Background: The roles of 17β-estradiol (E2) and estrogen receptor (ER) in skeletal muscles remains unclear.
Results: E2 inhibits myogenesis by increasing expression of ubiquitin-specific peptidase 19 (USP19), and depletion of ERα represses E2-increased USP19 expression.
Conclusion: USP19 plays an important role in E2-inhibited myogenesis.
Significance: The mechanism by which USP19 inhibits myogenesis is important for understanding the roles of E2 in myogenesis.
Keywords: Deubiquitination, Estrogen, Estrogen Receptor, Myogenesis, Skeletal Muscle
Abstract
Skeletal muscles express estrogen receptor (ER) α and ERβ. However, the roles of estrogens acting through the ERs in skeletal muscles remain unclear. The effects of 17β-estradiol (E2) on myogenesis were studied in C2C12 myoblasts. E2 and an ERα-selective agonist propylpyrazole-triol depressed myosin heavy chain (MHC), tropomyosin, and myogenin levels and repressed the fusion of myoblasts into myotubes. ER antagonist ICI 182,780 cancelled E2-repressed myogenesis. E2 induced ubiquitin-specific peptidase 19 (USP19) expression during myogenesis. E2 replacement increased USP19 expression in the gastrocnemius and soleus muscles of ovariectomized mice. Knockdown of USP19 inhibited E2-repressed myogenesis. Mutant forms of USP19 lacking deubiquitinating activity increased MHC and tropomyosin levels. E2 decreased ubiquitinated proteins during myogenesis, and the E2-decreased ubiquitinated proteins were increased by knockdown of USP19. Propylpyrazole-triol increased USP19 expression, and ICI 182,780 inhibited E2-increased USP19 expression. Overexpression of ERα or knockdown of ERβ enhanced the effects of E2 on the levels of USP19, MHC, and tropomyosin, whereas knockdown of ERα, overexpression of ERβ, or an ERβ-selective agonist diarylpropionitrile abolished their effects. A mutant form of ERα that is constitutively localized in the nucleus increased USP19 expression and decreased MHC and tropomyosin expression in the presence of E2. Furthermore, in skeletal muscle satellite cells, E2 inhibited myogenesis and increased USP19 expression, and diarylpropionitrile repressed E2-increased USP19 expression. These results demonstrate that (i) E2 induces USP19 expression through nuclear ERα, (ii) increased USP19-mediated deubiquitinating activity represses myogenesis, and (iii) ERβ inhibits ERα-activated USP19 expression.
Introduction
Estrogens function not only in reproductive tissues, but also in peripheral tissues such as brain, bone, and cardiovascular tissues. The biological activities of estrogens are mediated by two estrogen receptor (ER)2 isoforms, namely ERα and ERβ, which are members of the nuclear receptor superfamily of ligand-mediated transcriptional factors (1). ERα and ERβ share a high degree of structural similarity and are composed of three domains: a variable N-terminal domain, a highly conserved central DNA-binding domain, and a C-terminal ligand-binding domain. The binding of estrogen (17β-estradiol, E2) to the ligand-binding domain of ERα exerts genomic effects on DNA sequences called estrogen response elements (EREs), which regulate the transcription of target genes. On the other hand, ERβ acts as a transcription factor not only in the ligand-bound state, but also in the ligand-free state (2). ERα and ERβ do not redundantly act on gene expression, despite the similarity of their DNA-binding domains. Because they competitively bind to the same EREs or specifically bind to different EREs, they may have distinct functions in gene regulation. For example, ERα promotes cell proliferation in the development of the reproductive tract and mammary gland, whereas ERβ seems to have potent antiproliferative (3, 4) and anti-inflammatory properties (5). Recently, ERα and ERβ have been identified in skeletal muscle (6). Therefore, estrogens seem to act on skeletal muscle through ER isoforms. However, the roles of E2 and the two ER isoforms in skeletal muscle remain unclear.
Skeletal muscle is the most abundant tissue in the body. Body composition and body size are different between males and females. Sex hormones are thought to be responsible for gender-specific differences in skeletal muscle mass, but their specific effects remain poorly characterized. Satellite cells are the myogenic progenitor cells of postnatal skeletal muscle and reside at the periphery of myofiber in a quiescent state (7). Stimuli such as injury or stress allow satellite cells to proliferate and differentiate. The resultant mononucleated myoblasts proliferate, migrate, differentiate, and fuse into multinucleated myotubes and eventually into myofibers. Skeletal muscle formation (myogenesis) is controlled by regulatory factors, such as MyoD, myogenin, and Myf5, and skeletal muscles express structural muscle proteins such as myosin heavy chain (MHC) and tropomyosin (8).
Skeletal muscle mass and muscle proteins are decreased under various pathophysiological conditions such as disuse, fasting, diabetes, and prolonged bed rest. The protein loss results from an imbalance between synthesis and degradation of proteins. The ubiquitin-proteasome pathway controls the degradation of aberrant proteins and regulates the stability and function of the proteins. The conjugation of ubiquitin to muscle proteins is accomplished by a cascade of enzymes, ubiquitin-activating enzymes, ubiquitin-conjugating enzymes, and ubiquitin ligases, and subsequently the conjugated proteins are degraded by the proteasome. During muscle catabolism, ∼120 genes (called “atrogenes”) are induced or suppressed (9, 10). Atrogin-1/MAFbx (muscle atrophy F-box protein) and MuRF1 (muscle-specific RING finger protein) are induced early in the atrophy process and function as ubiquitin ligases (11). High expression of atrogin-1/MAFbx results in the loss of muscle weight (12), whereas knock-out of atrogin-1/MAFbx and MuRF1 results in resistance to atrophy (11). On the other hand, the ubiquitin-proteasome system is also modulated by deubiquitinating enzymes, which are called ubiquitin-specific peptidases (USPs). USPs release ubiquitins from ubiquitinated proteins, and the up-regulation of these enzymes presumably helps to recycle free ubiquitin when the ubiquitin-proteasome system is activated. Although many proteins that are predicted to be USPs based on their amino acid sequences are expressed in skeletal muscle (13, 14), it remains unclear whether they function as deubiquitinating enzymes and how their expression levels are regulated in skeletal muscle. Recently, the expression of ubiquitin-specific peptidase 19 (USP19), which is a member of USP family proteins, was found to be elevated in skeletal muscle during catabolic states (15). Depletion of USP19 increases not only the levels of myofibrillar proteins such as MHC, troponin T, and tropomyosin, but also the level of a muscle-specific transcription factor myogenin in L6 muscle cells (16). In the present paper, we report that E2 inhibits myogenic differentiation of mouse C2C12 myoblasts and mouse satellite cells and increases the expression of USP19 in skeletal muscle in vitro and in vivo. Furthermore, we demonstrate that USP19 is induced by the nuclear ERα and inhibits myogenesis by acting as a deubiquitinating enzyme. We also analyze the roles of ERβ in USP19 expression.
EXPERIMENTAL PROCEDURES
Reagents
Mouse anti-α-tubulin antibody (clone DM1A) and ICI 182,780 were obtained from Sigma, and propylpyrazole-triol (PPT; 1,3,5-tris(4-hydroxyphenyl)-4-propyl-1H-pyrazole) and diarylpropionitrile (DPN; 2,3-bis(4-hydroxyphenyl)propionitrile) were obtained from Tocris (Ballwin, MO). IC3-OSu, IC5-OSu, and HilyMAX were obtained from Dojindo Laboratories (Kumamoto, Japan). Rabbit polyclonal anti-ERα (MC-20) and anti-ERβ (H150) antibodies and mouse monoclonal anti-MyoD antibody (clone 5.8A) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-MHC (MF-20, supernatant for Western blotting and ascites for immunofluorescence analysis), anti-tropomyosin (CH1, supernatant), and anti-Pax7 antibodies were obtained from the Developmental Studies Hybridoma Bank, University of Iowa (Iowa City, IA). Mouse monoclonal anti-Myc (9B11) antibody and Alexa Fluor 488-conjugated anti-mouse IgG were obtained from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal anti-USP19 (ab93159) and rat monoclonal anti-HA (3F10) antibodies were obtained from Abcam (Cambridge, MA) and Roche Diagnostics, respectively. Mouse monoclonal anti-lamin B1 (L-5), anti-myogenin (F5D), and anti-ubiquitin (FK2) antibodies were obtained from Zymed Laboratories Inc. (San Francisco, CA), BD Biosciences (San Jose, CA), and Nippon Biotest Laboratories (Tokyo, Japan), respectively.
