Background: The combined effect of HuR and miR-331-3p on ERBB-2 expression in prostate cancer (PCa) is unknown.
Results: HuR regulated ERBB-2 expression and antagonized the repressive action of miR-331-3p.
Conclusion: HuR and miR-331-3p participate in overexpression of ERBB-2 in PCa.
Significance: Interplay between HuR and miR-331-3p regulates the post-transcriptional expression of ERBB-2 in PCa.
Keywords: Gene Expression, MicroRNA, Prostate Cancer, Receptor Tyrosine Kinase, RNA-binding Protein
Abstract
ERBB-2 overexpression is associated with the development and progression of cancer and mediates its resistance to therapy. It has been suggested that post-transcriptional mechanisms control the overexpression of ERBB-2 in prostate cancer (PCa). We recently demonstrated that the 3′-untranslated region (3′-UTR) of ERBB-2 mRNA contains two specific target sites for binding of the microRNA miR-331-3p and that miR-331-3p represses ERBB-2 expression and signaling in PCa cells. Here we investigate a U-rich element situated in close proximity to the distal miR-331-3p target site in the ERBB-2 3′-UTR. Specific binding of HuR to this U-rich element promotes ERBB-2 expression in PCa cells. We show that HuR antagonizes the repressive action of miR-331-3p on its distal ERBB-2 3′-UTR target site. These results support a model in which the interplay between RNA-binding proteins and microRNAs controls the post-transcriptional regulation of gene expression and suggest that both HuR and miR-331-3p participate in the overexpression of ERBB-2 observed in some PCas.
Introduction
Prostate cancer (PCa)3 is a leading cause of male cancer death (1). Although anti-androgen therapy is generally effective in the initial treatment of locally advanced or metastatic PCa, the disease ultimately transforms into a hormone-independent state that is associated with treatment resistance and a poor prognosis (2). A variety of agents have been tested for the treatment of hormone-independent PCa, but results to date have been disappointing, with only docetaxel-based chemotherapy being shown to improve the survival of this group of patients (3, 4). Consequently, there has been intense research toward understanding the molecular mechanisms that promote the development and progression of hormone-independent PCa and that mediate its aggressive behavior and treatment resistance. A recent meta-analysis of 5,976 PCa patients found a significant positive association between overexpression of the ERBB-2 receptor tyrosine kinase (also known as HER2/neu) and the risk of disease recurrence and death (5). In addition to its role as a prognostic indicator in PCa, ERBB-2 inhibitors have been evaluated in clinical trials for this disease, albeit with disappointing results. Very limited anti-tumor activity was observed in a phase II trial of trastuzumab involving 18 patients with advanced hormone-independent PCa (6), whereas a phase II trial of the dual ERBB-1/ERBB-2 inhibitor lapatinib in patients with advanced, hormone naive PCa did not produce disease regression (7). The precise mechanisms underlying the lack of response to ERBB-2 inhibitors in PCa are unclear; however, a majority of patients had tumors that did not express detectable levels of ERBB-2 protein. Nevertheless, it is possible that combining ERBB-2 inhibitors and cytotoxic agents might produce a better therapeutic effect. In support of this, trastuzumab potentiates the anti-tumor activity of docetaxel in a human PCa xenograft model (8), whereas a phase I trial of docetaxel, estramustine, and trastuzumab in metastatic androgen-independent PCa produced objective responses in a subset of patients (9). Similarly, strongly synergistic growth-inhibitory effects have been reported when inhibitors of the Hedgehog and ERBB-2 signaling pathways were used together in androgen-independent PCa cells (10). Together, these results highlight the importance of selecting patients with measurable tumor ERBB-2 expression for ERBB-2 inhibitor treatment and the need to more effectively inhibit downstream signaling pathways in those patients with low or undetectable tumor ERBB-2 expression.
ERBB-2 and activation of its downstream oncogenic signaling networks (e.g. PI3K/Akt pathway) is thought to promote persistent androgen receptor (AR) signaling in hormone-independent PCa (11). An understanding of the regulation of ERBB-2 expression in PCa might therefore enable the development of new strategies to effectively block ERBB-2 expression and downstream signaling, including cross-talk between ERBB-2 signaling and the AR signaling pathway. A recent study of more than 2,000 PCas found low level ERBB-2 overexpression in ∼20% of samples (12), although no significant difference in ERBB-2 expression was observed between hormone-dependent and hormone-independent tumors. Nevertheless, ERBB-2 gene amplification is extremely rare in PCa (12, 13), suggesting that ERBB-2 overexpression is likely to result from transcriptional and/or post-transcriptional mechanisms. Recently, we demonstrated that microRNA-331-3p (miR-331-3p) down-regulates ERBB-2 expression in PCa cells (14). microRNAs (miRNAs) are a family of short, endogenous, non-coding RNAs that bind with imperfect complementarity to the 3′-untranslated region (3′-UTR) of specific target mRNAs, resulting in degradation or translational repression (15, 16). An increasing number of studies implicate miRNAs in processes such as cell growth and differentiation (17, 18), and aberrant miRNA expression is found in diseases such as cancer (19). Several reports have described altered miRNA profiles in PCa (20–22), and it has been suggested that specific miRNAs might have potential as biomarkers or therapeutic targets in this disease (23–27). Our work demonstrated that expression of miR-331-3p is decreased in PCa cells relative to matched normal adjacent prostate tissue and that miR-331-3p regulates ERBB-2 expression via direct binding to two miR-331-3p target sites within the ERBB-2 mRNA 3′-UTR (14). As a result, miR-331-3p can also inhibit the AR signaling pathway in PCa cells, presumably at least in part via cross-talk between the PI3k/Akt pathway downstream of ERBB-2 and the AR pathway. Other studies have since confirmed the down-regulation of miR-331-3p in PCa (22, 28), providing additional evidence to support its role as a tumor suppressor miRNA in this system.
In this study, we investigated the potential interplay between miR-331-3p and RNA-binding proteins (RBPs) that simultaneously target the ERBB-2 3′-UTR. We hypothesized that a U-rich element (URE) adjacent to a miR-331-3p target site within the ERBB-2 3′-UTR might represent a target for binding of the RBP HuR and that this binding might alter the activity of miR-331-3p upon its target site and hence modulate the expression of ERBB-2 mRNA in PCa cells. HuR is a member of the Hu family of RBPs that has diverse biological functions, including cell development, growth and proliferation, and stress responses (29), and is overexpressed and/or accumulates in the cytoplasm in a range of cancers (30–36), including PCa (30, 37). In breast cancer cells, HuR binds the ERBB-2 mRNA 3′-UTR (38), and modulation of HuR expression alters the expression of ERBB-2 (39). Although the importance of transcriptional and post-transcriptional mechanisms in the control of ERBB-2 expression in PCa is known, the role of HuR in this system has not been determined. Furthermore, it is unclear whether HuR is able to modulate action of miR-331-3p upon the ERBB-2 mRNA. Here, we present evidence showing that the ERBB-2 3′-UTR URE is a specific target for binding of HuR in PCa cells and demonstrate that HuR promotes ERBB-2 expression by antagonizing the repressive action of miR-331-3p upon its distal ERBB-2 3′-UTR target site. Our findings support a model of post-transcriptional control of gene expression involving the combined action of RBPs and miRNAs and suggest that both HuR and miR-331-3p participate in the development and progression of PCa by regulating ERBB-2 expression in prostate tumors.
EXPERIMENTAL PROCEDURES
Cell Culture, siRNA, miRNA Precursors, and Plasmid DNA
LNCaP-FGC (LNCaP) cells were obtained from the American Type Culture Collection (ATCC) and cultured at 37 °C in 5% CO2 with RPMI 1640 supplemented with 10% fetal bovine serum. Synthetic siRNA molecules corresponding to human HuR (ELAV-1 Silencer® Select siRNA, catalog nos. S4608, S4609, and S4610) and Negative Control (negative control 1 Silencer® Select siRNA) were sourced (Ambion). In addition, an established HuR siRNA (si-HuR-Sully) (40) was obtained (Dharmacon). Synthetic miRNA precursor molecules corresponding to human miR-331-3p (pre-miR miRNA precursor product ID: PM10881) and a negative control miRNA (pre-miR miRNA precursor negative control 1 (miR-NC); catalog no. AM17110) were also sourced from Ambion.
