Background: Glioma-initiating cells are underlying causes of development and progression of glioblastoma.
Results: Depletion of Oct4 expression suppresses tumorigenic activity of glioma-initiating cells through down-regulation of Sox2.
Conclusion: Oct4 maintains tumorigenicity of glioma-initiating cells in cooperation with the Sox axis.
Significance: This study uncovers the transcriptional network of stemness genes in cancer-initiating cells.
Keywords: Brain tumors, Stem cells, Transcription factors, Transforming growth factor β (TGFβ), Tumor cell biology
Abstract
Although the concept of cancer stem cells or cancer-initiating cells had created a new paradigm for the treatment of malignant tumors, it remains unclear how cancer-initiating cells can be eradicated. We have previously reported that the transforming growth factor-β (TGF-β)-Sox4-Sox2 pathway is essential for glioma-initiating cells to retain their stemness, and inhibition of TGF-β signaling may lead to differentiation of glioma-initiating cells (Ikushima, H., Todo, T., Ino, Y., Takahashi, M., Miyazawa, K., and Miyazono, K. (2009) Cell Stem Cell 5, 504–514). Here we demonstrate that Oct4 plays essential roles in retention of the stemness properties of glioma-initiating cells through positive regulation of Sox2 expression. We also show that, in glioma-initiating cells, Oct4 is associated with Sox4 and that Oct4-Sox4 complexes cooperatively activate the enhancer activity of the SOX2 gene. In contrast, in fetal neural progenitor cells, Sox2 expression is enhanced by transcriptional complex containing Sox2 protein itself, and this self-reinforcing loop of Sox2 appears to be disrupted in glioma-initiating cells, suggesting that Sox2 expression in glioma-initiating cells is differently regulated from that in neural progenitor cells. Our findings reveal differences between glioma-initiating cells and fetal neural progenitor cells and may open the way to depriving glioma-initiating cells of tumorigenic activity without affecting normal tissues.
Introduction
Glioblastoma, also known as grade IV astrocytoma, is the most aggressive form of malignant glioma and is one of the most malignant human cancers, with an estimated median survival of only ∼1 year (1, 2). Despite past huge efforts, this statistic has not markedly improved over the past decades.
Cancer stem cells or cancer-initiating cells are tumor cells characterized by their ability to induce tumorigenesis and to self-renew (3). Similar to other types of tumor cells, glioma-initiating cells (or glioma stem cells) have been isolated from human glioblastoma tissues (4, 5). Following their identification, glioma-initiating cells have been intensively investigated and have been found to exhibit strong resistance to chemotherapy and radiotherapy (6, 7). It has been suggested that the failure to cure glioblastoma may be due to existing therapeutic strategies that affect only the tumor bulk and not glioma-initiating cells (8). These findings indicate the need for an innovative therapeutic strategy enabling functional eradication of glioma-initiating cells.
Although it has yet to be fully determined how the stemness of glioma-initiating cells is maintained, a few signaling pathways, including Hedgehog (9), bone morphogenetic protein 4 (10), and TGF-β (11–13), have been implicated to contribute to maintenance of the stemness properties of these cells. Although the transcriptional machinery required is under investigation, we have recently reported crucial roles for the Sox axis. Sox4 interacts with the SOX2 enhancer region to induce Sox2 expression, and this “Sox4-Sox2” axis maintains stemness properties of glioma-initiating cells under the control of TGF-β signaling (11).
The POU class 5 transcription factor Oct4 (also known as Pou5f1) is essential for establishing and maintaining the pluripotent state of embryonic stem cells (14, 15). Deletion of Oct4 from embryonic stem cells results in trophoblast differentiation (16). Introduction of Oct4 together with Sox2, Klf4, and c-Myc into human or mouse adult fibroblasts results in the generation of induced pluripotent stem cells (17, 18). In addition, Oct4 has been detected in high grade glioma and specific types of testicular germ cell tumors (19–21). However, the role of Oct4 in cancer stem cells has yet to be fully determined.
Here, we report that Oct4 is a factor of crucial importance for the maintenance of tumorigenic activity of glioma-initiating cells. We have previously reported that, in contrast to Sox4 and Sox2, the expression of Oct4 is not regulated by TGF-β signaling in glioma-initiating cells (11). However, inhibition of Oct4 expression in glioma-initiating cells resulted in suppression of sphere formation in vitro and tumor formation in vivo. Oct4 knockdown also potentiated sensitivity to conventional chemotherapy. We also demonstrated that Oct4 interacted with Sox4 and cooperatively activated the SOX2 enhancer region to maintain stemness properties of glioma-initiating cells. Notably, Sox2 expression in glioma-initiating cells was induced by the Oct4-Sox4 complex acting on the SOX2 enhancer region to maintain stemness properties, whereas that in fetal neural progenitor cells was regulated by a transcriptional complex containing Sox2 protein itself through a self-reinforcing regulatory loop. These findings indicate that Oct4 plays a role in the tumorigenic activity of glioblastoma and suggest that the stemness properties of glioma-initiating cells are regulated by mechanisms different from those of neural progenitor cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Primary grade IV glioblastoma samples were obtained during surgery from consenting patients, as approved by the Institutional Review Board of the University of Tokyo Hospital. Spheres were cultured in DMEM/F-12 serum-free medium (Invitrogen) supplemented with B27 (Invitrogen), 20 ng/ml EGF, and 20 ng/ml basic fibroblast growth factor (both from PeproTech). Characteristics of the glioma-initiating cells were evaluated in our previous study (11). Normal human fetal neural progenitor cells were obtained from Lonza and cultured in maintenance medium (NPMM, Lonza). U373MG cells were maintained in DMEM containing 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/ml streptomycin, sodium pyruvate (1 mm), and non-essential amino acids (0.1 mm). The antibodies used were as follows: anti-Musashi (Chemicon), anti-Nestin (Chemicon), anti-glial fibrillary acidic protein (Dako), anti-Tuj1 (Covance), anti-Oct4 (Santa Cruz Biotechnology), anti-Sox2 (R&D), anti-Sox4 (Santa Cruz Biotechnology), and anti-α-tubulin (Sigma-Aldrich).