Cell Culture
Murine C2C12 skeletal muscle myoblasts were obtained from the RIKEN Cell Bank (Ibaraki, Japan). C2C12 myoblasts were cultured in DMEM supplemented with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin (growth medium) at 37 °C in a 5% CO2, 95% air atmosphere at 100% humidity. To induce differentiation from myoblasts to myotubes, C2C12 myoblasts were grown to 90% confluence in the growth medium and shifted to phenol red-free DMEM supplemented with 2% dextran-coated charcoal-stripped horse serum and the above antibiotics (differentiation medium) in the presence or absence of 10 nm E2 unless otherwise indicated. The differentiation medium was replaced at 48-h intervals.
Animals
Female Kwl:ddY mice were obtained from Kiwa Laboratory Animals (Wakayama, Japan). These mice had free access to water and a diet and were kept at controlled temperature (23 ± 2 °C), humidity (60 ± 10%), and lighting (12-h light and 12-h dark cycle) conditions. All experimental procedures involving laboratory animals were approved by the Animal Care and Use Committee of Osaka Prefecture University.
Isolation of Satellite Cells
Satellite cells were isolated from the hind-limb muscles of neonatal (3–5 days old) or young (7–8 weeks old) female Kwl:ddY mice according to the method of Allen et al. (17). Satellite cells were cultured in DMEM supplemented with 15% FBS, 4.5 g/liter of d-glucose, and the above antibiotics at 37 °C in a 5% CO2, 95% air atmosphere at 100% humidity. To induce myogenic differentiation, cells at confluence were cultured in differentiation medium in the presence or absence of E2 as described above. The differentiation medium was replaced at 48-h intervals.
E2 Replacement
Female Kwl:ddY mice (7-week-old) were randomly divided into three groups (n = 5 per group). Two groups were ovariectomized, termed the OVX + V group and OVX + E2 group, and the other was sham-operated, termed the Sham group. After 1 week, the OVX + E2 group was intramuscularly injected with a single dose of estradiol valerate (0.1 mg/kg; Mochida Pharmaceutical, Tokyo, Japan). The OVX + V group and Sham group were intramuscularly injected with a single dose of vehicle (sesame oil). After 1 week, mice were sacrificed by exsanguination under anesthesia. The leg skeletal muscles (gastrocnemius and soleus muscles) and uterus were isolated, weighted, and immediately frozen in liquid nitrogen. The samples were stored at −80 °C until use.
Identification of Proteins by Two-dimensional Difference in Gel Electrophoresis (DIGE)
C2C12 cells were induced in differentiation medium in the presence or absence of E2 for 8 days. Cells were harvested, and cell lysates were prepared by sonicating cells in PBS. Guanidine hydrochloride was added to proteins from the E2-treated or vehicle-treated cell samples (each 250 μg) at a final concentration of 1 mm and mixed. The mixture was heated at 98 °C for 3 min, and put on ice. IC3-OSu and IC5-OSu at a final concentration of 4 μm were reacted with proteins from the E2-treated and vehicle-treated cell samples, respectively, for 1 h on ice, followed by incubation with lysine to stop the reaction. Proteins in each sample were precipitated by acetone, and the precipitates were dissolved in two-dimensional sample buffer (40 mm Tris base, 8 m urea, 4% CHAPS, and 2% mercaptoethanol). The two fluorescence-labeled samples were combined and subjected to first dimension isoelectric focusing on immobilized pH gradient strips (7 cm, pH 3–10, linear precast Immobiline gels, GE Healthcare AB, Uppsala, Sweden) according to the manufacturer's instructions. Second dimension separation was performed on 10% SDS-PAGE gels. Image scanning of difference spot patterns was performed by FLA-7000 (GE Healthcare). IC3-OSu- and IC5-OSu-labeled images were obtained using excitation/emission values of 532/580 and 635/670 nm, respectively; the green and red spots indicate differences in protein occurrence and abundance, whereas proteins with similar responses are yellow. The selected green spots, which were increased by E2, were subjected to in-gel trypsin digestion, followed by analysis with a hybrid linear ion trap/time-of-flight mass spectrometer (TOF-MS) (NanoFrontier L, Hitachi High-Technologies, Tokyo, Japan), and the proteins were identified by the Mascot MS/MS Ions Search algorithm.
Immunofluorescence Analysis
C2C12 cells were fixed in 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min as described previously (18). Cells were blocked and incubated with mouse monoclonal anti-MHC antibody, followed by Alexa Fluor 488-conjugated anti-mouse IgG. Cells were counterstained with DAPI. Fluorescent images were captured using a BIOREVO BZ-9000 fluorescence microscope (Keyence, Osaka, Japan). Five random fields were observed, and the number of nuclei within MHC-positive cells containing two or more nuclei was divided by the total number of nuclei in field, termed the fusion ratio. The fusion index was calculated as follows: the fusion ratio in reagent (e.g. E2)-treated cells was normalized to that in vehicle-treated cells. On the other hand, isolated satellite cells were evaluated by the phenotype as muscle progenitor cells as follows: isolated cells were fixed, permeated, and incubated with mouse monoclonal anti-Pax7 antibody as a maker of satellite cell or mouse monoclonal anti-MyoD antibody as a marker of myoblast, followed by reaction with Alexa Fluor 488-conjugated anti-mouse IgG. The nuclei were stained with DAPI, followed by inspection using a fluorescence microscope. Satellite cells were isolated with a high degree of myogenic purity (>80% Pax7 positive and >70% MyoD positive).
Western Blotting
C2C12 cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, 0.25% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, 1 mm EDTA, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 10 μg/ml of leupeptin, and 1 μg/ml of aprotinin) and sonicated. Cell lysates were subjected to SDS-PAGE and analyzed by Western blotting with following primary antibodies: rabbit polyclonal anti-ERα, anti-ERβ, anti-GAPDH (19), and anti-USP19 antibodies, mouse monoclonal anti-MHC, anti-tropomyosin, anti-myogenin, and anti-ubiquitin antibodies, and rat monoclonal anti-HA antibody. Immunoreactive proteins were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG, anti-mouse IgG, or anti-rat IgG and reacted with Super Signal West Femto Chemiluminescent Substrate (Pierce Biotechnology) or Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA), followed by detection with an LAS4000 imaging system (GE Healthcare). The intensities of immunoreactive proteins were quantified by densitometry using ImageJ (version 1.44, National Institutes of Health).
Subcellular Fractionation
Cells were induced in differentiation medium in the presence or absence of 10 nm E2 for 8 days. Cells were harvested, suspended in sucrose buffer (10 mm Hepes-NaOH, pH 7.5, containing 250 mm sucrose, 1 mm dithiothreitol, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 10 μg/ml of leupeptin, and 1 μg/ml of aprotinin), and homogenized by repeated passage through a 23-gauge needle, and whole cell lysate was prepared. The cytosolic, nuclear, and particulate fractions were separated by differential centrifugation as described previously (20). Proteins in each fraction were analyzed by Western blotting with rabbit polyclonal anti-ERα, anti-ERβ, and anti-triose-phosphate isomerase (19) antibodies and mouse monoclonal anti-lamin B1 antibody. Immunoreactive proteins were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG and anti-mouse IgG, respectively.