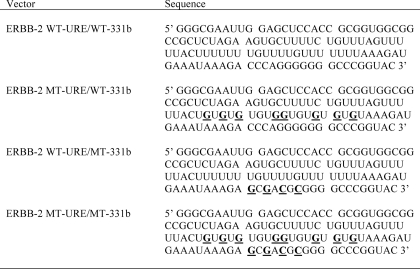
The dual luciferase reporter miRNA target clone for the ERBB-2 3′-UTR (HmiT004969) was sourced from Genecopoeia (Rockville, MD) and contained the full-length ERBB-2 3′-UTR inserted downstream of a firefly luciferase reporter gene as well as a Renilla luciferase gene for data normalization. pmiR-REPORT luciferase vectors were constructed by ligating synthetic DNA oligonucleotides corresponding to sequences within the ERBB-2 3′-UTR (wild type and mutant) into pmiR-REPORT (Ambion) using SpeI/HindIII restriction sites. Oligonucleotide sequences from the ERBB-2 3′-UTR are indicated in Table 1. The pGL3-ERBB-2 URE vector was constructed by cloning the ERBB-2 3′-UTR URE sequence using SpeI/ApaI restriction sites into a pGL3-control vector (Promega) that contained a multiple cloning site (MCS) downstream of the firefly luciferase gene (pGL3-MCS) (41). The pGL3-ERBB-2 URE deletion mutant vector was constructed by amplifying fragments on either side of the ERBB-2 3′-UTR URE with MseI restriction sites added. Amplified MseI fragments were digested with MseI and fused in the sense orientation to form an ERBB-2 3′-UTR deletion mutant, which was inserted (SpeI/ApaI) into the pGL3-MCS control vector. The empty pGL3-MCS vector was used as a negative control in IP-RT-PCR experiments (see “Immunoprecipitation (IP)-RT-PCR” and “Plasmid DNA, siRNA, and miRNA Precursor Transfections and Reporter Gene Assays”). For riboprobe synthesis, the ERBB-2 3′-UTR URE element was cloned into pBluescript (Stratagene) using BamHI/HindIII restriction sites. The sequences of all plasmids were verified by DNA sequencing.
TABLE 1.
RNA sequences of probes used
Probes were generated from Acc65I-linearized pBluescript vectors by in vitro transcription from the vector T7 promoter. Bases in ERBB-2 MT-URE, ERBB-2 WT-URE/MT-331b, or ERBB-2 MT-URE/MT-331b that are mutated in comparison with ERBB-2 WT-URE/WT-331b are underlined for ease of identification.
Immunoprecipitation (IP)-RT-PCR
IP-RT-PCR assays were performed as described previously (15) using extracts prepared from LNCaP cells. Antibodies used were against HuR (3A2; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), catalog no. sc-5261), anti-mouse IgG (NA931V; GE Healthcare), or hnRNP E1 as an unrelated antibody control (T18; Santa Cruz Biotechnology Inc., catalog no. sc-16504). PCR primers for ERBB-2 or LUC were as described above.
RT and PCR
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) and treated with DNase I (Promega) to eliminate contaminating genomic DNA. For IP-RT-PCR analysis of ERBB-2 or luciferase (LUC) mRNA expression, 1 μg of total RNA was reverse transcribed to cDNA with random hexamers and Thermoscript (Invitrogen). Semiquantitative PCR for ERBB-2 or luciferase RNA was performed using a PTC-100 programmable thermocycler (MJ Research Inc.) with Platinum Taq DNA polymerase (Invitrogen) and primers ERBB-2180-F (5′-GCT CCT CCT CGC CCT CTT GC-3′), ERBB-2670-R (5′-GCC AGC TGG TTG TTC TTG TG-3′), LUC-F (5′-TAC TGG GAC GAA GAC GAA CAC-3′), and LUC-R (5′-GTT CAC CGG CGT CAT CGT CG-3′).
Expression of HuR mRNA was determined by qRT-PCR analysis of HuR and six reference genes (GAPDH, β-actin, K-alpha-1, ALAS-1, HPRT-1, and 18S rRNA (42, 43)) that were selected using geNorm and NormFinder utilities in GENEX software (MultiD). Primer sequences are as follows: β-actin-F, 5′-GCC AAC ACA GTG CTG TCT GG-3′; β-actin-R, 5′-TAC TCC TGC TTG CTG ATC CA-3′; GAPDH-F, 5′-ATG GGG AAG GTG AAG GTC G-3′; GAPDH-R, 5′-GGG GTC ATT GAT GGC AAC AAT A-3′; K-alpha-1-F, 5′-TGG AAC CCA CAG TCA TTG ATG A-3′; K-alpha-1-R, 5′-TGA TCT CCT TGC CAA TGG TGT A-3′; HPRT-1-F, 5′-ACG AGC CCT CAG GCG AAC CT-3′; HPRT-1-R, 5′-AAT CAC GAC GCC AGG GCT GC-3′; ALAS-1-F, 5′-CGC TGT GGG GAC ACG ACC AC-3′; ALAS-1-R, 5′-ATG GGC AGC GGC GAA CAA CA-3′; 18 S-F, 5′-CAC TGT CAA CCC AAC ACA GG-3′; 18 S-R, 5′-GGC AGG TCA ATT TCA CTG GT-3′; HuR-F, 5′-CTC GGT TTG GGC GGA TCA TC-3′; HuR-R, 5′-TTC TGC CTC CGA CCG TTT G-3′. For qRT-PCR analysis of reference and target genes, 125 ng of total RNA was reverse transcribed to cDNA with random hexamers, oligo(dT) primers, and Quantiscript (Qiagen). Quantitative PCR for reference and target genes was performed on a Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science) using the Rotor-Gene SYBR Green PCR kit (Qiagen). Expression of HuR mRNA was normalized to GAPDH and β-actin mRNA expression, and relative HuR mRNA expression was determined using the 2−ΔΔCt method (44).
Plasmid DNA, siRNA, and miRNA Precursor Transfections and Reporter Gene Assays
LNCaP cells were transfected with siRNA to HuR (ELAV-1 Silencer® Select siRNA catalog no. S4608; ELAV-1 Silencer® Select siRNA catalog no. S4610 or si-HuR-Sully) or negative control (negative control 1 Silencer® Select siRNA) at a final concentration of 5 nm, using Lipofectamine 2000 (Invitrogen). Initial experiments confirmed that si-HuR-Sully was effective to silence HuR expression in LNCaP cells. ELAV-1 Silencer® Select siRNAs S4608 and s4610 were also confirmed as the most effective HuR siRNAs from a set of three ELAV-1 siRNAs (catalog nos. S4608, S4609, and S4610) in optimization experiments (data not shown).
miRNA transfection experiments were performed as described (45). Briefly, cells were transfected with miR-331-3p or miR-NC precursor at 2 nm (reporter gene assays, 24 h) or 30 nm (target validation, 3 days), respectively, using Lipofectamine 2000 (Invitrogen).
For reporter gene assays, 3 days after miRNA or siRNA transfection, LNCaP cells were retransfected using 100 ng of pmiR-REPORT vectors and a 5-ng Renilla luciferase vector. Lysates were assayed for firefly and Renilla luciferase activities 24 h after transfection using the Dual Luciferase reporter assay system (Promega) and a Fluostar OPTIMA microplate reader (BMG Labtech). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western Blotting
Cytoplasmic protein extracts were prepared as described (41), resolved on NuPAGE 4–12% BisTris gels or NuPAGE 10% BisTris gels (Invitrogen), and transferred to PVDF Plus membranes (Roche Applied Science). Membranes were blocked in 5% skim milk/TBST and probed with anti-β-actin mouse monoclonal antibody (1:10,000; Abcam catalog no. ab6276-100), anti-tubulin rat polyclonal antibody (1:1,000; Abcam catalog no. ab6161-100), anti-HuR (3A-2) (1:2,000, Santa Cruz Biotechnology, Inc., catalog no. sc-5261), anti-ERBB-2 (CB-11) mouse monoclonal antibody (1:1,000; Abcam catalog no. ab8054-1), or anti-AR (H-280) rabbit monoclonal antibody (1:1,000; Santa Cruz Biotechnology, Inc., catalog no. sc-13062). Secondary horseradish peroxidase-linked anti-mouse-IgG (NA931V; GE Healthcare), anti-rabbit-IgG (NA934V; GE Healthcare), or anti-rat-IgG (ab6734–1; Abcam) antibodies were used at 1:10,000 prior to detection with ECL Plus detection reagent and ECL-Hyperfilm (GE Healthcare). Protein bands of interest were quantitated by obtaining pixel densities of each band and correcting with a background pixel density using Quantity One software (Bio-Rad).
