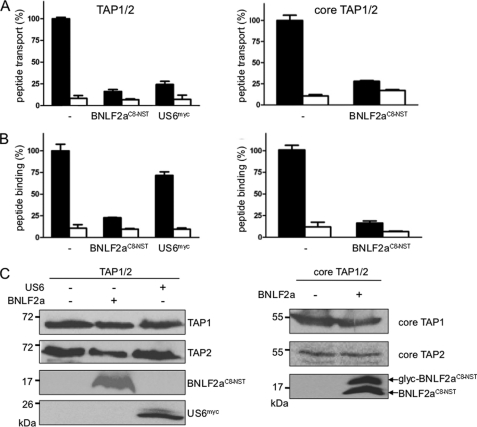
The core TAP complex has been demonstrated to be essential and sufficient for peptide translocation into the ER lumen (39). We therefore compared the inhibitory effect of BNLF2a on peptide translocation of the full-length and core TAP complex (Fig. 7A, right and left panel, respectively). BNLF2aC8-NST blocks peptide translocation into the ER lumen by full-length and core TAP. In contrast to herpesviral US6 (13, 41, 42), BNLF2aC8-NST blocks peptide binding to full-length and core TAP, indicating that the extra four N-terminal transmembrane helices of each TAP subunit are dispensable for BNLF2a-mediated TAP inhibition (Fig. 7B). Similar amounts of TAP were present in each binding and translocation assay (Fig. 7C). Taken together, our data illustrate that tail-anchored BNLF2a posttranslationally targets the core TAP complex, preventing peptide binding and conformation switch.
FIGURE 7.
BNLF2a targets the core TAP complex blocking peptide binding and transport. Full-length TAP (left panel) or core TAP (right panel) were expressed in Sf9 insect cells. BNLF2aC8-NST or US6myc were coexpressed as indicated. A, BNLF2a inhibits peptide transport by TAP. Equal amounts of membranes were incubated with RRYQNSTØL (1 μm, Ø, fluorescein-labeled cysteine) in the absence (apyrase, open bars) or presence of ATP (10 μm, filled bars) for 3 min at 32 °C. N-core glycosylated and thus translocated peptides were bound to ConA-beads and quantified by fluorescence (λex/em = 485/520 nm). Peptide transport by TAP was set to 100%. The means of at least three independent experiments are shown. Error bars indicate the S.D. B, BNLF2a inhibits peptide binding to TAP. Equal amounts of membranes were incubated with RRYØKSTEL (0.5 μm; Ø, fluorescein-labeled cysteine) for 15 min on ice (filled bars). A 100-fold excess of RRYQKSTEL was used to probe for unspecific binding (open bars). After washing on filter plates, the amount of membrane-associated peptide was quantified as described above. C, an equal amount of TAP in the membranes was confirmed by SDS-PAGE (10%) and immunoblotting with the corresponding antibodies. Glyc, glycosylated BNLF2a.
DISCUSSION
Members of the herpesviridae family have evolved a number of strategies to evade immune recognition (8, 43). For example, ICP47 of HSV-1 binds with high affinity to TAP from the cytosol, where it blocks peptide binding (9, 10). In contrast, US6 of HCMV targets TAP via its ER-lumenal domain and prevents ATP binding on the opposite side of the ER membrane (12, 13). However, only limited information is available regarding the recently described EBV BNLF2a (18, 32).
As demonstrated by in vitro translation, BNLF2a exhibits three distinct hallmarks of TA proteins: First, the lack of an N-terminal signal sequence; second, a C-terminal membrane anchor, which exposes its C terminus toward the ER lumen; and third, posttranslational insertion into the ER membrane. Thus, BNLF2a is unique among the viral TAP inhibitors with regard to its membrane insertion and inhibition mechanism. Although viral factors such as UL49.5 and US6 are inserted cotranslationally in the ER membrane via their cleavable N-terminal signal sequence, BNLF2a is targeted posttranslationally (Fig. 3B), most likely utilizing the cellular pathways used by endogenous TA proteins. The tail-anchored topology of BNLF2a was also confirmed by expression in insect cells, which lack the components of an adaptive immune system and components of the PLC.
Four main routes of TA protein targeting have been proposed (30, 44). Following release from the ribosome, TA proteins are either inserted into membranes without assistance of other factors or rely on one of three distinct chaperone-mediated pathways. One involves the chaperone machinery Hsp40/70 (45, 46), another is mediated by SRP (47), and a third pathway depends on Asna-1 (48) or its yeast homologue Get3 (34) and their membrane receptors WRB (38) or Get1/Get2 (34), respectively. According to a recently proposed model of tail-anchored protein insertion (35–37), binding of the rigid coiled-coil domain of Get1 to the Get3/tail-anchored protein cargo complex mediates substrate release and membrane insertion. Soluble fragments containing the coiled-coil region of WRB or Get1 are known to inhibit tail-anchored protein insertion (35, 38), and we observed a similar effect for BNLF2a insertion into the ER membrane (Fig. 4, A and B). Therefore, we conclude that BNLF2a can exploit the host Asna-1/WRB (Get3/Get1) pathway for ER membrane targeting and insertion.