Semi-quantitative RT-PCR and Quantitative Real-time RT-PCR
Total RNA was extracted and reverse transcribed. The resultant cDNAs were subjected to semi-quantitative RT-PCR (semi-qRT-PCR) or quantitative real-time RT-PCR (qRT-PCR) using the following specific primers: USP19 (forward primer, 5′-GCGGCACAAGATGAGAAATG-3′ and reverse primer, 5′-ACCAGGAACTTGATGGGCTT-3′); GAPDH (forward primer, 5′-AAAATGGTGAAGGTCGGTGT-3′ and reverse primer, 5′-TTTGATGTTAGTGGGGTCTC-3′); β-actin (forward primer, 5′-TTGCTGACAGGATGCAGAAG-3′ and reverse primer, 5′-GTACTTGCGCTCAGGAGGAG-3′); ERα (forward primer, 5′-ATGATTGGTCTCGTCTGGCGCT-3′ and reverse primer, 5′-AGCAGGTCATAGAGGGGCACAACG-3′); and ERβ (forward primer, 5′-GAGGCCTCCATGATGATGTCCCT-3′ and reverse primer, 5′-TCTGGAGCAAAGATGAGCTTGCC-3′). For GAPDH and β-actin, the PCR profiles consisted of denaturation at 95 °C for 1 min, primer annealing at 55 °C for 1 min, and primer extension at 72 °C for 30 s. The final primer extension was performed at 72 °C for 10 min. For USP19, the PCR profiles consisted of denaturation at 95 °C for 1 min, primer annealing at 57 °C for 1 min, and primer extension at 72 °C for 30 s. The final primer extension was performed at 72 °C for 10 min. For ERα and ERβ, the PCR profiles consisted of denaturation at 95 °C for 1 min, primer annealing at 60 °C for 1 min, and primer extension at 72 °C for 30 s. The final primer extension was performed at 72 °C for 10 min. The semi-qRT-PCR products were analyzed by electrophoresis on a 2% agarose gel and photographed under UV light. The relative expression levels of mRNAs were calculated by determining the ratio of the amount of each mRNA to that of GAPDH. The PCR in qRT-PCR was performed with Plexor One-Step qRT-PCR System (Promega, Madison, WI) on Thermal Cycler Dice, TP-800 (Takara, Shiga, Japan). Ct values were transformed into relative quantification data by the 2−ΔΔCt method, and data were normalized to β-actin or GAPDH.
Plasmids
Human ERα expression vector (pCAGGS-ERα) was described previously (18). The C-terminal Myc-tagged ERα (pCAGGS-ERα-Myc) was constructed. A ERα mutant cDNA encoding a siRNA-resistant form of ERα, designated ERα(mut), was synthesized using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA). Mutations were introduced into pCAGGS-ERα-Myc using mutation primers, 5′-aCTgCTcTTcGCcCCaAAtTTGCTCTTGGACAGGAACCAGGG-3′ and 5′-aTTtGGgGCgAAgAGcAGtTTCCCTGGGTGCTCCATGG-3′ (lowercase letters indicate mutation sites), followed by construction of pCAGGS-ERα(mut)-Myc. The expression vectors, pCAGGS-ERα-NLS-Myc and pCAGGS-ERα(mut)-NLS-Myc, were constructed by insertion of the DNA fragment encoding three tandem repeats of simian virus 40 large T antigen nuclear localization signal (NLS) in-frame to the BamHI sites of pCAGGS-ERα-Myc and pCAGGS-ERα(mut)-Myc, respectively, resulting in fusion proteins of ERα with NLS and the Myc tag. The cDNA encoding human ERβ (GenBankTM accession no. NM_001437) was amplified by two-sequential, nested PCR using human brain cDNA (cerebral cortex Marathon-ready cDNA, Clontech Laboratories, San Jose, CA) and subcloned into pCAGGS vector (21), which was kindly provided by Dr. J. Miyazaki (Osaka University, Osaka, Japan). The N-terminal HA-tagged ERβ expression vector (pCAGGS-HA-ERβ) was constructed. Murine USP19 cDNA was obtained from the Kazusa DNA Research Institute (Kisarazu, Japan). The USP19(C548A) and USP19(C548S) cDNAs encoding mutant forms of USP19 substituting Ala and Ser for Cys at position of 548 were generated by the QuikChange site-directed mutagenesis system using mutation primers for USP19(C548A) (forward, 5′-GGGAACACCgcCTTCATGAATAG-3′ and reverse, 5′-CTATTCATGAAGgcGGTGTTCCC-3′) and USP19(C548S) (forward, 5′-GGGAACACCTcCTTCATGAATAG-3′ and reverse, 5′-CTATTCATGAAGgAGGTGTTCCC-3′), respectively. Lowercase letters indicate mutation sites. The cDNAs encoding the wild-type and mutant forms of USP19 were subcloned into the pCAGGS-Myc vector, termed pCAGGS-Myc-USP19, pCAGGS-Myc-USP19(C548A), and pCAGGS-Myc-USP19(C548S). The cDNA encoding human ubiquitin (GenBankTM accession no. NM_018955) was amplified by two sequential, nested PCR using cDNA from human breast cancer MCF-7 cells. The N-terminal HA-tagged ubiquitin expression vector was constructed using pcDNA3.1-HA (22), termed pcDNA3.1-HA-Ub.
Transfection of siRNA
The siRNA duplexes targeting murine USP19 (product number, USP19 number 1 (siUSP19-1), SASI_Mm01_00116952, and USP19 number 2 (siUSP19-2), SASI_Mm01_00116956), murine ERα, and murine ERβ were purchased from Sigma. Sequences were as follows: USP19 siRNA-1 (5′-CCACUAUACUGCUUGUGCAdTdT-3′), USP19 siRNA-2 (5′-CCAAUGAACACUCAAACAAdTdT-3′), ERα (5′-GCUCCUGUUUGCUCCUAACdTdT-3′), and ERβ (5′-GUGCCAGCGAGCAGGUGCAdTdT-3′). Control siRNA (MISSION siRNA Universal Negative Control number 1) was obtained from Sigma. The siRNA duplexes (10 nm) were introduced into cells by Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's reverse transfection protocol.
Transfection of Plasmid for Mammalian Expression
C2C12 myoblasts were grown to 70–80% confluence in growth medium and transiently transfected with pCAGGS-ERα-Myc, pCAGGS-HA-ERβ, pCAGGS-Myc-USP19, pCAGGS-Myc-USP19(C548A), pCAGGS-Myc-USP19(C548S), or pcDNA3.1-HA-Ub using HilyMax reagent for 6 h. Cells were induced in differentiation medium in the presence or absence of E2.
Statistics
qRT-PCR data were logarithmically transformed to improve normality to compensate for unequal variance before statistics. Data from C2C12 myoblast cells and mouse satellite cells were analyzed by one-way or two-way analysis of variance with Tukey's post hoc testing. Data from the animal experiments were analyzed by Student's t tests. Statistical analysis was performed using JMP statistical software version 8.0.1 (SAS Institute, Cary, NC). Data are expressed as mean ± S.D., and differences were considered statistically significant at a p value of < 0.05.
RESULTS
E2 and ERα Agonist Inhibit Myogenesis
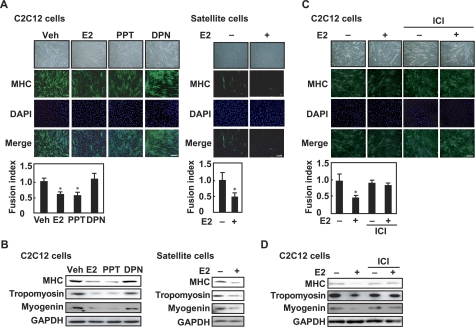
To study the effect of E2 on myogenic differentiation, C2C12 myoblasts were cultured in differentiation medium in the presence or absence of E2. The fusion index was calculated to estimate myotube formation. E2 inhibited the fusion of myoblasts into myotubes (Fig. 1A, left panel). Furthermore, to determine whether E2 inhibits myogenic differentiation of satellite cells, skeletal muscle satellite cells were isolated from young female mice and cultured in differentiation medium in the presence or absence of E2. In response to E2, myotube formation of satellite cells was inhibited (Fig. 1A, right panel). E2 decreased the protein levels of MHC and tropomyosin, which are dominant myofibrillar proteins in differentiated muscle, and the protein level of myogenin, which is an early skeletal muscle differentiation marker, in C2C12 cells (Fig. 1B, left panel) and satellite cells (Fig. 1B, right panel). When skeletal muscle satellite cells were isolated from neonatal female mice and cultured in differentiation medium, E2 inhibited myogenesis (supplemental Fig. S1A) and decreased the levels of MHC, tropomyosin, and myogenin (supplemental Fig. S1B). Likewise, an ERα agonist (PPT), but not an ERβ agonist (DPN), diminished myotube formation (Fig. 1A, left panel) and reduced the expression of MHC, tropomyosin, and myogenin in C2C12 cells (Fig. 1B, left panel). E2 and PPT had no effects on the expression of GAPDH. These results indicate that E2 and PPT inhibit myogenesis. Next, the inhibitory effect of E2 on myogenic differentiation was assessed in the presence of the ER antagonist (ICI 182,780). ICI 182,780 abolished E2-repressed myoblast fusion into myotubes (Fig. 1C) and rescued E2-decreased MHC, tropomyosin, and myogenin expression (Fig. 1D). These results suggest that ER is involved in E2-repressed myogenesis.