Sphere-forming Assay
Glioma-initiating cells were cultured in non-tissue-culture-treated flasks (BD Biosciences) with vented caps (BD Biosciences) for 7 days. Floating spheres in five fields per sample were counted under a microscope (magnification, ×40).
Limiting Dilution Assay
Sphere cells were dissociated and plated in 96-well plates in 200 μl of serum-free medium. After a 7-day culture, the percentage of wells not containing spheres for each cell plating density was calculated and plotted against the number of cells per well.
RNA Interference
siRNAs (see supplemental Table S1 for sequences) were purchased from Invitrogen and introduced into cells using Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions.
Immunostaining
Glioma-initiating cells were seeded on poly-l-ornithine (Sigma)- and fibronectin (Sigma)-coated slide glasses and cultured for 7 days with the indicated siRNA in serum-free medium. Cells were fixed with 3.7% paraformaldehyde, permeabilized with PBS containing 0.3% Triton X-100, and incubated with the indicated antibodies. Subsequently, samples were incubated with secondary antibodies and stained with propidium iodide (Molecular Probes) for nuclear staining. Stained cells were observed with a confocal microscope (LSM510, Carl Zeiss).
Cell Lysis and Immunoblotting
Cells were lysed with a buffer containing 1% Nonidet P-40, 20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm PMSF, 1% aprotinin, and 5 mm EDTA. Proteins in cleared cell lysates were subjected to SDS-PAGE and transferred to Fluoro Trans W membrane (Pall). Immunoblotting was performed using the indicated antibodies.
Quantitative Real-time PCR
Quantitative real-time reverse transcription-PCR was performed as described previously (22). All samples were run in triplicate in each experiment. The primers used are listed in supplemental Table S1. Values were normalized to that for glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
ChIP and ChIP Re-IP
Chromatin immunoprecipitation (ChIP)3 was performed as described previously (11). PCR primers are listed in supplemental Table S1. For ChIP re-immunoprecipitation (Re-IP) assays, protein-DNA complexes were eluted from immunoprecipitation by incubation with 10 mm DTT at 37 °C for 30 min and diluted 1:50 in buffer (20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100), followed by Re-IP with secondary antibodies.
Cell Viability Assay
Quantitation of cell viability was performed using a colorimetric assay for mitochondrial dehydrogenase activity (WST-8, Nacalai Tesque) after treatment with temozolomide (LKT Laboratories).
Luciferase Assay
The SOX2 enhancer region (+3553 through +4290) was cloned into a pGL4 vector (Promega, Madison, WI) with a minimal promoter, and a luciferase assay was performed as described previously (22). Values were normalized to Renilla luciferase activity under the control of thymidine kinase promoter.
Intracranial Proliferation Assay
Viable glioma-initiating cells (5 × 104) in 5 μl of DMEM/F-12 medium were injected stereotactically into the right cerebral hemisphere of 5-week-old female BALB/c nu/nu mice at a depth of 3 mm. All animal experimental protocols were performed in accordance with the policies of the Animal Ethics Committee of the University of Tokyo.
RESULTS
Oct4 Is an Essential Factor for Retention of Stemness of Glioma-initiating Cells in Vitro
The transcriptional network essential for maintenance of glioma-initiating cells has not been fully determined. We used glioma-initiating cells obtained from two patients with glioblastoma, termed TGS-01 and TGS-04, and cultured in serum-free medium to study this network. The glioma-initiating capacities of these cells were characterized in our previous studies (11).