By adding an N-glycosylation site to the C terminus of BNLF2a, we demonstrate by in vitro translation (Fig. 3B) as well as expression in human and insect cells (supplemental Fig. S1BB and Fig. 5) that the C terminus of BNLF2a faces the ER lumen. The hydrophilic N-terminal region of the viral factor faces the peptide and ATP-binding site of TAP. Because BNLF2a, but not its soluble cytosolic domain, inhibits both peptide and ATP binding to TAP (18), the BNLF2a topology might be a prerequisite for its functionality. In contrast, US6 targets TAP from the ER lumen and blocks ATP binding to the cytosolic NBDs of TAP (13). It was surprising that BNLF2a and US6, when expressed together, exclude each other from binding to the TAP complex (Fig. 5B). Steric competition between US6 and BNLF2a is unlikely because their active domains interact with TAP at different sides of the ER membrane.
It has been reported that viral inhibitors arrest TAP in distinct conformations. For example, binding of HSV-ICP47 to TAP inhibits peptide-induced conformational rearrangements of the transporter (12, 40). Remarkably, coexpression of BNLF2a and TAP blocks peptide-induced TAP1/2 cross-linking (Fig. 6A). Combined with the fact that the soluble part of BNLF2a (lacking its transmembrane domain) does not directly compete with substrate peptide binding to TAP (supplemental Fig. S5), we propose that BNLF2a prevents the conformational rearrangement of TAP that is normally induced by peptide binding (40). However, because of limitations of our assay, we are unable to distinguish whether BNLF2a indirectly prevents peptide binding to TAP, hence preventing conformational changes, or whether BNLF2a binding to TAP arrests the transporter in a conformation that is incompatible with peptide binding. In contrast, ICP47 acts directly as a competitive inhibitor of peptide binding to TAP (9, 10).
The PLC is composed of several proteins, all of them necessary to ascertain efficient peptide loading of MHC I, which ultimately results in the presentation of dominant epitopes. Within the PLC, the TAP heterodimer fulfills the important task of peptide translocation from the cytosol into the ER lumen. In detail, the core TAP complex, composed of 2 × 6 TMDs and two NBDs, is essential for peptide binding and translocation, whereas the extra N-terminal domain of each TAP subunit acts as an independent docking platform for tapasin (39). To narrow down the interaction sites of TAP and BNLF2a, we performed peptide transport and peptide binding experiments with full-length and core TAP. Our data show that BNLF2a is able to bind to core TAP and fully inhibits peptide transport by blocking peptide binding to core TAP (Fig. 7).
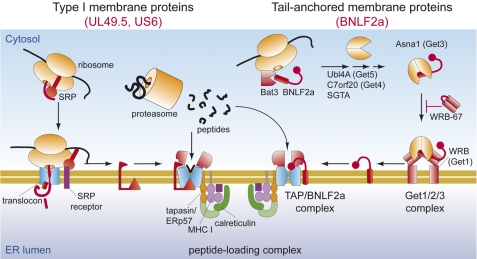
In conclusion, we propose the following inhibition mechanism of BNLF2a (Fig. 8). After release from free ribosomes, BNLF2a is posttranslationally inserted into the ER membrane using the host Asna-1/WRB (Get3/Get1) machinery for targeting TA proteins. After insertion into the ER membrane, BNLF2a interacts directly with the core TAP complex, arresting a conformation that is incompatible with peptide binding. It will be important to precisely map the BNLF2a interaction sites of the core TAP subunits to reveal details of how BNLF2a prevents TAP-mediated peptide transport. This may aid the development of small therapeutic compounds that modulate immune surveillance.
FIGURE 8.
Distinct insertion and inhibition mechanisms of viral TAP inhibitors. As type I membrane proteins, US6 and UL49.5 are cotranslationally inserted into the ER membrane where they subsequently inhibit TAP function. BNLF2a exploits the host Asna-1/WBR (Get3/Get1) insertion machinery for TA-anchored insertion to subsequently arrest TAP in a translocation-incompetent conformation.
Acknowledgments
We thank R. Abele (Goethe University Frankfurt) for helpful discussions on the manuscript and M. Mariappan and R. S. Hegde (Medical Research Council, Cambridge, UK) for generously providing the WRB-67 inhibitory peptide.