FIGURE 1.
Inhibitory effects of E2 and ERα agonist on myogenesis. A, C2C12 myoblasts (left panel) or satellite cells from young mice (right panel) were cultured in differentiation medium in the presence of E2, PPT, or DPN (each 10 nm) for 8 days. Fixed cells were visualized with anti-MHC antibody and fluorescent-labeled secondary antibody. The nuclei were counterstained with DAPI, and the fusion index was calculated. B, C2C12 myoblasts (left panel) or satellite cells (right panel) were cultured in differentiation medium in the presence of E2, PPT, or DPN (each 10 nm) for 8 days. Cell lysates were subjected to Western blotting with anti-MHC, anti-tropomyosin, anti-myogenin, and anti-GAPDH antibodies. C, C2C12 myoblasts were induced in differentiation medium in the presence or absence of 10 nm E2 and/or 1 μm ICI 182,780 (ICI) for 8 days. Immunoreaction with anti-MHC antibody was performed, followed by incubation with fluorescent-labeled secondary antibody. The nuclei were counterstained with DAPI, and the fusion index was calculated. D, C2C12 myoblasts were induced in differentiation medium in the presence or absence of 10 nm E2 and/or 1 μm ICI 182,780 for 8 days. Cell lysates were analyzed by Western blot with anti-MHC, anti-tropomyosin, anti-myogenin, and anti-GAPDH antibodies. A and C, values are indicated as mean ± S.D. Statistically significant differences compared with the vehicle (Veh) are indicated by *, p < 0.05. Scale bar represents 150 μm. All experiments were performed in triplicate, and each graph is representative of three independent experiments.
USP19 Expression Is Increased during E2-repressed Myogenic Differentiation
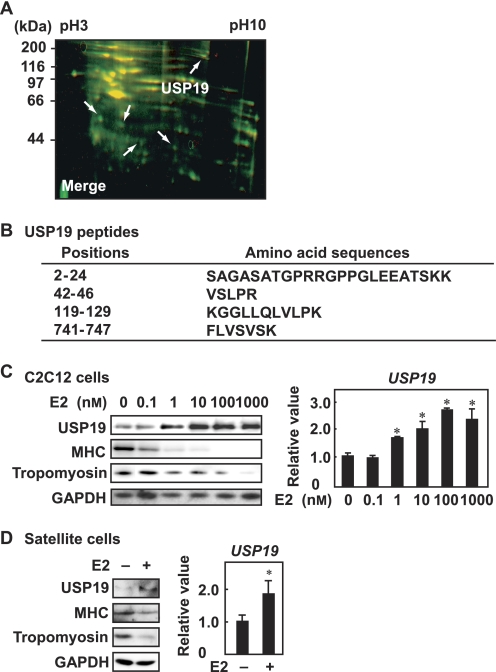
To identify the quantitatively altered proteins during E2-repressed myogenic differentiation, C2C12 cells were cultured in differentiation medium in the presence and absence of E2. Proteins in lysates from vehicle- and E2-treated cells were labeled with IC5-OSu and IC3-OSu, respectively, and analyzed by two-dimensional DIGE. Differentially expressed proteins are shown in Fig. 2A. Five spots (green color) were increased in E2-treated cell lysates and analyzed by linear ion trap/TOF-MS. Peptide sequences predicted by the mass spectrometric analysis were blasted against the protein databases. Four peptides from one of the five spots matched USP19, which is a deubiquitinating enzyme (Fig. 2B). When C2C12 cells were treated with various concentrations of E2 during myogenic differentiation, the expression level of the USP19 protein was increased in a dose-dependent manner (Fig. 2C, left panel). However, the expression levels of MHC and tropomyosin were decreased. E2 also enhanced USP19 expression at mRNA levels in a dose-dependent manner (Fig. 2C, right panel). When satellite cells were cultured in differentiation medium in the presence or absence of E2, USP19 expression was increased at the protein and mRNA levels (Fig. 2D). These results indicate that USP19 is up-regulated by E2 at the transcriptional level.
FIGURE 2.
E2-induced USP19 expression during myogenesis. A, C2C12 myoblasts were induced to differentiate in the presence or absence of 10 nm E2 for 8 days. The overlapping image of two-dimensional DIGE analyses is shown. Differentially expressed proteins are marked with arrows. B, amino acid sequences of peptide fragments were obtained with linear ion trap/TOF-MS and Mascot MS/MS Ions Search algorithm. C, C2C12 cells were cultured in differentiation medium containing various concentrations of E2 for 8 days. Left panel, Western blot analyses with anti-USP19, anti-MHC, anti-tropomyosin, and anti-GAPDH antibodies are shown. Right panel, qRT-PCR was performed for USP19 mRNA. Data were normalized to GAPDH as the endogenous control. D, satellite cells were induced to differentiate in the presence or absence of 10 nm E2. Left panel, cell lysates were analyzed by Western blotting with anti-USP19, anti-MHC, anti-tropomyosin, and anti-GAPDH antibodies. Right panel, qRT-PCR was performed for USP19 mRNA. Data were normalized to GAPDH as the endogenous control. C and D, values are indicated as mean ± S.D. Statistically significant differences are indicated by *, p < 0.05. The result is representative of three independent experiments.
USP19 Expression Is Regulated by E2 in Skeletal Muscle in Vivo
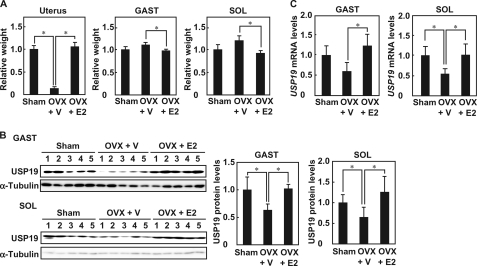
To determine the effects of E2 on USP19 expression in gastrocnemius and soleus muscles, female mice were ovariectomized. The ratios of skeletal muscle masses to body weight tended to be greater in ovariectomized mice than in control sham mice, although the difference was not statistically significant (gastrocnemius, p = 0.09; soleus, p = 0.09) (Fig. 3A). Treatment of ovariectomized mice with E2 replacement decreased the ratios of skeletal muscle masses to body weight. The ratio of uterine mass to body weight in ovariectomized mice was significantly decreased, and was recovered by E2 replacement. In ovariectomized mice, USP19 protein levels in the soleus and gastrocnemius muscles were decreased and were restored by E2 replacement (Fig. 3B). Ovariectomy resulted in a decreased USP19 mRNA level in the soleus muscle, but not in the gastrocnemius muscle (p = 0.06) (Fig. 3C). E2 replacement increased USP19 mRNA levels in the gastrocnemius and soleus muscles. These results indicate that E2 up-regulates USP19 expression in skeletal muscles in vivo.
FIGURE 3.
USP19 expression in skeletal muscle of ovariectomized mice with E2 replacement. Ovariectomized mice were intramuscularly injected with a single dose of vehicle (OVX + V)) or estradiol valerate (OVX + E2). Sham-operated mice were injected with vehicle (Sham). A, the ratios of wet weight of the uterus, gastrocnemius muscle (GAST), and soleus muscle (SOL) to body weight were determined. B, left panel, Western blot analyses were performed with anti-USP19 and anti-α-tubulin antibodies. Right panel, the intensities of immunoreactive bands were quantified, and data were normalized to α-tubulin. C, total RNA was prepared from gastrocnemius muscle and soleus muscle. The USP19 and β-actin mRNA levels were analyzed by qRT-PCR. Data were normalized to β-actin. In all experiments, values are indicated as mean ± S.D., and statistically significant differences are indicated by *, p < 0.05.