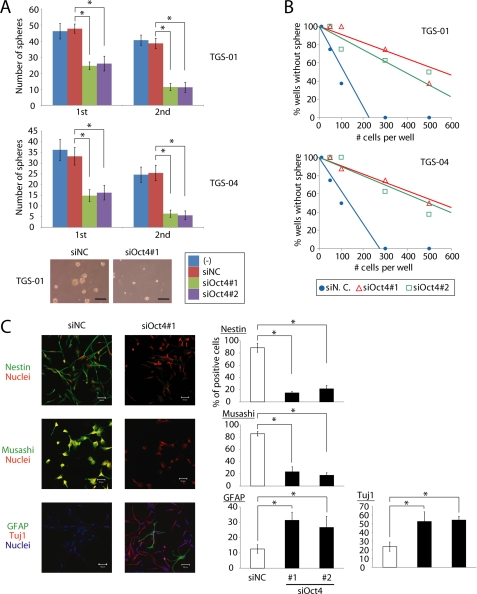
Oct4 is known to be one of the most crucial self-renewal gene products and to play pivotal roles in maintaining stemness of embryonic stem cells and neural stem cells. We have demonstrated that expression of Oct4 is not affected by TGF-β signaling in glioma-initiating cells (11). To study the role of Oct4 in glioma-initiating cells, we first examined the effects of Oct4 knockdown on their biological properties. After Oct4 expression was knocked down (supplemental Fig. S1), glioma-initiating cells exhibited marked reduction of sphere-forming ability in serial sphere-forming assay (Fig. 1A), suggesting that Oct4 is required for self-renewal of glioma-initiating cells. In limiting dilution assay, TGS-01 or TGS-04 with Oct4 siRNA also showed less capacity for self-renewal than control cells (Fig. 1B). Similar results were obtained with the use of glioma-initiating cells, TGS-02, TGS-03, and TGS-05, derived from other patients with glioblastoma (supplemental Fig. S2). We also examined the effects of Oct4 knockdown on proliferation and apoptosis of glioma-initiating cells. Treatment of siRNA against Oct4 did not significantly induce apoptosis but reduced proliferation of TGF-01 and TGS-04 cells (supplemental Fig. S3).
FIGURE 1.
Oct4 is essential for retention of stemness of glioma-initiating cells. A, TGS-01 and TGS-04 cells were dissociated into single cell populations, transfected with control (NC) or Oct4 siRNA duplex, and cultured for 7 days (1st). After the 7-day culture, spheres were dissociated into single cell populations and equal numbers of cells were cultured for another 7 days (2nd). Values are the number of glioma spheres formed (means ± S.E. of five fields). *, p < 0.001. Scale bars, 100 μm. B, knockdown of Oct4 expression by siRNA in TGS-01 and TGS-04 cells resulted in a decrease of self-renewal capacity in limiting dilution assay. C, immunostaining of TGS-01 cells. Spheres were disaggregated, seeded on poly-l-ornithine- and fibronectin-coated slide glasses, and cultured in serum-free medium with control (NC) or Oct4 siRNA duplex for 7 days. Quantification of Nestin-, Musashi-, Tuj1-, or glial fibrillary acidic protein (GFAP)-positive cells is shown in the right graphs. *, p < 0.01. Scale bars, 50 μm.
Glioma-initiating cells have been reported to express neural precursor cell markers, but to only minimally express neural or glial differentiation markers (11). To examine the expression of these marker proteins in each type of cell, spheres in serum-free medium were disaggregated and seeded on poly-l-ornithine- and fibronectin-coated slide glasses. Knockdown of Oct4 expression by siRNA decreased the number of cells positive for Nestin or Musashi (neural precursor cell markers) and increased that for glial fibrillary acidic protein (astrocyte differentiated marker) or Tuj1 (neuronal marker) (Fig. 1C). These results indicate that Oct4 is required for maintenance of the stemness properties of glioma-initiating cells in vitro.
Knockdown of Oct4 Expression Decreases Tumorigenicity of Glioma-initiating Cells in Vivo
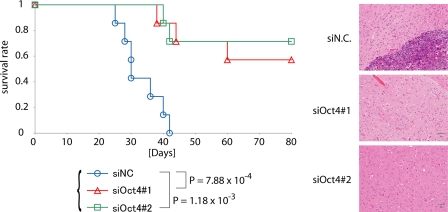
To study the role of Oct4 in the tumorigenic activity of glioma-initiating cells in vivo, we next examined the effects of Oct4 knockdown on intracranial growth of glioma-initiating cells. We treated dissociated glioma-initiating cells with siRNA against Oct4. Cells from the newly formed glioma spheres were orthotopically inoculated into the brains of immunocompromised mice. The growth of glioma-initiating cells was inhibited by pretreatment with siRNA against Oct4, and the mice inoculated with the pretreated glioma-initiating cells survived significantly longer than those inoculated with control cells (Fig. 2). We also examined tumor formation in the brain 30 days after transplantation. Whereas mice with control glioma-initiating cells displayed large tumors in the brain, those with the pretreated glioma-initiating cells exhibited no evidence of tumor on histopathologic examination (Fig. 2). These results suggest that Oct4 is essential for the maintenance of tumorigenicity of glioma-initiating cells and that loss of tumorigenicity by Oct4 knockdown is an irreversible process.
FIGURE 2.
Development of brain tumors after intracerebral transplantation of 5 × 104 TGS-01 cells pretreated with control (NC) or Oct4 siRNA duplex for 7 days. Survival of mice (n = 7 mice for each condition) was evaluated by Kaplan-Meier analysis (left graph). p values were calculated by the log-rank test. The right panels show the results of histological examination of the samples dissected at 30 days after intracerebral transplantation. Tissue sections were stained with hematoxylin-eosin. Scale bars, 50 μm. Experiments were repeated twice with essentially similar results.