Addendum
While this paper was undergoing revision, a report by Host et al. (49) was published that also describes data relevant to tail-anchored insertion of BNLF2a.
This work was supported by German Research Foundation Grants SFB 807, Transport and Communication across Biological Membranes, and TA157/7 (to R. T.), by a grant from the European Drug Initiative on Channels and Transporters (EDICT) funded by the European Commission Seventh Framework Program (to R. T.). This work was also supported, in whole or in part, by National Institutes of Health Grants GM53457 and DK51818 (to W. R. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MHC I
- major histocompatibility complex class I
- TAP
- transporter associated with antigen processing
- ER
- endoplasmic reticulum
- PLC
- peptide-loading complex
- HCMV
- human cytomegalovirus
- NBD
- nucleotide-binding domain
- SRP
- signal recognition particle
- TA
- tail-anchored
- IRES
- internal ribosome entry site
- NST
- N-glycosylation site
- EndoH
- endoglycosidase H
- WRB
- tryptophan-rich basic protein.
REFERENCES
- 1. Abele R., Tampé R. (2009) Curr. Opin. Cell Biol. 21, 508–515 [DOI] [PubMed] [Google Scholar]
- 2. Cresswell P., Ackerman A. L., Giodini A., Peaper D. R., Wearsch P. A. (2005) Immunol. Rev. 207, 145–157 [DOI] [PubMed] [Google Scholar]
- 3. Rock K. L., Goldberg A. L. (1999) Annu. Rev. Immunol. 17, 739–779 [DOI] [PubMed] [Google Scholar]
- 4. Yewdell J. W. (2007) Curr. Opin. Immunol. 19, 79–86 [DOI] [PubMed] [Google Scholar]
- 5. Koch J., Tampé R. (2006) Cell Mol. Life Sci. 63, 653–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wearsch P. A., Cresswell P. (2008) Curr. Opin. Cell Biol. 20, 624–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yewdell J. W., Hill A. B. (2002) Nat. Immunol. 3, 1019–1025 [DOI] [PubMed] [Google Scholar]
- 8. Parcej D., Tampé R. (2010) Nat. Chem. Biol. 6, 572–580 [DOI] [PubMed] [Google Scholar]
- 9. Ahn K., Meyer T. H., Uebel S., Sempé P., Djaballah H., Yang Y., Peterson P. A., Früh K., Tampé R. (1996) EMBO J. 15, 3247–3255 [PMC free article] [PubMed] [Google Scholar]
- 10. Tomazin R., Hill A. B., Jugovic P., York I., van Endert P., Ploegh H. L., Andrews D. W., Johnson D. C. (1996) EMBO J. 15, 3256–3266 [PMC free article] [PubMed] [Google Scholar]
- 11. Aisenbrey C., Sizun C., Koch J., Herget M., Abele R., Bechinger B., Tampé R. (2006) J. Biol. Chem. 281, 30365–30372 [DOI] [PubMed] [Google Scholar]
- 12. Hewitt E. W., Gupta S. S., Lehner P. J. (2001) EMBO J. 20, 387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kyritsis C., Gorbulev S., Hutschenreiter S., Pawlitschko K., Abele R., Tampé R. (2001) J. Biol. Chem. 276, 48031–48039 [DOI] [PubMed] [Google Scholar]
- 14. Boname J. M., May J. S., Stevenson P. G. (2005) Eur. J. Immunol. 35, 171–179 [DOI] [PubMed] [Google Scholar]
- 15. Lybarger L., Wang X., Harris M. R., Virgin H. W., 4th, Hansen T. H. (2003) Immunity 18, 121–130 [DOI] [PubMed] [Google Scholar]
- 16. Koppers-Lalic D., Reits E. A., Ressing M. E., Lipinska A. D., Abele R., Koch J., Marcondes Rezende M., Admiraal P., van Leeuwen D., Bienkowska-Szewczyk K., Mettenleiter T. C., Rijsewijk F. A., Tampé R., Neefjes J., Wiertz E. J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5144–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loch S., Klauschies F., Schölz C., Verweij M. C., Wiertz E. J., Koch J., Tampé R. (2008) J. Biol. Chem. 283, 13428–13436 [DOI] [PubMed] [Google Scholar]
- 18. Hislop A. D., Ressing M. E., van Leeuwen D., Pudney V. A., Horst D., Koppers-Lalic D., Croft N. P., Neefjes J. J., Rickinson A. B., Wiertz E. J. (2007) J. Exp. Med. 204, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kutok J. L., Wang F. (2006) Annu. Rev. Pathol. 1, 375–404 [DOI] [PubMed] [Google Scholar]
- 20. Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. (1984) Nature 310, 207–211 [DOI] [PubMed] [Google Scholar]