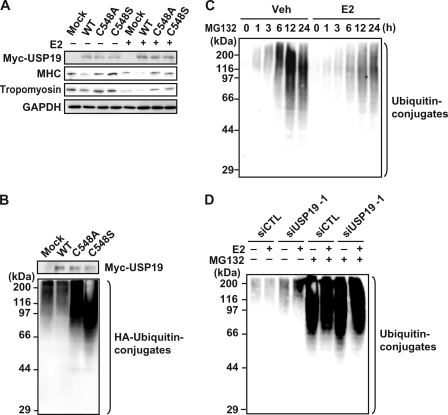
USP19 Is Involved in E2-repressed Myogenic Differentiation as a Deubiquitinating Enzyme
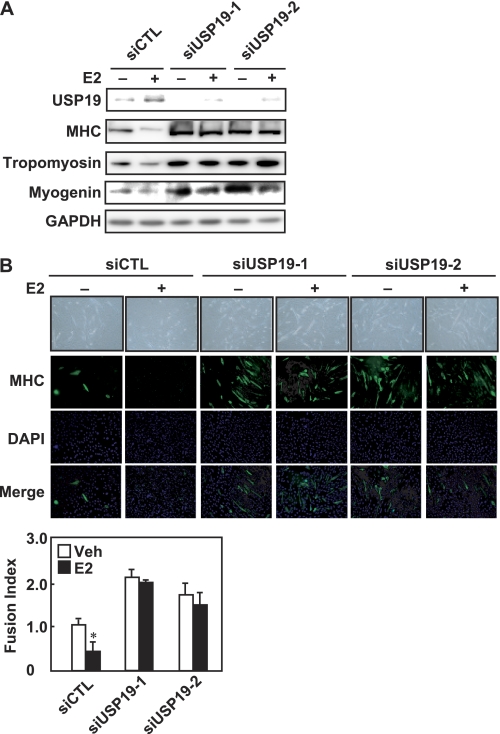
To determine whether USP19 is involved in E2-repressed myogenic differentiation, C2C12 myoblasts were transfected with USP19 siRNA (either USP19 number 1 or USP19 number 2), followed by induction of differentiation. USP19 siRNA specifically knocked down expression of the USP19 protein, but not the GAPDH protein, in the presence or absence of E2, indicating the specificity and effectiveness of the designed USP19 siRNAs (Fig. 4A). Knockdown of USP19 increased the expression levels of MHC, tropomyosin, and myogenin not only in E2-treated cells, but also in vehicle-treated cells. In fact, knockdown of USP19 promoted myogenesis in vehicle-treated cells and rescued E2-inhibited myotube formation (Fig. 4B). In contrast, when C2C12 cells exogenously overexpressed the wild-type form of USP19 during differentiation, the expression levels of MHC and tropomyosin were decreased in the presence or absence of E2 (Fig. 5A). To assess whether the inhibitory effect of USP19 on myogenesis is required for the deubiquitinating activity, mutant forms of USP19 lacking deubiquitinating activity were constructed, termed USP19(C548A) and USP19(C548S). These mutants replacing Ala and Ser for Cys at position 548 of USP19 are inactive (23). As shown in Fig. 5B, overexpression of wild-type USP19 decreased the amounts of ubiquitinated proteins, whereas the mutant form of USP19 (USP19(C548A) or USP19(C548S)) dominant-negatively had the opposite effect. Therefore, the effects of these mutants on the expression of the myogenic differentiation marker proteins were examined. The ability of USP19 to reduce the expressions of MHC and tropomyosin was lost by the mutation in the presence or absence of E2 (Fig. 5A). Next, we examined the effect of E2 on ubiquitination of intracellular proteins in C2C12 cells. When C2C12 cells were cultured in differentiation medium in the presence of MG132, the levels of ubiquitinated proteins were increased in a time-dependent manner (Fig. 5C). Exposure of C2C12 cells to E2 repressed the ubiquitinated protein levels. To determine the effect of USP19 knockdown on ubiquitinated protein levels, C2C12 cells were transfected with USP19 siRNA, followed by culture in the presence or absence of E2. Knockdown of USP19 increased ubiquitinated protein levels in the presence or absence of E2 and/or the proteasome inhibitor MG132 (Fig. 5D). These results indicate that USP19 is involved in E2-repressed myogenic differentiation and that the deubiquitinating activity of USP19 is required for repression of myoblast fusion in both the presence and absence of E2.
FIGURE 4.
Involvement of USP19 in E2-repressed myogenesis. A, C2C12 cells were transfected with USP19 siRNA (either siUSP19-1 or siUSP19-2) or negative control siRNA (siCTL) for 24 h, followed by culturing in differentiation medium in the presence or absence of 10 nm E2 for an additional 4 days. Cell lysates were analyzed by Western blotting with anti-USP19, anti-MHC, anti-tropomyosin, anti-myogenin, and anti-GAPDH antibodies. B, the siRNA duplexes were introduced into C2C12 cells for 24 h. C2C12 myoblasts were induced to differentiate in the presence or absence of 10 nm E2 for 4 days. The fixed cells were incubated with anti-MHC antibody, and the nuclei were counterstained with DAPI. The fusion index was calculated. Scale bar represents 150 μm. Values are indicated as mean ± S.D. Statistically significant differences compared with the vehicle (Veh) are indicated by *, p < 0.05. All experiments were performed in triplicate, and the graph is representative of three independent experiments.
FIGURE 5.
DUB activity of USP19 and E2-repressed myogenesis. A, C2C12 cells were transfected with pCAGGS-Myc-USP19, pCAGGS-Myc-USP19(C548A), or pCAGGS-Myc-USP19(C548S) for 24 h, followed by culturing in differentiation medium in the presence or absence of 10 nm E2 for an additional 4 days. Cell lysates were analyzed by Western blotting with anti-Myc, anti-MHC, anti-tropomyosin, and anti-GAPDH antibodies. B, pcDNA3.1-HA-Ub was transfected into C2C12 cells with pCAGGS-Myc-USP19, pCAGGS-Myc-USP19(C548A), or pCAGGS-Myc-USP19(C548S) for 24 h, followed by induction of differentiation for an additional 4 days. Cell lysates were analyzed by Western blotting with anti-Myc and anti-HA antibodies. C, C2C12 myoblasts were cultured in differentiation medium containing 5 μm MG132 in the presence or absence of 10 nm E2. Cell lysates were analyzed by Western blotting with anti-ubiquitin antibody. D, C2C12 cells were transfected with USP19 siRNA (siUSP19-1) or control siRNA (siCTL) for 24 h. Cells were induced to differentiate in the presence or absence of 5 μm MG132 and/or 10 nm E2 for 12 h, and cell lysates were subjected to Western blot analysis with anti-ubiquitin antibody. The graph is representative of three independent experiments.
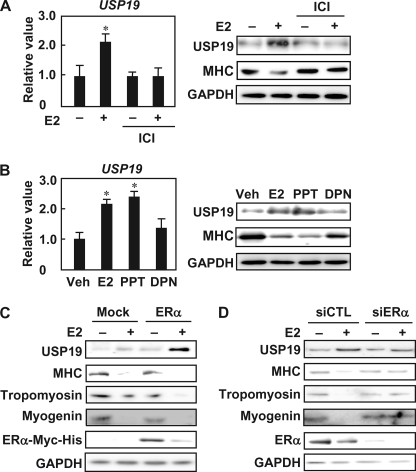
E2 Up-regulates USP19 Expression through ERα
To determine whether ERα is involved in the mechanisms by which E2 increases USP19 expression, the enhancing effect of E2 on USP19 expression during myogenic differentiation was assessed in the presence of the ER antagonist. ICI 182,780 abolished E2-increased USP19 expression at the mRNA and protein levels and restored the E2-decreased MHC protein level (Fig. 6A). Furthermore, when C2C12 myoblasts were cultured in differentiation medium in the presence of either of the ER-selective agonists, PPT and E2 were equally effective in increasing USP19 mRNA and protein levels but DPN had no significant effect (Fig. 6B). PPT, but not DPN, decreased the MHC protein level. Overexpression of exogenous ERα in the presence of E2 increased the expression of USP19, but decreased the expressions of MHC, tropomyosin, and myogenin (Fig. 6C). Knockdown of ERα with ERα siRNA decreased the E2-increased USP19 level and restored E2-decreased MHC, tropomyosin, and myogenin levels (Fig. 6D). These results indicate that E2 increases USP19 expression and represses myogenesis through ERα.