RNA Electromobility Shift Assay (REMSA)
Overexpression and purification of GST-HuR was performed as described previously (41). REMSA and RNase footprinting were performed as described previously (46)4 for supershifts using HuR antibody (3A2; Santa Cruz Biotechnology catalog no. sc-5261) or non-related hnRNP E1 antibody (T18; Santa Cruz Biotechnology, Inc., catalog no. sc-16504). DNA analogues used to probe miR-331-3p interactions with the ERBB-2 URE were as follows: 331-3p DNA (5′-GCC CCT GGG CCT ATC CTA GAA-3′) and URE-Comp DNA (a perfectly complementary DNA to the ERBB-2 miR-331b site; 5′-CCC CCT GGG TCT TTA TTT CAT-3′). A 10-fold excess of DNA was mixed with probe RNA in water, heated to 75 °C for 5 min prior to dilution to 2× final concentration in 2× binding buffer, and incubated at 37 °C for 20 min prior to the addition to binding reactions. RNase H digestions (0.01 unit of RNase H from Invitrogen/reaction) were performed in a manner comparable with other RNase digestions.4
Statistical Analysis
The statistical significance of differences in luciferase expression data were evaluated using paired (one-tailed or two-tailed, as indicated) Student's t test. Synergy in combined reporter mutations was assessed according to the method of Bliss (48), where synergy occurs when EBliss < EObserved. Statistical analysis of qRT-PCR data was performed using GENEX software (MultiD). All analyses were performed at a minimum confidence interval of 95% (CI = 0.95) and normality of data were confirmed by the Kolmogorov-Smirnov test.
RESULTS
HuR Interacts with a U-rich Element in Close Proximity to an Established miR-331-3p Target Site within the 3′-UTR of ERBB-2 mRNA in PCa Cells
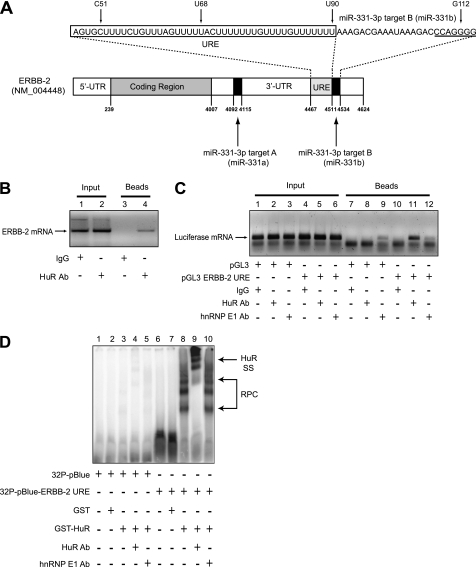
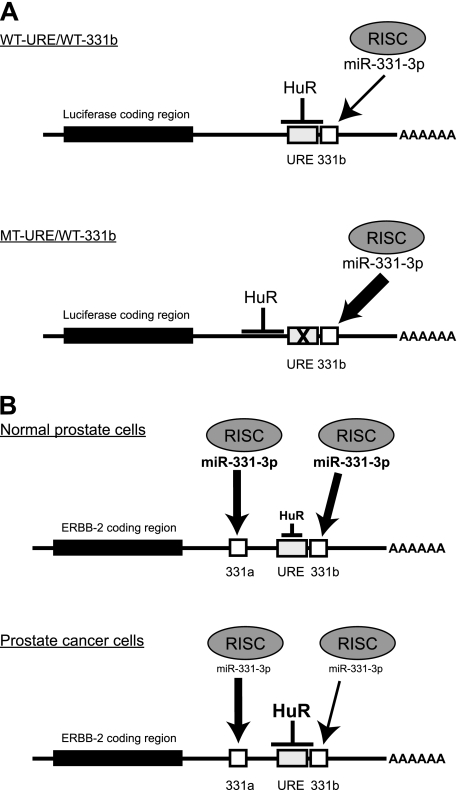
Previously, we demonstrated that the ERBB-2 mRNA 3′-UTR contains two specific target sites for direct binding of miR-331-3p (14). Further analysis of the ERBB-2 3′-UTR sequence identified a URE (nucleotides 4467–4511 of GenBankTM accession number NM_004448) immediately adjacent to the distal miR-331-3p target site (331b; Fig. 1A), raising the possibility that this U-rich sequence was a target for specific binding of HuR, an AU- and U-rich RBP implicated in regulating expression of oncogenic mRNAs (49, 50), including ERBB-2 in breast cancer (38, 39). To investigate the interaction, we performed IP-RT-PCR assays using LNCaP cells (Fig. 1B). These demonstrated co-immunopurification of ERBB-2 mRNA with HuR (Fig. 1B, lane 4), providing evidence for an in vivo interaction between HuR and ERBB-2 mRNA in PCa cells.
FIGURE 1.
HuR binds the ERBB-2 mRNA 3′-UTR URE in vitro and in vivo. A, the ERBB-2 mRNA 3′-UTR contains two miR-331-3p binding sites (miR-331-3p target A and miR-331-3p target B) as well as a URE. The nucleotide sequences of the URE (boxed) and the miR-331-3p target B (seed region is underlined) are provided, and the individual nucleotide positions are indicated (also refer to Fig. 5). B, co-immunopurification of ERBB-2 mRNA with HuR in PCa cells. LNCaP protein lysates were immunoprecipitated with IgG (control) or HuR antibody, and RT-PCR was used to detect co-purification of ERBB-2 mRNA with endogenous HuR. C, co-immunopurification of HuR with chimeric ERBB-2 3′-UTR URE-luciferase RNA in LNCaP cells. After transfection of LNCaP cells with pGL3-control or pGL3-ERBB-2 URE plasmid DNA, protein lysates were immunoprecipitated with IgG (control), HuR, or hnRNP E1 (unrelated) antibody. RT-PCR was used to detect co-purification of ERBB-2 URE-luciferase RNA with endogenous HuR, demonstrating an in vivo association between HuR and the ERBB-2 URE in PCa cells. D, recombinant GST-HuR binds the ERBB-2 URE in REMSA. GST fusion proteins were incubated with 32P-pBluescript (vector control) or 32P-pBluescript-ERBB-2 URE riboprobes. RPCs were resolved by non-denaturing PAGE and visualized by autoradiography. The addition of HuR antibody to binding reactions produced a specific supershift (SS) of RPCs containing GST-HuR and 32P-pBluescript-ERBB-2 URE RNA. Lane numbers are shown above all gels.
To determine whether the ERBB-2 3′-UTR URE was a specific target for HuR binding, we transfected LNCaP cells with pGL3-control or pGL3-ERBB-2-URE luciferase reporter constructs, immunoprecipitated cell lysates with HuR or unrelated antibody (mouse IgG or hnRNP E1), and used RT-PCR to detect exogenous luciferase mRNA. Marked enrichment of luciferase mRNA was observed from HuR immunoprecipitates of LNCaP cells transfected with pGL3-ERBB-2-URE (Fig. 1C, lane 11) but not with pGL3-control-transfected cells (Fig. 1C, lanes 7–9) or with unrelated antibodies (Fig. 1C, lanes 10 and 12). This suggested that HuR was directly associated with the ERBB-2 URE element in PCa cells.
To confirm this finding, we performed REMSA experiments with recombinant GST-HuR and 32P-labeled riboprobes (Fig. 1D). RNA-protein complexes (RPCs) formed when GST-HuR was incubated with 32P-pBlue-ERBB-2-URE (lane 8) but not with 32P-pBlue (lane 3). GST did not bind to either riboprobe (lanes 2 and 7), validating the specificity of the GST-HuR/32P-pBlue-ERBB-2-URE interaction. Furthermore, an anti-HuR antibody shifted the GST-HuR/32P-pBlue-ERBB-2-URE RPCs, whereas an unrelated (hnRNP E1) antibody did not (lane 9 versus lane 10). Together, these results indicate that HuR binds directly and specifically to the ERBB-2 3′-UTR URE in PCa cells.
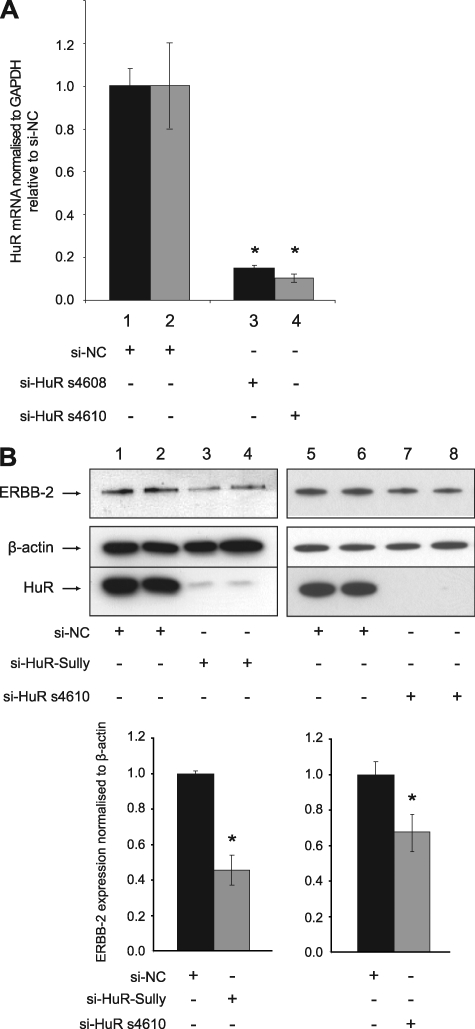
HuR Regulates ERBB-2 Expression in PCa Cells
HuR siRNAs (si-HuR) were transfected into LNCaP cells to assess the functional significance of HuR on ERBB-2 expression in PCa. This resulted in a significant HuR mRNA knockdown assessed by qRT-PCR for two different siRNA molecules (Fig. 2A). Immunoblotting confirmed the reduction in HuR expression and revealed decreased levels of ERBB-2 protein, which was quantified by densitometry (Fig. 2B). In contrast, si-HuR did not alter expression of β-actin. These data suggest that binding of HuR to the ERBB-2 URE regulates the post-transcriptional expression of ERBB-2 in PCa cells.