Knockdown of Oct4 Expression in Glioma-initiating Cells Affects Sensitivity to Chemotherapy
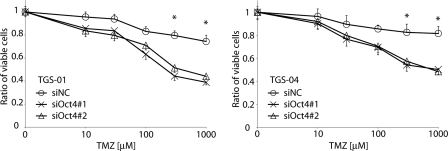
As suggested by the cancer stem cell model, the resistance of glioma-initiating cells to conventional chemotherapy may be a major cause of the low cure rate for glioblastoma (3). Although oral administration of temozolomide, a new alkylating agent, has shown efficacy in patients with glioblastoma, it was found that glioma-initiating cells were resistant to temozolomide-induced cell death, causing tumors to recur (7). We examined the effects of Oct4 knockdown on the sensitivity of glioma-initiating cells to temozolomide-induced cell death. Control TGS-01 and TGS-04 cells exhibited low sensitivities to temozolomide treatment (Fig. 3 and supplemental Fig. S4). In contrast, treatment with increasing concentrations of temozolomide suppressed the viability of TGS-01 and TGS-04 cells pretreated with Oct4 siRNA in dose-dependent fashion. These results suggest that Oct4 is involved in acquisition of drug-resistance properties of glioblastoma.
FIGURE 3.
Knockdown of Oct4 expression enhances sensitivity to chemotherapy in glioma-initiating cells. TGS-01 and TGS-04 cells with control (NC) or Oct4 siRNA duplex were seeded in 96-well plates and treated with temozolomide (0, 10, 30, 100, 300, and 1000 μm) for 72 h. Cell viability was assessed by using a WST-8 assay. *, p < 0.01 (siNC versus siOct4#1 and siNC versus siOct4#2).
Oct4 Directly Induces Sox2 Expression to Maintain Stemness Properties of Glioma-initiating Cells
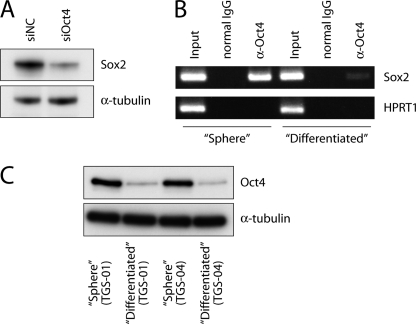
We next studied the molecular mechanisms that underlie the putative pathological roles of Oct4 in glioma-initiating cells. Oct4 regulates stemness properties of embryonic and neural stem cells via several mechanisms (23). Among them, Sox2, which has recently been reported to be a critical regulator of the stemness of glioma-initiating cells (11, 24), is known to be one of the major downstream targets of Oct4 in embryonic stem cells (25). We therefore examined whether Sox2 acts downstream of Oct4 in glioma-initiating cells. Oct4 knockdown in glioma-initiating cells resulted in down-regulation of Sox2 expression (Fig. 4A), indicating that Oct4 positively regulates Sox2 expression in glioma-initiating cells. To examine whether this regulation is directly mediated at the transcriptional level, we performed a ChIP assay using an antibody to Oct4. It has been demonstrated that the enhancer element located in the 3′ flanking region of the SOX2 gene is important for the regulation of Sox2 expression in embryonic stem cells (25, 26). Recruitment of Oct4 to the SOX2 enhancer element was observed in glioma-initiating cells (Fig. 4B). In contrast, Oct4 was only minimally associated with the SOX2 enhancer element in matched “differentiated” cells that were derived from the same patient but were cultured in media containing 10% fetal bovine serum to induce differentiation. These results appear to be due to the lower levels of expression of Oct4 in the “differentiated” cells compared with the “sphere” cells (Fig. 4C). These findings together indicate that Oct4 induces Sox2 expression in glioma-initiating cells through direct binding to the SOX2 enhancer region.
FIGURE 4.
Oct4 is associated with the SOX2 enhancer region to up-regulate expression levels of Sox2 in glioma-initiating cells. A, effects of Oct4 knockdown on expression of Sox2. Amounts of Sox2 protein were determined after treatment with control (NC) or Oct4 siRNA #1 duplex for 24 h. α-Tubulin was used as a loading control. B, association of Oct4 with the SOX2 enhancer region. ChIP analysis was performed using TGS-01 cells (“Sphere”) and matched “Differentiated” cells. Eluted DNAs were subjected to conventional RT-PCR. The first intron of hypoxanthine phosphoribosyltransferase 1 (HPRT1) was used as a negative control. Input: 1%. C, levels of expression of Oct4 protein in “Sphere” cells and “Differentiated” cells. α-Tubulin was used as a loading control.
Oct4 Induces Sox2 Expression Cooperatively with Sox4 and Activates the Sox4-Sox2 Cascade in Glioma-initiating Cells
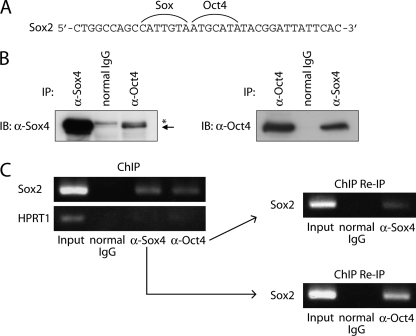
In our previous study, another transcription factor, Sox4, was shown to associate with the SOX2 enhancer region and maintain the stemness and tumorigenicity of glioma-initiating cells (11). In addition, consensus sequences of Sox proteins and Oct4 exist proximally in the SOX2 enhancer region (Fig. 5A). These findings prompted us to study the interaction of the Sox axis and Oct4 in the maintenance of stemness properties of glioma-initiating cells. First, we examined the interaction between Sox4 and Oct4. As shown in Fig. 5B, endogenous Oct4 physically interacted with endogenous Sox4 in glioma-initiating cells. Moreover, ChIP Re-IP experiments demonstrated that Sox4 and Oct4 exist in the same transcription complex on the SOX2 enhancer region (Fig. 5C).