FIGURE 6.
Involvement of ERα in E2-induced USP19 expression. A, C2C12 myoblasts were differentiated in differentiation medium in the presence of 10 nm E2 and/or 1 μm ICI 182,780 (ICI) for 8 days. Left panel, qRT-PCR was performed, and the USP19 mRNA level was normalized to GAPDH mRNA level. Right panel, cell lysates were subjected to Western blot analyses with anti-USP19, anti-MHC, and anti-GAPDH antibodies. B, C2C12 myoblasts were cultured in differentiation medium with E2, PPT, or DPN (each 10 nm) for 8 days. Left panel, qRT-PCR was performed for USP19 and GAPDH mRNAs. Data were normalized to GAPDH. Right panel, cell lysates were subjected to Western blot analyses with anti-USP19, anti-MHC, and anti-GAPDH antibodies. A and B, values are indicated as mean ± S.D. Statistically significant differences compared with vehicle in the absence of E2 are indicated by *, p < 0.05. C, C2C12 myoblasts were transfected with pCAGGS-ERα-Myc for 24 h, followed by culturing in differentiation medium in the presence or absence of 10 nm E2 for an additional 4 days. Cell lysates were analyzed by Western blotting with anti-USP19, anti-MHC, anti-tropomyosin, anti-myogenin, anti-Myc, and anti-GAPDH antibodies. D, after 24 h of transfection with ERα siRNA (siERα) or negative control siRNA (siCTL), C2C12 myoblasts were differentiated in the presence or absence of 10 nm E2 for 4 days. Cell lysates were analyzed by Western blotting with anti-USP19, anti-MHC, anti-tropomyosin, anti-ERα, and anti-GAPDH antibodies. In all experiments, the result is representative of three independent experiments.
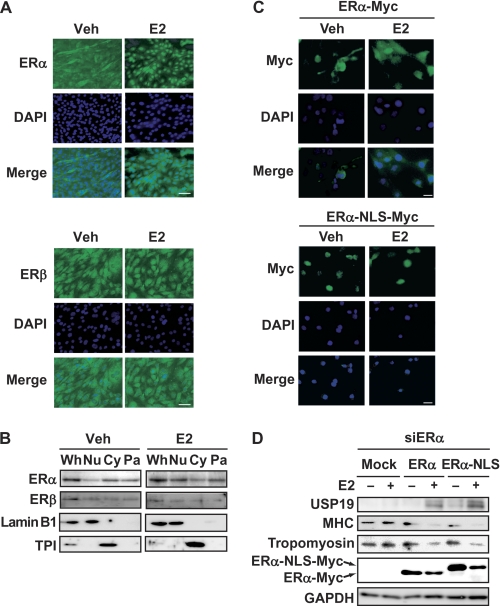
Nuclear ERα Is Involved in E2-increased USP19 Expression
Intracellular localization of ER isoforms was determined by immunofluorescence microscopy. C2C12 myoblasts were induced to differentiate into myotubes in the presence or absence of E2, fixed, and incubated with anti-ERα or anti-ERβ antibody. ERα was localized in the cytoplasm and nucleus in the absence of E2, and E2 stimulated nuclear accumulation of ERα (Fig. 7A). In contrast, ERβ was localized in the cytoplasm and nucleus in the presence or absence of E2. Differential centrifugation revealed that ERα and ERβ were distributed in the nuclear, cytosolic, and particulate fractions in the presence or absence of E2 and that nuclear ERα was increased by E2 (Fig. 7B). Therefore, we examined whether nuclear ERα is involved in up-regulation of USP19 expression in the presence of E2. First, we constructed a recombinant fusion protein with NLS and Myc tag at the C terminus of ERα, termed ERα-NLS-Myc. Exogenous ERα-NLS-Myc was constitutively localized in the nucleus in the presence or absence of E2 (Fig. 7C). Intracellular localization of exogenous ERα-NLS-Myc was different from that of exogenous ERα-Myc, which was a recombinant protein with only a Myc tag at the C terminus. Because ERα-NLS-Myc and ERα-Myc were knocked down by ERα siRNA, we further constructed siRNA-resistant forms of ERα-NLS-Myc and ERα-Myc, termed ERα(mut)-NLS-Myc and ERα(mut)-Myc, respectively. When endogenous ERα was knocked down and exogenous ERα(mut)-Myc was overexpressed in C2C12 cells, E2 increased the USP19 level and decreased the MHC and tropomyosin levels (Fig. 7D). Likewise, in the cells that had been knocked down by endogenous ERα, overexpression of ERα(mut)-NLS-Myc resulted in an increased USP19 level and decreased MHC and tropomyosin levels in the presence of E2. These results indicate that nuclear ERα is involved in the up-regulation of USP19 expression in the presence of E2.
FIGURE 7.
E2-induced USP19 expression through nuclear ERα. A, C2C12 myoblasts were cultured in differentiation medium in the presence or absence of 10 nm E2 for 8 days, followed by fixation. Cells were incubated with anti-ERα or anti-ERβ antibody and fluorescent-labeled secondary antibody. The nuclei were stained with DAPI. Scale bar represents 150 μm. B, C2C12 myoblasts were cultured in differentiation medium in the presence or absence of 10 nm E2 for 8 days. Whole cell lysates were prepared (Wh). The nuclear (Nu), cytosolic (Cy), and particulate (Pa) fractions were separated by differential centrifugation. Each fraction was subjected to Western blot analyses for ERα, ERβ, lamin B1 (nuclear marker protein), and triose-phosphate isomerase (TPI, cytosolic marker protein). C, C2C12 myoblasts were transfected with pCAGGS-ERα-Myc or pCAGGS-ERα-NLS-Myc for 24 h. Cells were cultured in differentiation medium in the presence or absence of 10 nm E2 for an additional 24 h. The fixed cells were visualized with anti-Myc antibody and fluorescence-labeled secondary antibody. The nuclei were stained with DAPI. Scale bar represents 100 μm. D, C2C12 myoblasts were transfected with ERα siRNA (siERα) for 6 h, followed by further transfection with pCAGGS-ERα(mut)-Myc or pCAGGS-ERα(mut)-NLS-Myc for 24 h. Cells were cultured in differentiation medium in the presence or absence of 10 nm E2 for an additional 4 days. Cell lysates were analyzed by Western blotting for USP19, MHC, tropomyosin, c-Myc, and GAPDH. In all experiments, the result is representative of three independent experiments.
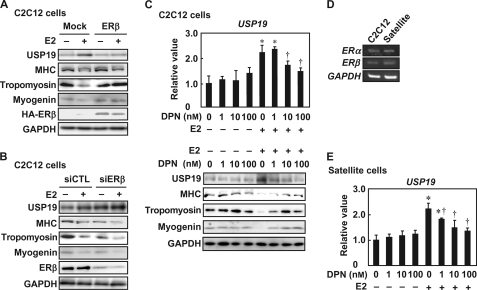
ERβ or ERβ Agonist Inhibits E2-increased USP19 Expression
The role of ERβ in E2-increased USP19 expression was assessed in C2C12 cells during myogenesis in the presence of E2. Overexpression of ERβ attenuated the E2-increased USP19 level and restored the E2-decreased MHC, tropomyosin, and myogenin levels (Fig. 8A). In contrast, knockdown of ERβ increased the USP19 level and reduced MHC, tropomyosin, and myogenin levels in the presence or absence of E2 (Fig. 8B). Furthermore, the effect of DPN on E2-increased USP19 expression was determined in C2C12 cells during myogenesis. DPN repressed the E2-increased USP19 mRNA and protein levels in a dose-dependent manner (Fig. 8C, upper panel and lower panel, respectively). In contrast, E2-repressed MHC and tropomyosin expression levels were restored by DPN. Furthermore, satellite cells expressed ERα and ERβ mRNAs at levels similar to those in C2C12 cells (Fig. 8D), and DPN attenuated E2-increased USP19 mRNA levels in a dose-dependent manner (Fig. 8E). In satellite cells from neonatal mice, USP19 mRNA was increased by E2, and DPN repressed the expression of E2-increased USP19 mRNA (supplemental Fig. S2). Together, these results indicate that ERβ or DPN inhibit E2-increased USP19 expression during myogenesis.
FIGURE 8.