FIGURE 2.
HuR regulates ERBB-2 expression in LNCaP cells. A, qRT-PCR assays of HuR mRNA from LNCaP cells transfected with HuR siRNAs (si-HuR s4608 and s4610) or si-NC for 24 h. HuR mRNA expression was normalized to GAPDH mRNA expression and is shown as a ratio of si-HuR-transfected cells to si-NC-transfected cells using the 2−ΔΔCt method and GENEX statistical software. The asterisks indicate a significant difference from si-NC-transfected control (*, p < 0.01). Error bars represent confidence intervals (CI = 0.95). B, immunoblotting detection of ERBB-2, β-actin, and HuR expression using protein extracts harvested from LNCaP cells 3 days after transfection with HuR siRNAs (si-HuR Sully and s4610) or si-NC. ERBB-2 expression was quantitated by densitometry measurement using Quantity One software. Asterisks indicate a significant difference from si-NC-transfected control (*, p < 0.05). Error bars, S.D.
HuR Antagonizes the Action of miR-331-3p on ERBB-2 mRNA
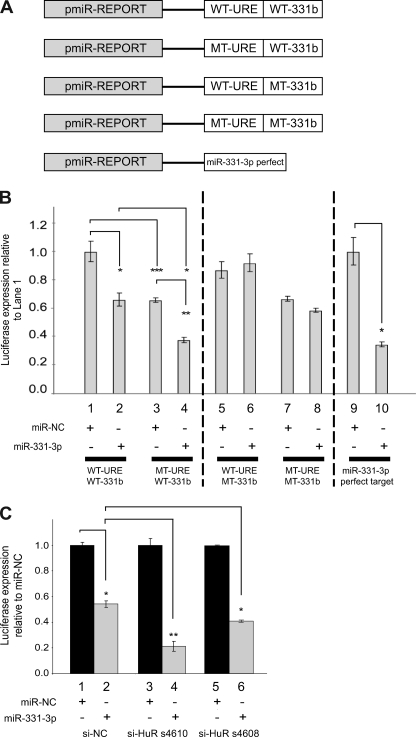
Our previous work indicated that miR-331-3p regulates ERBB-2 expression in PCa cells via its interaction with two specific target sites in the ERBB-2 3′-UTR (14), one of which is immediately distal to the ERBB-2-URE (miR-331b; Fig. 1A). Interestingly, whereas miR-331-3p expression is down-regulated in PCa cells (14, 22, 28), HuR is typically overexpressed and/or aberrantly accumulates in the cytoplasm in various cancers (15, 16, 30, 31, 51–55), including PCa (37). We investigated whether HuR could antagonize the ability of miR-331-3p to down-regulate ERBB-2 expression in PCa cells. To address this, we focused our analysis on the ERBB-2 URE and its distal miR-331-3p site (miR-331b; Fig. 1A). In addition to the wild type ERBB-2 URE and wild type miR-331-3p site (WT-URE/WT-331b), we generated luciferase reporter constructs that contained mutants of the URE and/or the miR-331b site (Fig. 3A) for transfection studies using LNCaP cells together with synthetic miR-331-3p precursor RNA.
FIGURE 3.
HuR antagonizes the action of miR-331-3p on ERBB-2 mRNA. A, schematic representation of firefly luciferase reporter constructs for wild type or mutant ERBB-2 URE (WT-URE or MT-URE) together with wild type or mutant miR-331-3p target B (WT-331b or MT-331b). A firefly luciferase plasmid that contained a perfect miR-331-3p target site was included as a positive control for miR-331-3p activity (14). B, LNCaP cells were co-transfected with reporter constructs containing wild type or mutant ERBB-2 URE and miR-331-3p site B and miR-NC or miR-331-3p and assayed for firefly and Renilla luciferase activities after 24 h. Relative luciferase expression (firefly normalized to Renilla) values are expressed as a ratio of WT-URE/WT-331b + miR-NC-transfected cells (i.e. relative to lane 1). Results are representative of three independent experiments. Error bars, S.E. Asterisks represent statistically significant differences (*, p < 0.05; **, p < 0.005; ***, p < 0.0005) and are from one-tailed t tests except for the comparison between WT-URE/WT-331b + miR-331-3p and MT-URE/WT-331b + miR-331-3p, which is two-tailed. Synergy for the combined effect of URE mutation and transfection of miR-331-3p was assessed using Bliss analysis (48), where EBliss = 0.6857 − 0.1175 = 0.5682, and EObserved = 0.6265. C, LNCaP cells were co-transfected with a luciferase plasmid containing full-length ERBB-2 3′-UTR, si-NC or si-HuR (HuR siRNA s4610 or s4608), and miR-NC or miR-331-3p. At 24 h, lysates were assayed for firefly and Renilla luciferase activities and relative luciferase expression (firefly normalized to Renilla) indicated as a ratio of miR-NC-transfected cells. Error bars, S.D. Asterisks indicate a significant difference from miR-NC (*, p < 0.05; **, p < 0.01).
Co-transfection of LNCaP cells with miR-331-3p and pmiR-REPORT-WT-URE/WT-331b resulted in repression of reporter gene activity relative to the same vector with miR-NC (Fig. 3B, lane 2 versus lane 1), consistent with our earlier finding (14). In LNCaP cells transfected with pmiR-REPORT-MT-URE/WT-331b, co-transfection with miR-NC reduced basal reporter expression relative to the WT-URE construct with miR-NC (lane 3 versus lane 1), an effect that was increased when cells were instead co-transfected with miR-331-3p (lane 4 versus lane 1). These results suggested that HuR can antagonize the action of miR-331-3p on ERBB-2 mRNA because altering HuR binding (MT-URE; lanes 3 and 4) lowers basal reporter activity and increases the repressive effect of miR-331-3p on reporter activity. To determine whether the combined effect of altered HuR binding (MT-URE) and co-transfection with miR-331-3p was synergistic, we performed Bliss analysis (48). For the effect to be synergistic, the observed fractional inhibition for the combination of altered HuR binding and miR-331-3p transfection (lane 4) would exceed the sum of the individual effects. We observed a fractional inhibition for the combination of 0.6265 (EObserved) that is greater than the expected fractional inhibition (EBliss; 0.5682), indicating that the combined effect is synergistic.
When the miR-331b site was mutated (pmiR-REPORT WT-URE/MT-331b), the repressive action of miR-331-3p was blocked (Fig. 3B, compare lanes 5 and 6 with lanes 1 and 2), supporting its role as a negative regulator of ERBB-2 expression. The double mutant construct (MT-URE/MT-331b) showed reduced expression consistent with reduced HuR binding, and this effect was not substantially increased by the addition of miR-331-3p (lanes 7 and 8). Finally, we confirmed that the observed miR-331-3p repression was due to miR-331-3p, by co-transfection of miR-331-3p with a luciferase reporter vector containing a perfect miR-331-3p target site. Co-transfection of miR-331-3p resulted in significant reduction of activity of this reporter compared with co-transfection of miR-NC (lane 10 versus lane 9).
To support these observations, we used si-HuR to first deplete HuR expression in LNCaP cells. Co-transfection of these cells with miR-331-3p and a full-length ERBB-2 3′-UTR reporter construct showed significantly reduced luciferase expression in si-HuR-transfected cells relative to cells transfected with a non-targeting siRNA (si-NC) (Fig. 3C, lanes 4 and 6 versus lane 2). Because a large number of proteins could potentially interact with the URE (56), this result indicates that it is the binding of HuR to the ERBB-2-URE that prevents miR-331-3p from targeting the miR-331b site.