FIGURE 5.
Oct4 physically interacts with Sox4 on SOX2 enhancer region. A, the sequences of Oct4-binding element and Sox-binding element in the SOX2 enhancer region. B, physical interaction of endogenous Oct4 with endogenous Sox4 in TGS-01 cells. Cell lysates were subjected to immunoprecipitation with anti-Oct4 antibody followed by immunoblotting with anti-Sox4 (left) or with anti-Sox4 antibody followed by immunoblotting with anti-Oct4 (right). Asterisk: nonspecific band. C, recruitment of the Oct4-Sox4 complex to the SOX2 enhancer region. Soluble chromatin was prepared from TGS-01 cells, and ChIP analysis was performed using anti-Sox4 and anti-Oct4 antibodies. Subsequently, ChIP Re-IP of protein-DNA complex eluted from the first immunoprecipitation was performed. Eluted DNAs were subjected to conventional RT-PCR.
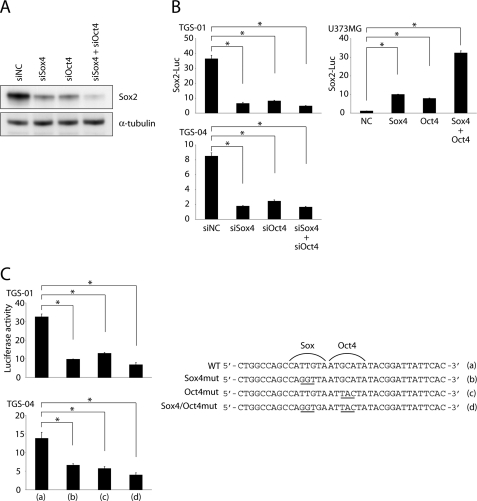
We next studied the effects of the Oct4-Sox4 complex on Sox2 expression in glioma-initiating cells. When Oct4 and Sox4 were both knocked down, Sox2 expression was more strongly down-regulated than it was by separate knockdown of Oct4 or Sox4 (Fig. 6A). Suppression of SOX2 enhancer activity by knockdown of Oct4 or Sox4 was also confirmed in luciferase assay using TGS-01 and TGS-04 cells (Fig. 6B). Moreover, the enhancer activity was synergistically activated by Oct4 and Sox4 overexpression in glioblastoma cell line U373MG (Fig. 6B), in which the levels of expression of Oct4 and Sox4 were significantly lower than in glioma-initiating cells (data not shown).
FIGURE 6.
Oct4 acts in concert with Sox4 to potentiate SOX2 enhancer activity. A, effects of Oct4 and/or Sox4 knockdown on expression of Sox2. Amount of Sox2 protein was determined after treatment with indicated siRNA duplex for 24 h. B, roles of Oct4 and Sox4 in activation of the SOX2 enhancer region. Effects of Oct4 and/or Sox4 knockdown on SOX2 enhancer activity were examined in TGS-01 and TGS-04 cells (left graphs). Effects of Oct4 and/or Sox4 overexpression on SOX2 enhancer activity were examined in U373MG cells (right graph). Error bars represent ±S.E. *, p < 0.001. C, TGS-01 or TGS-04 cells were transfected with luciferase constructs containing wild-type or mutated SOX2 enhancer region. The cells were collected 24 h after transfection, and luciferase activity was examined. *, p < 0.001. The right panel indicates the sequence of the SOX2 enhancer region and corresponding mutations (underlined) used in this study.
To confirm a direct association of Oct4 and Sox4 with the SOX2 enhancer region, we generated luciferase constructs with mutated Oct4 and/or Sox4 binding elements in the SOX2 enhancer region (Fig. 6C). Mutation of one of the two elements led to a reduction of enhancer activity compared with the wild-type enhancer. When both binding elements were mutated, the enhancer activity was more strongly reduced. These results indicate that both Oct4 and Sox4 directly interact with the SOX2 enhancer region and synergistically induce Sox2 expression.