Effects of ERβ knockdown and ERβ-selective agonist on E2-induced USP19 expression. A, C2C12 cells were transfected with pCAGGS-HA-ERβ for 24 h, followed by incubation in differentiation medium in the presence or absence of 10 nm E2 for 4 days. Cell lysates were subjected to Western blot analyses with anti-USP19, anti-MHC, anti-tropomyosin, anti-myogenin, anti-HA, and anti-GAPDH antibodies. B, C2C12 cells were transfected with ERβ siRNA (siERβ) and control siRNA (siCTL) for 24 h, followed by incubation in differentiation medium in the presence or absence of 10 nm E2 for an additional 4 days. Cell lysates were analyzed by Western blotting for USP19, MHC, tropomyosin, myogenin, ERβ, and GAPDH. C, C2C12 cells were differentiated in differentiation medium containing various concentrations of DPN in the presence or absence of 10 nm E2 for 8 days. Upper panel, qRT-PCR was performed, and USP19 mRNA level was normalized to GAPDH mRNA level. Lower panel, cell lysates were subjected to Western blot analyses for USP19, MHC, tropomyosin, myogenin, and GAPDH. D, satellite cells and C2C12 cells were cultured in differentiation medium in the presence of 10 nm E2 for 8 days. Semi-qRT-PCRs for ERα and ERβ mRNAs are shown. E, satellite cells were cultured in differentiation medium containing various concentrations of DPN in the presence or absence of 10 nm E2 for 8 days. USP19 and GAPDH mRNA levels were determined by qRT-PCR. Data were normalized to GAPDH. C and E, values are indicated as mean ± S.D. Statistically significant differences compared with the vehicle in the absence of E2 are indicated by *, p < 0.05. In the E2-treated cells, statistically significant differences compared with the vehicle are indicated by †, p < 0.05. In all experiments, data are representative of three independent experiments.
DISCUSSION
Estrogens, which are defined as female reproductive hormones, function in nonreproductive tissues in addition to reproductive tissues irrespective of sex. Skeletal muscle is expected to be a target tissue for estrogens because two ER isoforms are expressed in skeletal muscle (6). In the present study, E2 induced USP19 expression in vitro and in vivo, and E2-induced USP19 inhibited myogenesis. In rat skeletal muscle, USP19 expression is increased by catabolic stimuli such as fasting, streptozotocin-induced diabetes, dexamethasone treatment, and cancer, and is inversely proportional to muscle mass (15). Depletion of USP19 increases the expression levels of MHC, tropomyosin, and myogenin during myogenesis of rat L6 cells, although it remains unclear whether USP19 functions as a deubiquitinating enzyme (16). USP19 expression is increased by dexamethasone, which induces skeletal muscle atrophy, and knockdown of USP19 attenuates dexamethasone-repressed MHC expression (16). These results indicate that E2 and dexamethasone are potent inducers of USP19 expression and that USP19 functions as a repressor of myotube formation.
Both E2 and USP19 diminished the levels of ubiquitinated proteins. The ubiquitin-proteasome system is a major intracellular nonlysosomal protein degradation mechanism for repairing or removing abnormal proteins. The specificity of protein ubiquitination is determined by ubiquitin ligases. However, intracellular levels of ubiquitinated proteins depend on the balance between the ubiquitination and deubiquitination reactions in the ubiquitin-proteasome system. Although the effects of E2 on the expression levels of ubiquitin ligases in skeletal muscle remain unclear, the ubiquitin-proteasome system during E2-inhibited skeletal muscle formation is strongly biased toward the deubiquitination reaction. These results, together with the finding that E2 enhances USP19 expression, indicate that the deubiquitinating activity of USP19 decreases the levels of ubiquitinated proteins during skeletal muscle formation and acts to repress myogenesis. On the contrary, under various catabolic conditions, the expression levels of muscle-specific ubiquitin ligases, such as atrogin-1/MAFbx, MuRF1, and Cbl-b, are increased (11, 24), and the steady-state levels of ubiquitinated proteins are increased in muscle wasting, indicating that the ubiquitin-proteasome system during skeletal muscle atrophy is biased toward the ubiquitination reaction (25, 26). These results suggest that the balance in the ubiquitin-proteasome system during repression of skeletal muscle formation is different from that during skeletal muscle atrophy.
Mutant forms of USP19 lacking the deubiquitinating activity inhibited E2-decreased MHC and tropomyosin expression and resulted in increased ubiquitinated proteins during myogenesis. Although many proteins that appear to be deubiquitinating enzymes based on their amino acid sequences are expressed in skeletal muscle, their functions are unknown (13, 14). In addition to USP19 (15), two other deubiquitinating enzymes, USP2 (27) and USP14 (10), are expressed during myogenesis. USP2 is expressed as two alternatively spliced variants (UBP45 and UBP69). UBP45 and UBP69 are expressed at different stages of myoblast fusion and antagonistically regulate the morphological differentiation of myoblasts. On the other hand, USP14 expression is induced during muscle wasting (10). Because USP14 interacts with proteasomes, USP14 may remove and recycle ubiquitin from the polyubiquitin chain of substrate proteins on the proteasome (28, 29). In addition to recycling of ubiquitin, removal of the ubiquitin chain from specific substrate proteins inhibits their degradation. USP19 does not appear to associate with the proteasome (16). Therefore, USP19 may regulate myogenesis through deubiquitination of specific proteins rather than through recycling of ubiquitin on the proteasome in skeletal muscle.
E2 and PPT, but not DPN, inhibited myogenesis and increased USP19 expression. In the presence of E2, ERα decreased the levels of MHC and tropomyosin and increased the level of USP19, whereas ERβ had the opposite effects. E2 promotes proliferation not only in the normal mammary gland, but also in mammary tumors through ERα. However, ERβ inhibits the growth of breast cancer cells (4) and the presence of ERβ is linked to survival in patients with breast cancer (30), indicating that ERβ has an anti-proliferative effect. Furthermore, ERβ inhibits ERα-regulated gene transcription (31). Taken together, our data indicate that ERβ down-regulates USP19 expression as a repressor of ERα action and represses the inhibitory effect of ERα on myogenesis in the presence of E2 in skeletal muscle.
Knockdown of ERβ activated USP19 expression even in the absence of E2. Although ERα results in significant expression changes of numerous genes only in the presence of E2, ERβ up- and down-regulates the expression of three classes of target genes (classes I, II, and III) in the presence or absence of E2 (2). Class I genes are regulated by ligand-free ERβ, class II genes are regulated by ligand-bound ERβ, and class III genes were regulated by both types of ERβ. Because endogenous ERβ appears to occur and function on the ERβ-binding sites in the USP19 gene in the absence of E2, the USP19 gene may be down-regulated by ERβ as a class III gene.
ERα-NLS-Myc increased USP19 expression and decreased MHC and tropomyosin expression in the presence of E2 in endogenous ERα-depleted cells. Ligand-activated ERs have both genomic and nongenomic effects. The former refer to transcriptional activation of target genes through binding of ERs to EREs of their promoters in the nucleus (on a time scale of hours), whereas the latter refer to the effects of extranuclear ERs such as membrane-associated or cytoplasmic ERs, which elicit a rapid cellular response (on a time scale of seconds to minutes) such as nitric oxide production and Ca2+ flux in the extranuclear compartment (32, 33). When the murine USP19 gene sequence is analyzed by rVISTA, which is a program that finds potential regulatory elements in noncoding regions of the genome, there is one putative half-ERE (termed dhERE) in intron 1 of the USP19 gene. Chromatin immunoprecipitation assays showed that binding of ERα to dhERE was increased in the presence of E2 and that DPN inhibited the binding of ERα to dhERE (data not shown). Taken together, we suggest that E2 affects USP19 expression by a genomic rather than a nongenomic mechanism, i.e. it up-regulates USP19 expression at the transcriptional level by inducing the binding of ERα to the promoter region of the USP19 gene.
E2 replacement decreased the ratios of skeletal muscle mass to body weight and increased USP19 levels in vivo, supporting the idea that E2 has an inhibitory effect on myogenesis and up-regulates USP19 expression in vitro. The present experiments were performed in ovariectomized young female mice. E2 replacement was found to reduce soleus muscle fiber size in 7-week-old ovariectomized female rats (34, 35). Furthermore, estrogen injection was found to reduce soleus and gastrocnemius muscle weights of both sexes (36). Ovariectomy increases the incidence of small (<500 μm2) fibers, which is a morphological characteristic of newly formed myofibers, in soleus muscle, and E2 replacement diminishes this increase (37). These results suggest that E2 represses skeletal muscle formation through activating USP19 in young females.