The ERBB-2 MT-URE Is Still Bound by HuR
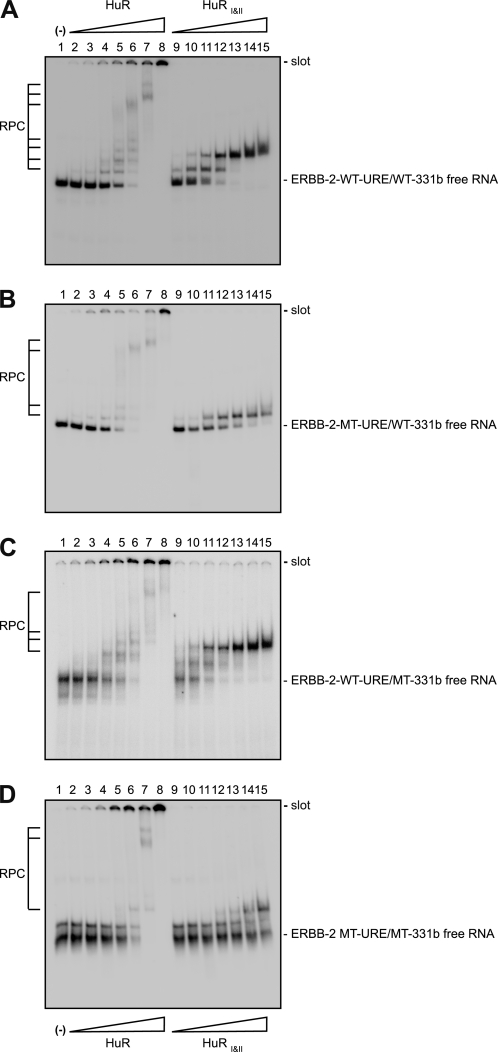
To confirm that the MT-URE mutation affects HuR binding and that the MT-331b mutation does not, we performed REMSA experiments with purified full-length HuR or with a truncated form of the protein, HuRI&II, that contains only the first two RRM domains (50, 57) and target RNAs corresponding to the relevant portion of the luciferase constructs used in Fig. 3B (Table 1). As shown in Fig. 4 and Table 2, HuR and HuRI&II bind to ERBB-2 WT-URE/WT-331b RNA: HuR with an affinity comparable with that of other target RNAs evaluated previously and HuRI&II with a higher affinity than HuR, a situation contrary to that seen with other target RNAs.4 Surprisingly, given the dramatic effects on HuR binding caused by introducing guanine residues in other RNA targets (50),4 the MT-URE mutation (MT-URE/WT-331b) had a modest effect on HuR binding (Fig. 4B) with a less than 2-fold reduction in Kd (Table 2), although a greater effect is discernible for HuRI&II binding. There is also an apparent reduction in the mobility shift of the bound (retarded) band in both cases. As expected, mutations within the miR-331-3p seed region (WT-URE/MT-331b) did not affect HuR binding (Fig. 4C and Table 2), whereas the combined mutations (MT-URE/MT-331b) were bound by HuR in an equivalent manner to the MT-URE/WT-331b RNA (Fig. 4D and Table 2). The addition of competitor tRNA did not significantly alter HuR or HuRI&II binding (supplemental Fig. S1).
FIGURE 4.
Mutations within the ERBB-2 URE affect HuR binding. REMSAs were performed with purified HuR or HuRI&II and several different probes: ERBB-2 WT-URE/WT-331b (A), ERBB-2 MT-URE/WT-331b (B), ERBB-2 WT-URE/MT-331b (C), or ERBB-2 MT-URE/MT-331b (D). Target sequences are shown in Table 1. Lane numbers are indicated above each gel. The binding reaction for lane 1 contained no protein. Binding reactions for lanes 2–8 contained the following: 1 × 10−8 m, 2 × 10−8 m, 5 × 10−8 m, 1 × 10−7 m, 2 × 10−7 m, 5 × 10−7 m, or 1 × 10−6 m HuR (calculated per monomer), respectively. Binding reactions for lanes 9–15 contained amounts of HuRI&II equivalent to the amounts of HuR in lanes 2–8. All binding reactions contained 1 × 10−8 m concentrations of the relevant target RNA. −, absence of protein. Wedges labeled with HuR or HuRI&II indicate increasing concentrations of each protein above the other lanes. The positions of the free RNA (unbound) and slot (origin) are indicated to the right of each gel. Kd values calculated by non-linear regression analysis of multiple repeats of each gel are shown in Table 2.
TABLE 2.
Apparent Kd values for the initial binding of HuR or HuRI&II to the ERBB-2 WT-URE/WT-331b or mutant RNAs shown in Fig. 4
| Target RNA |
Kd |
|
|---|---|---|
| HuR | HuRI&II | |
| m | ||
| WT-URE/WT-331b | 8.2 ± 1.1 × 10−8 | 4.1 ± 0.4 × 10−8 |
| MT-URE/WT-331b | 1.1 ± 0.3 × 10−7 | 2.0 ± 0.5 × 10−7 |
| WT-URE/MT-331b | 8.3 ± 1.1 × 10−8 | 4.3 ± 0.7 × 10−8 |
| MT-URE/MT-331b | 1.2 ± 0.3 × 10−7 | 3.4 ± 1.9 × 10−7 |
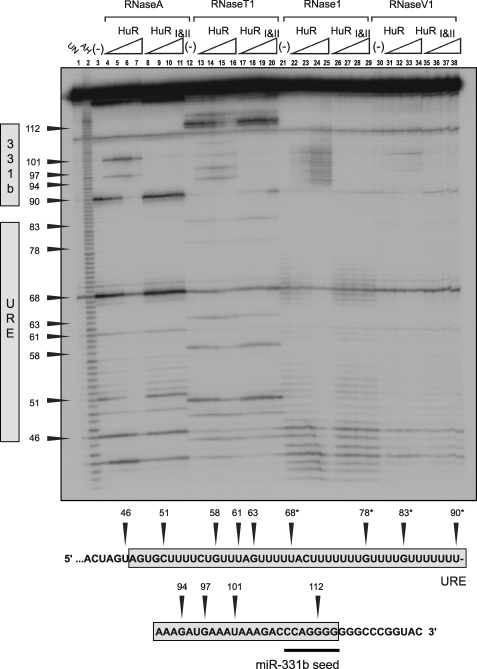
Given the results obtained in Fig. 4, B and D, we determined the position of the primary binding sites for HuR in ERBB-2 WT-URE/WT-331b (Fig. 5) and ERBB-2 MT-URE/WT-331b (Fig. 6) using RNase footprint assays. As noted previously for c-Fos ARE, TNFα ARE, and MTA1 URE RNAs,4 a marked preference of RNase A for the distal uracil in a string of uracils in the ERBB-2 URE makes assignment of protected bases difficult in these regions. However, weak digestion of unbound RNA at bases immediately distal to U68 by RNase 1 and to C70 by RNase A is absent at the lowest concentration of added HuR (Fig. 5, lanes 5 and 22). Protection is also apparent at positions proximal to G63 but only at higher concentrations of added protein. The appearance of RNase hypersensitivity (Fig. 5, lanes 4–7, 13–16, 25–29, and 31–34) with increasing HuR or HuRI&II concentration at bases distal to U90 indicates the absence of bound protein in this region as well as potential changes to RNA secondary structure upon HuR binding.
FIGURE 5.
RNase footprint analysis of HuR binding to ERBB-2 WT-URE/WT-331b RNA. Lane 1 contains “untreated” ERBB-2 WT-URE/WT-331b RNA (mock-digested) and is also labeled UN. Lane 2 contains ERBB-2 WT-URE/WT-331b RNA subjected to partial alkaline hydrolysis in order to generate a ladder corresponding to consecutive bases and is also labeled AH. Lanes 3–11 are partially digested with RNase A (which cleaves after U or C bases), and binding reactions contain the following: no protein (lane 3); 2 × 10−8 m HuR (lane 4); 5 × 10−8 m HuR (lane 5); 1 × 10−7 m HuR (lane 6); 2 × 10−7 m HuR (lane 7); 5 × 10−8 m HuRI&II (lane 8); 1 × 10−7 m HuRI&II (lane 9); 2 × 10−7 m HuRI&II (lane 10); or 5 × 10−7 m HuRI&II (lane 11). The presence or absence of protein is indicated as previously. RNase A digestion is also indicated by a label above lanes 3–11. Lanes 12–20 are RNase T1-digested (which cleaves after G bases); lanes 21–29 are RNase 1-digested (which cleaves preferentially after non-base-paired residues); and lanes 30–38 are RNase V1-digested (which cleaves preferentially after base-paired residues). Bands in the gel are assigned to the ERBB-2 WT-URE/WT-331b target RNA sequence below the gel by numbering. *, putative HuR binding site.