- 21. Skach W. R., Lingappa V. R. (1993) J. Biol. Chem. 268, 23552–23561 [PubMed] [Google Scholar]
- 22. Favaloro V., Spasic M., Schwappach B., Dobberstein B. (2008) J. Cell Sci. 121, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herget M., Kreissig N., Kolbe C., Schölz C., Tampé R., Abele R. (2009) J. Biol. Chem. 284, 33740–33749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Meyer T. H., van Endert P. M., Uebel S., Ehring B., Tampé R. (1994) FEBS Lett. 351, 443–447 [DOI] [PubMed] [Google Scholar]
- 25. McCormick P. J., Miao Y., Shao Y., Lin J., Johnson A. E. (2003) Mol. Cell 12, 329–341 [DOI] [PubMed] [Google Scholar]
- 26. Stam N. J., Spits H., Ploegh H. L. (1986) J. Immunol. 137, 2299–2306 [PubMed] [Google Scholar]
- 27. Krieg U. C., Johnson A. E., Walter P. (1989) J. Cell Biol. 109, 2033–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crowley K. S., Reinhart G. D., Johnson A. E. (1993) Cell 73, 1101–1115 [DOI] [PubMed] [Google Scholar]
- 29. Kim J., Hamamoto S., Ravazzola M., Orci L., Schekman R. (2005) J. Biol. Chem. 280, 7758–7768 [DOI] [PubMed] [Google Scholar]
- 30. Rabu C., Schmid V., Schwappach B., High S. (2009) J. Cell Sci. 122, 3605–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borgese N., Brambillasca S., Colombo S. (2007) Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 32. Horst D., van Leeuwen D., Croft N. P., Garstka M. A., Hislop A. D., Kremmer E., Rickinson A. B., Wiertz E. J., Ressing M. E. (2009) J. Immunol. 182, 2313–2324 [DOI] [PubMed] [Google Scholar]
- 33. Saksena S., Shao Y., Braunagel S. C., Summers M. D., Johnson A. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12537–12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H. D., Schwappach B., Weissman J. S. (2008) Cell 134, 634–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mariappan M., Mateja A., Dobosz M., Bove E., Hegde R. S., Keenan R. J. (2011) Nature 477, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stefer S., Reitz S., Wang F., Wild K., Pang Y. Y., Schwarz D., Bomke J., Hein C., Löhr F., Bernhard F., Denic V., Dötsch V., Sinning I. (2011) Science 333, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang F., Whynot A., Tung M., Denic V. (2011) Mol. Cell 43, 738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vilardi F., Lorenz H., Dobberstein B. (2011) J. Cell Sci. 124, 1301–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koch J., Guntrum R., Heintke S., Kyritsis C., Tampé R. (2004) J. Biol. Chem. 279, 10142–10147 [DOI] [PubMed] [Google Scholar]
- 40. Lacaille V. G., Androlewicz M. J. (1998) J. Biol. Chem. 273, 17386–17390 [DOI] [PubMed] [Google Scholar]
- 41. Ahn K., Gruhler A., Galocha B., Jones T. R., Wiertz E. J., Ploegh H. L., Peterson P. A., Yang Y., Früh K. (1997) Immunity 6, 613–621 [DOI] [PubMed] [Google Scholar]
- 42. Hengel H., Koopmann J. O., Flohr T., Muranyi W., Goulmy E., Hämmerling G. J., Koszinowski U. H., Momburg F. (1997) Immunity 6, 623–632 [DOI] [PubMed] [Google Scholar]
- 43. Horst D., Verweij M. C., Davison A. J., Ressing M. E., Wiertz E. J. (2011) Curr. Opin. Immunol. 23, 96–103 [DOI] [PubMed] [Google Scholar]
- 44. Borgese N., Fasana E. (2011) Biochim. Biophys. Acta. 1808, 937–946 [DOI] [PubMed] [Google Scholar]
- 45. Abell B. M., Rabu C., Leznicki P., Young J. C., High S. (2007) J. Cell Sci. 120, 1743–1751 [DOI] [PubMed] [Google Scholar]
- 46. Rabu C., Wipf P., Brodsky J. L., High S. (2008) J. Biol. Chem. 283, 27504–27513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Abell B. M., Pool M. R., Schlenker O., Sinning I., High S. (2004) EMBO J. 23, 2755–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stefanovic S., Hegde R. S. (2007) Cell 128, 1147–1159 [DOI] [PubMed] [Google Scholar]
- 49. Horst D., Favaloro V., Vilardi F., van Leeuwen H. C., Garstka M. A., Hislop A. D., Rabu C., Kremmer E., Rickinson A. B., High S., Dobberstein B., Ressing M. E., Wiertz E. J. (2011) J. Immunol. 186, 3594–3605 [DOI] [PubMed] [Google Scholar]