Transcription Factor Complexes on the SOX2 Enhancer Region in Glioma-initiating Cells Are Distinct from Those in Neural Progenitor Cells
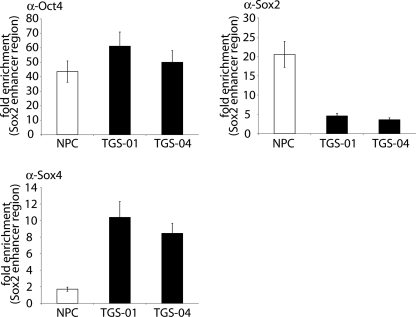
As demonstrated here using glioma-initiating cells, Sox2 expression is also induced by Oct4 through interaction of Oct4 with the SOX2 enhancer region in embryonic stem cells. Furthermore, in embryonic stem cells, Sox2 is associated with Oct4 and the Oct4-Sox2 complex cooperatively activates the SOX2 enhancer region to form a positive regulatory loop (25, 26). To determine whether this regulatory loop exists in neural progenitor cells and glioma-initiating cells, we examined recruitment of these transcription factors to the SOX2 enhancer region in a ChIP assay (Fig. 7). Anti-Oct4 antibody enriched the DNA fragments of SOX2 enhancer region equally well in fetal neural progenitor cells and glioma-initiating cells. In addition, anti-Sox2 antibody immunoprecipitated this region in neural progenitor cells. However, strong enrichment of the same region by anti-Sox2 antibody was not observed in glioma-initiating cells. These results indicate that transcription factor complex on the SOX2 enhancer region does not contain Sox2 in glioma-initiating cells and that Sox2 expression in glioma-initiating cells is regulated by mechanisms different from those in fetal neural progenitor cells.
FIGURE 7.
The partner of Oct4 on SOX2 enhancer region in glioma-initiating cells is distinct from that in neural progenitor cells. Soluble chromatin was prepared from glioma-initiating cells (TGS-01 and TGS-04) and neural progenitor cells (NPC). ChIP analysis was performed using anti-Oct4, anti-Sox2, and anti-Sox4 antibodies. Eluted DNAs were subjected to quantitative real-time PCR analysis. Values were normalized to the amount of the first intron of HPRT1. Error bars represent ±S.E.
We next examined recruitment of Sox4 to the SOX2 enhancer region in neural progenitor cells and glioma-initiating cells. In contrast to the experiment using anti-Sox2 antibody, anti-Sox4 antibody immunoprecipitated the DNA fragments of the SOX2 enhancer region in glioma-initiating cells, whereas the enrichment observed in fetal neural progenitor cells was much weaker. These findings together indicate that Sox2 expression in glioma-initiating cells is potentiated by the Oct4-Sox4 complex acting on the SOX2 enhancer region to maintain tumorigenic activity, whereas that in neural progenitor cells may be promoted by transcriptional complex containing Sox2 protein itself through a positive regulatory loop.
DISCUSSION
Although the origin of glioma stem cells (or glioma-initiating cells) is controversial (27), several studies have suggested that glioma-initiating cells share characteristics with neural or glial stem/progenitor cells (28, 29). Glioma stem cells express neural stem cell markers, including Nestin, Musashi, and Prominin-1 (CD133). Like normal neural stem cells, glioma stem cells are located in specific niches surrounding the tumor vasculature. A recent study has shown that the perivascular niches control self-renewal of glioma stem cells through endothelial cell-derived factors (30). However, in terms of transcription factor complexes, the similarities and differences between glioma stem cells and neural stem cells have not been clearly determined.
Here we have shown that Oct4 expression is required for the maintenance of the self-renewal capacity of glioma-initiating cells. In addition, transient suppression of Oct4 by siRNA abolished the induction of Sox2 by TGF-β4 and decreased the tumorigenic activity of glioma-initiating cells (Fig. 2), suggesting that impairment of stemness properties via Oct4 knockdown may be an irreversible process. We also demonstrated that Oct4 knockdown increases sensitivity to the chemotherapeutic alkylating agent, temozolomide.
Oct4 is essential for establishing and maintaining the pluripotent state of stem cells (14, 15). Moreover, Oct4 is one of the key factors in the generation of induced pluripotent stem cells (17, 18). However, the role of Oct4 in the development and progression of malignant tumors has not been fully determined. Our findings indicate that Oct4 is an essential factor for glioma-initiating cells and plays roles similar to those in embryonic stem cells.
One of the most intensively investigated topics in current cancer research is the identification of specific therapeutic compounds that can effectively eliminate cancer-initiating cells. Recent studies have identified factors essential for retention of cancer-initiating cells, including several growth factor signaling pathways such as Wnt, Hedgehog, Notch, PI3K-mTOR, TGF-β, and LIF (9, 31–36). Although new therapeutic targets have been intensively sought based on findings related to these pathways, one problem is that almost all of these signaling pathways are also indispensable for normal stem cells. Inhibitors of these signaling pathways may affect the characteristics of normal stem cells and impair maintenance of normal tissues. Thus, from a clinical standpoint, it is important to identify factors not only essential for the maintenance of cancer-initiating cells but also different from those present in normal stem cells.