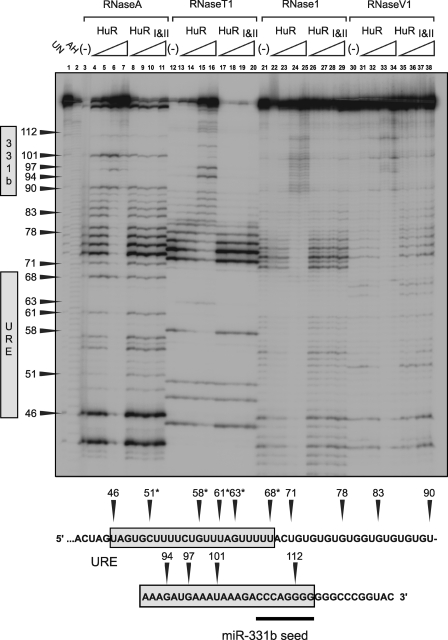
FIGURE 6.
RNase footprint analysis of HuR binding to ERBB-2 MT-URE/WT-331b target RNA. Lane labeling, protein concentrations, and RNase designations are identical to the gel shown in Fig. 5 except that the target RNA used is the ERBB-2 MT-URE/WT-331b target RNA. *, putative HuR binding site.
The introduction of multiple guanines in ERBB-2 WT-URE produced clearer RNase digestion patterns. Nevertheless, RNase 1 gives the clearest indication of protection at the lowest concentration of HuR, which occurs between C51 and U68 (Fig. 6, lane 22). Some protection, notably at the highest HuR concentration examined, also occurs between U68 and U90, and RNase hypersensitivity is apparent distal to U90, again raising the possibility of alterations to RNA secondary structure upon HuR binding. This result confirms that a shift in the primary binding site for HuR has occurred, from between U68 and U90 for ERBB-2 WT-URE (Fig. 5) to between C51 and U68 for ERBB-2 MT-URE (Fig. 6).
HuR Binding Does Not Directly Inhibit miR-331-3p Binding to the ERBB-2 URE
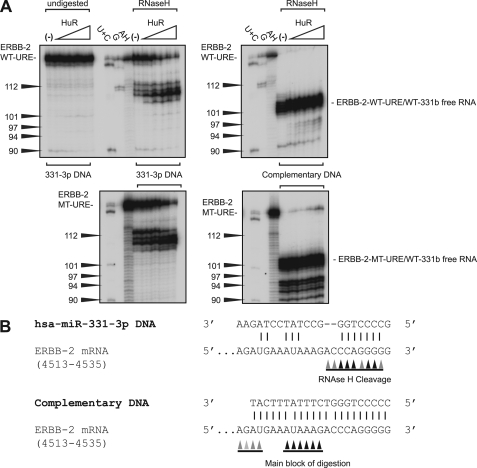
Because the primary HuR binding site in the ERBB-2 URE shifts upon introduction of the MT-URE mutation, we hypothesized that the synergy between introduction of the MT-URE mutation and the addition of miR-331-3p by co-transfection (Fig. 3B) could be due to removal of steric hindrance of miR-331-3p binding by bound HuR and designed an in vitro RNase H digestion assay to test this. First, ERBB-2 WT-URE/WT-331b or ERBB-2 MT-URE/WT-331b RNA was hybridized with either a DNA analog of miR-331-3p or a DNA fully complementary to the miR-331b region. A mobility shift was apparent for the latter but not the former upon hybridization to both RNAs (supplemental Fig. S2), indicating that if a complex does form between 331-3p DNA and the miR-331b site, it is at best transient. Interestingly, although significant alterations in the pattern of band retardation by HuR are not apparent (supplemental Fig. S2), the presence of the fully complementary URE-Comp DNA leads to an approximate 2-fold tightening of HuR binding for both target RNAs (Table 3). Probing the observed complexes for DNA-RNA hybrids using RNase H digestion reveals that 331-3p DNA forms a complex with both ERBB-2 WT-URE/WT-331b and ERBB-2 MT-URE/WT-331b RNAs and that this complex appears to be strengthened by HuR binding (Fig. 7A). The main sites of digestion observed for both RNA targets are within the miR-331b seed sequence, indicating that the miR-331-3p DNA analog is faithfully mimicking miR-331-3p binding (Fig. 7B). The URE-Comp DNA complex does not alter in intensity with increasing HuR as expected, with the major sites of RNase H digestion occurring around the middle of the hybridized DNA. Thus, HuR binding to the wild-type ERBB-2 URE does not prevent miR-331-3p binding to its adjacent seed (miR-331b), although the spacing between the two sites may be critical for the regulatory outcome (Fig. 3B).
TABLE 3.
Apparent Kd values for the initial binding of HuR to the ERBB-2 WT-URE/WT-331b and MT-URE/WT-331b RNAs in the presence of hybridizing DNA shown in Fig. 7
| Target RNA | Hybridizing DNA | HuR Kd (M) |
|---|---|---|
| WT-URE/WT-331b | 331-3p DNA | 7.4 ± 2.0 × 10−8 |
| WT-URE/WT-331b | Complementary DNA | 3.7 ± 0.8 × 10−8 |
| MT-URE/WT-331b | 331-3p DNA | 1.4 ± 0.4 × 10−7 |
| MT-URE/WT-331b | Complementary DNA | 7.6 ± 3.0 × 10−8 |
FIGURE 7.
RNase H digestion assay reveals that miR-331-3p can bind both ERBB-2 WT-URE/WT-331b and ERBB-2 MT-URE/WT-331b RNAs. A, 331-3p DNA (left) or URE-Comp DNA (right) was hybridized to ERBB-2 WT-URE/WT-331b RNA (top) or ERBB-2 MT-URE/WT-331b RNA (bottom) for binding reactions with HuR, followed by RNase H digestion. RNase A (U + C) and RNase T1 (G) digests were done in the absence of hybridizing DNA. Protein and RNA concentrations and labeling are the same as described previously. B, a schematic summarizing the digestion results obtained from A.
Finally, to confirm the validity of our ERBB-2 3′-UTR URE mutation (MT-URE) with respect to miR-331-3p action on its miR-331b site, we deleted the entire URE from the ERBB-2 full-length 3′-UTR and tested its repression by exogenous miR-331-3p in LNCaP cells. The addition of miR-331-3p yielded a significantly greater repression of luciferase expression from the ERBB-2 full-length 3′-UTR-URE deletion mutant reporter than from the ERBB-2 full-length 3′-UTR reporter (supplemental Fig. S3). This result indicates that loss of HuR binding to the ERBB-2 3′-UTR URE increases the repressive action of miR-331-3p upon ERBB-2 mRNA.
DISCUSSION
In this study, we have investigated the coordinate roles of HuR and miR-331-3p in regulating ERBB-2 expression in PCa cells. We have shown that HuR is able to bind the ERBB-2 3′-UTR both in vivo and in vitro. Scott and co-workers (39) demonstrated the binding of HuR to the 3′-UTR of ERBB-2 in SkBr3 breast cancer cells; our work complements this finding in PCa. We have found that RNAi-mediated knockdown of HuR in the LNCaP PCa cell line reduces ERBB-2 expression. Observing the close proximity between the HuR binding site and the distal miR-331-3p binding site in the ERBB-2 3′-UTR, we hypothesized that altering HuR levels may impact on accessibility of miR-331-3p to its distal 3′-UTR binding site. The RNase H assay we used to probe this interaction indicated, however, that naked miR-331-3p is able to hybridize to the miR-331b site even when HuR is bound and that moving the relative position of the bound HuR in relation to the miR-331b site does not alter miR-331-3p binding (Fig. 8A). However, the combined effect on reporter constructs (moving the HuR binding site by mutation and the addition of exogenous miR-331-3p) is synergistic in LNCaP cells. This suggests that HuR may be sterically hindering miR-331-3p/RISC association with the miR-331b site in the ERBB-2 3′-UTR.
FIGURE 8.
Model for the interaction between HuR, miR-331-3p, and the ERBB-2 mRNA 3′-UTR. A, in vitro luciferase transfection and RNase footprinting studies suggest that HuR binding to the ERBB-2 mRNA 3′-UTR URE partially protects the distal miR-331-3p site (miR-331b) from the action of miR-331-3p via its associated RISC. Mutation of the URE (MT-URE/WT-331b) results in HuR binding at an upstream site and promotes the action of miR-331-3p upon the miR-331b target site. B, normal prostate cells express miR-331-3p, whereas HuR is expressed to normal levels and is predominantly nuclear. As a consequence, miR-331-3p fine tunes ERBB-2 expression to normal levels via its interactions with two specific mRNA 3′-UTR target sites (miR-331a and miR-331b). In some PCa cells, elevated expression and cytoplasmic accumulation of HuR promotes ERBB-2 overexpression, at least in part by binding to the URE and reducing the action of miR-331-3p-associated RISC on its distal 3′-UTR target site (331b). This effect is exacerbated by the decreased expression of miR-331-3p in PCa cells.
Moving the HuR binding site has a greater effect than HuR depletion (hence the observed synergy) upon miR-331-3p-mediated down-regulation, raising the possibility that HuR could even aid miR-331-3p/RISC association if the two contacted sites are sufficiently separated to avoid steric clashes. IL-1β mRNA, with an ARE-containing 3′-UTR, has been shown to be destabilized by HuR binding (53). Bioinformatic predictions (TargetScan release 5.1) suggest a seed for miR-1178 10 bases to the 5′-end of the ARE (not shown). In contrast, the well characterized TNFα ARE has multiple miRNA seed sequences overlapping its 5′-end, raising the possibility that the conversion of HuR into a prodegradation factor via repositioning of its binding site in relation to close miRNA seed sequences could be a general property. We are currently testing this hypothesis.
Deregulated expression of miRNAs in cancer may be associated with gain or loss of chromosomal regions because many miRNA genes are located in fragile sites that are frequently altered in cancer (58, 59). In addition, aberrant miRNA expression can occur through epigenetic mechanisms or by defects in miRNA biogenesis (60–62). We previously observed decreased expression of both primary and mature miR-331-3p in PCa tissue relative to normal adjacent prostate tissue (14), suggesting that reduced miR-331-3p expression in PCa cells does not result from abnormal processing of pre-miR-331-3p. The human miR-331 gene is located at 12q22, a chromosomal region that is not commonly altered in PCa. One possible explanation for decreased miR-331-3p expression in PCa is that there is reduced miR-331 gene transcription, resulting in less primary, and therefore mature, miR-331-3p being produced. We propose that in the normal prostate, the net effect of a relative abundance of miR-331-3p and low cytoplasmic HuR maintains low steady state expression of ERBB-2 mRNA. In contrast, down-regulation of miR-331-3p combined with increased levels of cytoplasmic HuR in PCa (37) facilitates stabilization of ERBB-2 mRNA (Fig. 8B) and increased ERBB-2 protein expression in the absence of ERBB-2 gene amplification. Of interest, recent reports have demonstrated regulation of HuR by miR-16 in breast cancer cells (63) and by miR-519 in lung, ovarian, colon, and kidney cancer cells (64, 65). It is unclear whether HuR overexpression in PCa is the result of a loss of miRNA-mediated repression.
Several recent studies have raised the possibility of interplay between miRNAs and RBPs (47, 66–71). HuR binding to the 3′-UTR of CAT1 counteracts the effects of miR-122 in hepatocellular carcinoma cells (47). Similarly, Dead End 1 (Dnd1) RBP affects the function of several miRNAs in human cells and in primordial germ cells of zebrafish (69). In these instances, an RBP interferes with the association between a miRNA and its associated silencing complex and a target mRNA. Our findings suggest that HuR acts in a similar manner. In other words, HuR may not prevent miR-331-3p binding to its distal ERBB-2 3′-UTR binding site; rather, it may reduce the association between ERBB-2 mRNA and the RNA silencing complex. Furthermore, these findings emphasize the importance of considering the contribution of both miRNA and RBP binding sites when studying the posttranscriptional regulation of gene expression (56).
The combined effects of increasing cytoplasmic HuR and decreased miR-331-3p could lead to the increase of ERBB-2 expression seen in the absence of gene amplification in some PCas (12). Counteracting these combined effects to reduce ERBB-2 expression represents a possible approach for the treatment of hormone-refractory PCa.
Acknowledgments
We thank Dr. Andrew Thomson for helpful discussions regarding the manuscript, Dr. Susan Magdaleno (Invitrogen) for the generous gift of HuR siRNA reagents, and Dr. Henry Furneaux for the GST-HuR expression construct.
This work was supported by the National Health and Medical Research Council of Australia and the Prostate Cancer Foundation of Australia.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
A. Barker, M. R. Epis, C. J. Porter, B. R. Hopkins, M. C. Wilce, J. A. Wilce, K. M. Giles, and P. J. Leedman, manuscript in preparation.
- PCa
- prostate cancer
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR
- URE
- U-rich element
- RBP
- RNA-binding protein
- pre-miR
- precursor miRNA
- AR
- androgen receptor
- ARE
- AR element
- miR-NC
- negative control miRNA
- si-NC
- non-targeting siRNA
- si-HuR
- HuR siRNA
- RISC
- RNA-induced silencing complex
- CI
- confidence interval
- RPC
- RNA-protein complex
- LNCaP
- LNCaP-FGC
- MCS
- multiple cloning site
- LUC
- luciferase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- REMSA
- RNA electromobility shift assay
- hnRNP
- heterogeneous nuclear ribonucleoprotein.
REFERENCES
- 1. Australian Institute of Health and Welfare (2010) Cancer in Australia: An Overview, Cancer Series Number 60, Australian Institute of Health and Welfare, Canberra [Google Scholar]
- 2. Feldman B. J., Feldman D. (2001) Nat. Rev. Cancer 1, 34–45 [DOI] [PubMed] [Google Scholar]
- 3. Stavridi F., Karapanagiotou E. M., Syrigos K. N. (2010) Cancer Treat. Rev. 36, 122–130 [DOI] [PubMed] [Google Scholar]
- 4. Yap T. A., Zivi A., Omlin A., de Bono J. S. (2011) Nat. Rev. Clin. Oncol. 8, 597–610 [DOI] [PubMed] [Google Scholar]
- 5. Neto A. S., Tobias-Machado M., Wroclawski M. L., Fonseca F. L., Teixeira G. K., Amarante R. D., Wroclawski E. R., Del Giglio A. (2010) J. Urol. 184, 842–850 [DOI] [PubMed] [Google Scholar]
- 6. Ziada A., Barqawi A., Glode L. M., Varella-Garcia M., Crighton F., Majeski S., Rosenblum M., Kane M., Chen L., Crawford E. D. (2004) Prostate 60, 332–337 [DOI] [PubMed] [Google Scholar]
- 7. Sridhar S. S., Hotte S. J., Chin J. L., Hudes G. R., Gregg R., Trachtenberg J., Wang L., Tran-Thanh D., Pham N. A., Tsao M. S., Hedley D., Dancey J. E., Moore M. J. (2010) Am. J. Clin. Oncol. 33, 609–613 [DOI] [PubMed] [Google Scholar]
- 8. Legrier M. E., Oudard S., Judde J. G., Guyader C., de Pinieux G., Boyé K., de Cremoux P., Dutrillaux B., Poupon M. F. (2007) Br. J. Cancer 96, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Small E. J., Bok R., Reese D. M., Sudilovsky D., Frohlich M. (2001) Semin. Oncol. 28, Suppl. 15, 71–76 [DOI] [PubMed] [Google Scholar]
- 10. Shaw G., Prowse D. M. (2008) Cancer Cell Int. 8, 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Craft N., Shostak Y., Carey M., Sawyers C. L. (1999) Nat. Med. 5, 280–285 [DOI] [PubMed] [Google Scholar]
- 12. Minner S., Jessen B., Stiedenroth L., Burandt E., Köllermann J., Mirlacher M., Erbersdobler A., Eichelberg C., Fisch M., Brümmendorf T. H., Bokemeyer C., Simon R., Steuber T., Graefen M., Huland H., Sauter G., Schlomm T. (2010) Clin. Cancer Res. 16, 1553–1560 [DOI] [PubMed] [Google Scholar]
- 13. Bubendorf L., Kononen J., Koivisto P., Schraml P., Moch H., Gasser T. C., Willi N., Mihatsch M. J., Sauter G., Kallioniemi O. P. (1999) Cancer Res. 59, 803–806 [PubMed] [Google Scholar]
- 14. Epis M. R., Giles K. M., Barker A., Kendrick T. S., Leedman P. J. (2009) J. Biol. Chem. 284, 24696–24704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 16. Humphreys D. T., Westman B. J., Martin D. I., Preiss T. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 16961–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006) Nat. Genet. 38, 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng A. M., Byrom M. W., Shelton J., Ford L. P. (2005) Nucleic Acids Res. 33, 1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang B., Pan X., Cobb G. P., Anderson T. A. (2007) Dev. Biol. 302, 1–12 [DOI] [PubMed] [Google Scholar]