Here, we have demonstrated that Oct4-Sox4 complex activates the enhancer region of SOX2 genes to sustain stemness properties of glioma-initiating cells. Oct4 and Sox2 are also important for the maintenance of normal stem cells, and Oct4-Sox2 complex activates the SOX2 enhancer region to form a positive regulatory loop. However, in glioma-initiating cells, Sox2 is not predominantly present in the transcription factor complex on the SOX2 enhancer region. Instead, Sox4 forms a transcriptional complex with Oct4 in glioma-initiating cells to activate the enhancer region of SOX2, a gene essential for the maintenance of tumorigenicity of glioma-initiating cells. These findings suggest that Sox2 expression in glioma-initiating cells can be potentiated via up-regulation of Sox4, whereas Sox2 expression in neural progenitor cells is regulated by a self-reinforcing regulatory loop and is relatively self-contained (Fig. 7 and supplemental Fig. S5). In other words, the positive regulatory loop of Sox2 is not active in glioma-initiating cells, and alternatively, Oct4 acts with Sox4 to enhance Sox2 expression. We also confirmed that, in neural progenitor cells, Sox2 is only weakly induced by TGF-β stimulation (supplemental Fig. S6), whereas this cytokine activates the Sox4-Sox2 cascade in glioma-initiating cells (11). Loss of the regulatory loop of Sox2 expression may thus cause glioma-initiating cells to become susceptible to exogenous stimuli. However, we should bear in mind that our glioma-initiating cells were obtained from adult tumors, whereas neural progenitor cells were from a fetus. Further studies in neural progenitor cells from adults may be important to elucidate the differences between glioma-initiating cells and normal neural progenitor cells.
We examined combined effects of siRNAs against Sox4 and Oct4 in a limiting dilution assay but failed to observe any significant synergistic effects (supplemental Fig. S7). It may be because a defect of either factor in the Sox4-Oct4 complex results in significant inactivation of the SOX2 enhancer and/or because a single effect of siSox4 or siOct4 is strong enough to reduce sphere-forming ability of glioma-initiating cells.
It remains to be determined why the common Oct4-binding sequence and Sox-binding elements are differently regulated in neural progenitor cells and glioma-initiating cells. Upon differentiation of erythroid precursors into mature erythrocytes, GATA-binding protein 2 (GATA2) on some promoter regions is replaced by GATA1 (37). This process is termed the “GATA switch” and is an essential step in the maturation of erythrocytes and the expression of α-globin. One of the crucial mediators of this switching is Friend of GATA1 (FOG-1, also known as Zfpm1), a multi-zinc-finger protein critical for the development of erythrocytes and megakaryocytes (38, 39), and GATA-FOG interaction is believed to be required for “GATA switch” (40). Like the “GATA switch,” the Sox-binding element on SOX2 enhancer region in glioma-initiating cells is differently regulated from that in neural progenitor cells, although the mechanism responsible for this remains to be determined.
Although Sox4 plays a crucial role in the retention of tumorigenicity of glioma-initiating cells through up-regulation of Sox2 expression (11), Sox4−/− mice exhibit no neurological defects (41). This finding suggests that the mechanism of action of Sox4 in glioma-initiating cells is distinct from that in neural stem/precursor cells. Because the self-renewal and proliferation of normal stem cells are likely strictly regulated, perhaps by genetic or epigenetic programs, the uncontrolled expansion of cancer-initiating cells may result from deregulation of such strict programs. In support of this conclusion, we found that the self-regulatory loop of Sox2 expression observed in neural progenitor cells was disrupted in glioma-initiating cells. This finding may enable the determination of a novel molecular target and eventually yield a therapeutic approach to eradication of glioblastoma without affecting the normal brain.
Acknowledgments
We are grateful to Yasuyuki Morishita and Daisuke Itoh for skilled technical assistance.
This work was supported in part by KAKENHI (grant-in-aid for scientific research) and the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant-in-aid for Cancer Research for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health, Labour, and Welfare of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Table S1.
H. Ikushima and K. Miyazono, unpublished observation.
- ChIP
- chromatin immunoprecipitation
- Re-IP
- re-immunoprecipitation
- Tuj1
- βIII-tubulin
- GATA1
- -2, GATA-binding proteins 1 and 2.
REFERENCES
- 1. Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J., Belanger K., Brandes A. A., Marosi C., Bogdahn U., Curschmann J., Janzer R. C., Ludwin S. K., Gorlia T., Allgeier A., Lacombe D., Cairncross J. G., Eisenhauer E., Mirimanoff R. O.; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. (2005) N. Engl. J. Med. 352, 987–996 [DOI] [PubMed] [Google Scholar]
- 2. Tanaka M., Ino Y., Nakagawa K., Tago M., Todo T. (2005) Lancet Oncol. 6, 953–960 [DOI] [PubMed] [Google Scholar]
- 3. Reya T., Morrison S. J., Clarke M. F., Weissman I. L. (2001) Nature 414, 105–111 [DOI] [PubMed] [Google Scholar]