- 20. Ambs S., Prueitt R. L., Yi M., Hudson R. S., Howe T. M., Petrocca F., Wallace T. A., Liu C. G., Volinia S., Calin G. A., Yfantis H. G., Stephens R. M., Croce C. M. (2008) Cancer Res. 68, 6162–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozen M., Creighton C. J., Ozdemir M., Ittmann M. (2008) Oncogene 27, 1788–1793 [DOI] [PubMed] [Google Scholar]
- 22. Wang L., Tang H., Thayanithy V., Subramanian S., Oberg A. L., Cunningham J. M., Cerhan J. R., Steer C. J., Thibodeau S. N. (2009) Cancer Res. 69, 9490–9497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gandellini P., Folini M., Zaffaroni N. (2010) Discov. Med. 9, 212–218 [PubMed] [Google Scholar]
- 24. Barnabas N., Xu L., Savera A., Hou Z., Barrack E. R. (2011) Prostate 71, 857–871 [DOI] [PubMed] [Google Scholar]
- 25. Liu C., Kelnar K., Liu B., Chen X., Calhoun-Davis T., Li H., Patrawala L., Yan H., Jeter C., Honorio S., Wiggins J. F., Bader A. G., Fagin R., Brown D., Tang D. G. (2011) Nat. Med. 17, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Long Q., Johnson B. A., Osunkoya A. O., Lai Y. H., Zhou W., Abramovitz M., Xia M., Bouzyk M. B., Nam R. K., Sugar L., Stanimirovic A., Williams D. J., Leyland-Jones B. R., Seth A. K., Petros J. A., Moreno C. S. (2011) Am. J. Pathol. 179, 46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peng X., Guo W., Liu T., Wang X., Tu X., Xiong D., Chen S., Lai Y., Du H., Chen G., Liu G., Tang Y., Huang S., Zou X. (2011) PLoS One 6, e20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu G., Wu J., Zhou L., Chen B., Sun Z., Zhao F., Tao Z. (2010) PLoS One 5, e15519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hinman M. N., Lou H. (2008) Cell Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barbisan F., Mazzucchelli R., Santinelli A., Lopez-Beltran A., Cheng L., Scarpelli M., Montorsi F., Montironi R. (2009) Eur. Urol. 56, 105–112 [DOI] [PubMed] [Google Scholar]
- 31. Cho N. P., Han H. S., Soh Y., Son H. J. (2007) J. Oral Pathol. Med. 36, 297–303 [DOI] [PubMed] [Google Scholar]
- 32. Denkert C., Weichert W., Pest S., Koch I., Licht D., Köbel M., Reles A., Sehouli J., Dietel M., Hauptmann S. (2004) Cancer Res. 64, 189–195 [DOI] [PubMed] [Google Scholar]
- 33. Denkert C., Weichert W., Winzer K. J., Müller B. M., Noske A., Niesporek S., Kristiansen G., Guski H., Dietel M., Hauptmann S. (2004) Clin. Cancer Res. 10, 5580–5586 [DOI] [PubMed] [Google Scholar]
- 34. Dixon D. A., Tolley N. D., King P. H., Nabors L. B., McIntyre T. M., Zimmerman G. A., Prescott S. M. (2001) J. Clin. Invest. 108, 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hostetter C., Licata L. A., Witkiewicz A., Costantino C. L., Yeo C. J., Brody J. R., Keen J. C. (2008) Cancer Biol. Ther. 7, 1496–1506 [DOI] [PubMed] [Google Scholar]
- 36. Kang M. J., Ryu B. K., Lee M. G., Han J., Lee J. H., Ha T. K., Byun D. S., Chae K. S., Lee B. H., Chun H. S., Lee K. Y., Kim H. J., Chi S. G. (2008) Gastroenterology 135, 2030–2042, 2042.e1–3 [DOI] [PubMed] [Google Scholar]
- 37. Niesporek S., Kristiansen G., Thoma A., Weichert W., Noske A., Buckendahl A. C., Jung K., Stephan C., Dietel M., Denkert C. (2008) Int. J. Oncol. 32, 341–347 [PubMed] [Google Scholar]
- 38. Mehta A., Trotta C. R., Peltz S. W. (2006) Genes Dev. 20, 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Scott G. K., Marx C., Berger C. E., Saunders L. R., Verdin E., Schäfer S., Jung M., Benz C. C. (2008) Mol. Cancer Res. 6, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sully G., Dean J. L., Wait R., Rawlinson L., Santalucia T., Saklatvala J., Clark A. R. (2004) Biochem. J. 377, 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Giles K. M., Daly J. M., Beveridge D. J., Thomson A. M., Voon D. C., Furneaux H. M., Jazayeri J. A., Leedman P. J. (2003) J. Biol. Chem. 278, 2937–2946 [DOI] [PubMed] [Google Scholar]
- 42. Huggett J., Dheda K., Bustin S., Zumla A. (2005) Genes Immun. 6, 279–284 [DOI] [PubMed] [Google Scholar]
- 43. Ohl F., Jung M., Xu C., Stephan C., Rabien A., Burkhardt M., Nitsche A., Kristiansen G., Loening S. A., Radonić A., Jung K. (2005) J. Mol. Med. 83, 1014–1024 [DOI] [PubMed] [Google Scholar]
- 44. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 45. Giles K. M., Barker A., Zhang P. M., Epis M. R., Leedman P. J. (2011) Methods Mol. Biol. 676, 147–163 [DOI] [PubMed] [Google Scholar]
- 46. Thomson A. M., Rogers J. T., Walker C. E., Staton J. M., Leedman P. J. (1999) BioTechniques 27, 1032–1039, 1042 [DOI] [PubMed] [Google Scholar]
- 47. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Cell 125, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 48. Bliss C. (1939) Ann. Appl. Biol. 26, 585–615 [Google Scholar]
- 49. Barreau C., Paillard L., Méreau A., Osborne H. B. (2006) Biochimie 88, 515–525 [DOI] [PubMed] [Google Scholar]
- 50. Yeap B. B., Voon D. C., Vivian J. P., McCulloch R. K., Thomson A. M., Giles K. M., Czyzyk-Krzeska M. F., Furneaux H., Wilce M. C., Wilce J. A., Leedman P. J. (2002) J. Biol. Chem. 277, 27183–27192 [DOI] [PubMed] [Google Scholar]
- 51. Figueroa A., Cuadrado A., Fan J., Atasoy U., Muscat G. E., Muñoz-Canoves P., Gorospe M., Muñoz A. (2003) Mol. Cell Biol. 23, 4991–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gorospe M. (2003) Cell Cycle 2, 412–414 [PubMed] [Google Scholar]
- 53. Katsanou V., Papadaki O., Milatos S., Blackshear P. J., Anderson P., Kollias G., Kontoyiannis D. L. (2005) Mol. Cell 19, 777–789 [DOI] [PubMed] [Google Scholar]
- 54. López de Silanes I., Lal A., Gorospe M. (2005) RNA Biol. 2, 11–13 [DOI] [PubMed] [Google Scholar]
- 55. Yaman I., Fernandez J., Sarkar B., Schneider R. J., Snider M. D., Nagy L. E., Hatzoglou M. (2002) J. Biol. Chem. 277, 41539–41546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jacobsen A., Wen J., Marks D. S., Krogh A. (2010) Genome Res. 20, 1010–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ma W. J., Cheng S., Campbell C., Wright A., Furneaux H. (1996) J. Biol. Chem. 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 58. Cho N. H., Kang S., Hong S., An H. J., Choi Y. H., Jeong G. B., Choi H. K. (2006) Cancer Lett. 232, 170–178 [DOI] [PubMed] [Google Scholar]
- 59. López de Silanes I., Fan J., Galbán C. J., Spencer R. G., Becker K. G., Gorospe M. (2004) Gene Expr. 12, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bueno M. J., Pérez de Castro I., Gómez de Cedrón M., Santos J., Calin G. A., Cigudosa J. C., Croce C. M., Fernández-Piqueras J., Malumbres M. (2008) Cancer Cell 13, 496–506 [DOI] [PubMed] [Google Scholar]
- 61. Bueno M. J., Pérez de Castro I., Malumbres M. (2008) Cell Cycle 7, 3143–3148 [DOI] [PubMed] [Google Scholar]
- 62. Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu F., Zhang X., Lei Y., Liu X., Liu Z., Tong T., Wang W. (2010) J. Cell Biochem. 111, 727–734 [DOI] [PubMed] [Google Scholar]
- 64. Abdelmohsen K., Kim M. M., Srikantan S., Mercken E. M., Brennan S. E., Wilson G. M., de Cabo R., Gorospe M. (2010) Cell Cycle 9, 1354–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abdelmohsen K., Srikantan S., Kuwano Y., Gorospe M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20297–20302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Cold Spring Harb. Symp. Quant. Biol. 71, 513–521 [DOI] [PubMed] [Google Scholar]
- 67. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 68. Kedde M., Agami R. (2008) Cell Cycle 7, 899–903 [DOI] [PubMed] [Google Scholar]
- 69. Kedde M., Strasser M. J., Boldajipour B., Oude Vrielink J. A., Slanchev K., le Sage C., Nagel R., Voorhoeve P. M., van Duijse J., Ørom U. A., Lund A. H., Perrakis A., Raz E., Agami R. (2007) Cell 131, 1273–1286 [DOI] [PubMed] [Google Scholar]
- 70. Ketting R. F. (2007) Cell 131, 1226–1227 [DOI] [PubMed] [Google Scholar]
- 71. Nielsen C. B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C. B. (2007) RNA 13, 1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]