- 4. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 5. Singh S. K., Clarke I. D., Terasaki M., Bonn V. E., Hawkins C., Squire J., Dirks P. B. (2003) Cancer Res. 63, 5821–5828 [PubMed] [Google Scholar]
- 6. Bao S., Wu Q., McLendon R. E., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., Rich J. N. (2006) Nature 444, 756–760 [DOI] [PubMed] [Google Scholar]
- 7. Liu G., Yuan X., Zeng Z., Tunici P., Ng H., Abdulkadir I. R., Lu L., Irvin D., Black K. L., Yu J. S. (2006) Mol. Cancer 5, 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou B. B., Zhang H., Damelin M., Geles K. G., Grindley J. C., Dirks P. B. (2009) Nat. Rev. Drug Discov. 8, 806–823 [DOI] [PubMed] [Google Scholar]
- 9. Clement V., Sanchez P., de Tribolet N., Radovanovic I., Ruiz i Altaba A. (2007) Curr. Biol. 17, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piccirillo S. G., Reynolds B. A., Zanetti N., Lamorte G., Binda E., Broggi G., Brem H., Olivi A., Dimeco F., Vescovi A. L. (2006) Nature 444, 761–765 [DOI] [PubMed] [Google Scholar]
- 11. Ikushima H., Todo T., Ino Y., Takahashi M., Miyazawa K., Miyazono K. (2009) Cell Stem Cell 5, 504–514 [DOI] [PubMed] [Google Scholar]
- 12. Peñuelas S., Anido J., Prieto-Sánchez R. M., Folch G., Barba I., Cuartas I., García-Dorado D., Poca M. A., Sahuquillo J., Baselga J., Seoane J. (2009) Cancer Cell 15, 315–327 [DOI] [PubMed] [Google Scholar]
- 13. Ikushima H., Miyazono K. (2010) Cancer Sci. 101, 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 15. Niwa H., Miyazaki J., Smith A. G. (2000) Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 16. Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005) Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- 17. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 18. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 19. Du Z., Jia D., Liu S., Wang F., Li G., Zhang Y., Cao X., Ling E. A., Hao A. (2009) Glia 57, 724–733 [DOI] [PubMed] [Google Scholar]
- 20. Cheng L., Sung M. T., Cossu-Rocca P., Jones T. D., MacLennan G. T., De Jong J., Lopez-Beltran A., Montironi R., Looijenga L. H. (2007) J. Pathol 211, 1–9 [DOI] [PubMed] [Google Scholar]
- 21. Jones T. D., Ulbright T. M., Eble J. N., Cheng L. (2004) Clin. Cancer Res. 10, 8544–8547 [DOI] [PubMed] [Google Scholar]
- 22. Ikushima H., Komuro A., Isogaya K., Shinozaki M., Hellman U., Miyazawa K., Miyazono K. (2008) EMBO J. 27, 2955–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boiani M., Schöler H. R. (2005) Nat. Rev. Mol. Cell Biol. 6, 872–884 [DOI] [PubMed] [Google Scholar]
- 24. Gangemi R. M., Griffero F., Marubbi D., Perera M., Capra M. C., Malatesta P., Ravetti G. L., Zona G. L., Daga A., Corte G. (2009) Stem Cells 27, 40–48 [DOI] [PubMed] [Google Scholar]
- 25. Chew J. L., Loh Y. H., Zhang. W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tomioka M., Nishimoto M., Miyagi S., Katayanagi T., Fukui N., Niwa H., Muramatsu M., Okuda A. (2002) Nucleic Acids Res. 30, 3202–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanai N., Alvarez-Buylla A., Berger M. S. (2005) N. Engl. J. Med. 353, 811–822 [DOI] [PubMed] [Google Scholar]
- 28. Stiles C. D., Rowitch D. H. (2008) Neuron 58, 832–846 [DOI] [PubMed] [Google Scholar]
- 29. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A. (2008) Nat. Genet. 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calabrese C., Poppleton H., Kocak M., Hogg T. L., Fuller C., Hamner B., Oh E. Y., Gaber M. W., Finklestein D., Allen M., Frank A., Bayazitov I. T., Zakharenko S. S., Gajjar A., Davidoff A., Gilbertson R. J. (2007) Cancer Cell 11, 69–82 [DOI] [PubMed] [Google Scholar]
- 31. Zhao C., Blum J., Chen A., Kwon H. Y., Jung S. H., Cook J. M., Lagoo A., Reya T. (2007) Cancer Cell 12, 528–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao C., Chen A., Jamieson C. H., Fereshteh M., Abrahamsson A., Blum J., Kwon H. Y., Kim J., Chute J. P., Rizzieri D., Munchhof M., VanArsdale T., Beachy P. A., Reya T. (2009) Nature 458, 776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Visvader J. E., Lindeman G. J. (2008) Nat. Rev. Cancer 8, 755–768 [DOI] [PubMed] [Google Scholar]
- 34. Sato A., Sunayama J., Matsuda K., Tachibana K., Sakurada K., Tomiyama A., Kayama T., Kitanaka C. (2010) Neurosci. Lett. 470, 115–120 [DOI] [PubMed] [Google Scholar]
- 35. Ikushima H., Miyazono K. (2010) Nat. Rev. Cancer 10, 415–424 [DOI] [PubMed] [Google Scholar]
- 36. Lin B., Madan A., Yoon J. G., Fang X., Yan X., Kim T. K., Hwang D., Hood L., Foltz G. (2010) PLoS One 5, e10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grass J. A., Boyer M. E., Pal S., Wu J., Weiss M. J., Bresnick E. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsang A. P., Fujiwara Y., Hom D. B., Orkin S. H. (1998) Genes Dev. 12, 1176–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. (2005) EMBO J. 24, 2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jing H., Vakoc C. R., Ying L., Mandat S., Wang H., Zheng X., Blobel G. A. (2008) Mol. Cell 29, 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schilham M. W., Oosterwegel M. A., Moerer P., Ya J., de Boer P. A., van de Wetering M., Verbeek S., Lamers W. H., Kruisbeek A. M., Cumano A., Clevers H. (1996) Nature 380, 711–714 [DOI] [PubMed] [Google Scholar]