The estrogen signaling pathways involving the two ERs played opposite roles in skeletal muscle formation. Because the balance between ERα and ERβ expression levels would regulate USP19 expression, it is of interest to know how the expressions of ERs are regulated in skeletal muscle. Furthermore, the mechanism by which USP19 removes ubiquitin from ubiquitinated proteins remains to be elucidated. Skeletal muscle formation should be promoted not only by repression of USP19 expression and inhibition of USP19 deubiquitinating activity, but also by a blockade of interaction between USP19 and substrate proteins.
Acknowledgments
We thank Dr. Jun-ichi Miyazaki (Osaka University, Osaka, Japan) for pCAGGS vector. We also thank Yoshihiro Kariya and Mana Ishikawa for helpful technical support.
This work was supported by Grant-in-aid 23580182 for scientific research (to R. Y.) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ER
- estrogen receptor
- DPN
- diarylpropionitrile
- E2
- 17β-estradiol
- EREs
- estrogen response elements
- MHC
- myosin heavy chain
- NLS
- nuclear localization signal
- PPT
- propylpyrazole-triol
- qRT-PCR
- quantitative real-time PCR
- DIGE
- difference in gel electrophoresis
- USP
- ubiquitin-specific peptidase.
REFERENCES
- 1. Heldring N., Pike A., Andersson S., Matthews J., Cheng G., Hartman J., Tujague M., Ström A., Treuter E., Warner M., Gustafsson J. A. (2007) Physiol. Rev. 87, 905–931 [DOI] [PubMed] [Google Scholar]
- 2. Vivar O. I., Zhao X., Saunier E. F., Griffin C., Mayba O. S., Tagliaferri M., Cohen I., Speed T. P., Leitman D. C. (2010) J. Biol. Chem. 285, 22059–22066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayashi S. I., Eguchi H., Tanimoto K., Yoshida T., Omoto Y., Inoue A., Yoshida N., Yamaguchi Y. (2003) Endocr. Relat. Cancer 10, 193–202 [DOI] [PubMed] [Google Scholar]
- 4. Lazennec G., Bresson D., Lucas A., Chauveau C., Vignon F. (2001) Endocrinology 142, 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saijo K., Collier J. G., Li A. C., Katzenellenbogen J. A., Glass C. K. (2011) Cell 145, 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wiik A., Ekman M., Johansson O., Jansson E., Esbjörnsson M. (2009) Histochem. Cell Biol. 131, 181–189 [DOI] [PubMed] [Google Scholar]
- 7. Hawke T. J., Garry D. J. (2001) J. Appl. Physiol. 91, 534–551 [DOI] [PubMed] [Google Scholar]
- 8. Charbonnier F., Gaspera B. D., Armand A. S., Van der Laarse W. J., Launay T., Becker C., Gallien C. L., Chanoine C. (2002) J. Biol. Chem. 277, 1139–1147 [DOI] [PubMed] [Google Scholar]
- 9. Jagoe R. T., Lecker S. H., Gomes M., Goldberg A. L. (2002) FASEB J. 16, 1697–1712 [DOI] [PubMed] [Google Scholar]
- 10. Lecker S. H., Jagoe R. T., Gilbert A., Gomes M., Baracos V., Bailey J., Price S. R., Mitch W. E., Goldberg A. L. (2004) FASEB J. 18, 39–51 [DOI] [PubMed] [Google Scholar]
- 11. Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., Pan Z. Q., Valenzuela D. M., DeChiara T. M., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- 12. Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quesada V., Díaz-Perales A., Gutiérrez-Fernández A., Garabaya C., Cal S., López-Otín C. (2004) Biochem. Biophys. Res. Commun. 314, 54–62 [DOI] [PubMed] [Google Scholar]
- 14. Altun M., Besche H. C., Overkleeft H. S., Piccirillo R., Edelmann M. J., Kessler B. M., Goldberg A. L., Ulfhake B. (2010) J. Biol. Chem. 285, 39597–39608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Combaret L., Adegoke O. A., Bedard N., Baracos V., Attaix D., Wing S. S. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E693–700 [DOI] [PubMed] [Google Scholar]
- 16. Sundaram P., Pang Z., Miao M., Yu L., Wing S. S. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1283–1290 [DOI] [PubMed] [Google Scholar]
- 17. Allen R. E., Temm-Grove C. J., Sheehan S. M., Rice G. (1997) Methods Cell Biol. 52, 155–176 [DOI] [PubMed] [Google Scholar]
- 18. Harada N., Yasunaga R., Higashimura Y., Yamaji R., Fujimoto K., Moss J., Inui H., Nakano Y. (2007) J. Biol. Chem. 282, 22651–22661 [DOI] [PubMed] [Google Scholar]
- 19. Yamaji R., Fujita K., Takahashi S., Yoneda H., Nagao K., Masuda W., Naito M., Tsuruo T., Miyatake K., Inui H., Nakano Y. (2003) Biochim. Biophys. Acta 1593, 269–276 [DOI] [PubMed] [Google Scholar]
- 20. Yamaji R., Chatani E., Harada N., Sugimoto K., Inui H., Nakano Y. (2005) Biochim. Biophys. Acta 1726, 261–271 [DOI] [PubMed] [Google Scholar]
- 21. Niwa H., Yamamura K., Miyazaki J. (1991) Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 22. Harada N., Ohmori Y., Yamaji R., Higashimura Y., Okamoto K., Isohashi F., Nakano Y., Inui H. (2008) Biochem. Biophys. Res. Commun. 373, 373–377 [DOI] [PubMed] [Google Scholar]
- 23. Hassink G. C., Zhao B., Sompallae R., Altun M., Gastaldello S., Zinin N. V., Masucci M. G., Lindsten K. (2009) EMBO Rep. 10, 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakao R., Hirasaka K., Goto J., Ishidoh K., Yamada C., Ohno A., Okumura Y., Nonaka I., Yasutomo K., Baldwin K. M., Kominami E., Higashibata A., Nagano K., Tanaka K., Yasui N., Mills E. M., Takeda S., Nikawa T. (2009) Mol. Cell. Biol. 29, 4798–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Medina R., Wing S. S., Haas A., Goldberg A. L. (1991) Biomed. Biochim. Acta 50, 347–356 [PubMed] [Google Scholar]
- 26. Attaix D., Ventadour S., Codran A., Béchet D., Taillandier D., Combaret L. (2005) Essays Biochem. 41, 173–186 [DOI] [PubMed] [Google Scholar]
- 27. Park K. C., Kim J. H., Choi E. J., Min S. W., Rhee S., Baek S. H., Chung S. S., Bang O., Park D., Chiba T., Tanaka K., Chung C. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9733–9738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 29. Peth A., Besche H. C., Goldberg A. L. (2009) Mol. Cell 36, 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakopoulou L., Lazaris A. C., Panayotopoulou E. G., Giannopoulou I., Givalos N., Markaki S., Keramopoulos A. (2004) J. Clin. Pathol. 57, 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindberg M. K., Movérare S., Skrtic S., Gao H., Dahlman-Wright K., Gustafsson J. A., Ohlsson C. (2003) Mol. Endocrinol. 17, 203–208 [DOI] [PubMed] [Google Scholar]
- 32. Picotto G., Massheimer V., Boland R. (1996) Mol. Cell. Endocrinol. 119, 129–134 [DOI] [PubMed] [Google Scholar]
- 33. Chen Z., Yuhanna I. S., Galcheva-Gargova Z., Karas R. H., Mendelsohn M. E., Shaul P. W. (1999) J. Clin. Invest. 103, 401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCormick K. M., Burns K. L., Piccone C. M., Gosselin L. E., Brazeau G. A. (2004) J. Muscle Res. Cell Motil. 25, 21–27 [DOI] [PubMed] [Google Scholar]
- 35. Tsai W. J., McCormick K. M., Brazeau D. A., Brazeau G. A. (2007) Exp. Biol. Med. 232, 1314–1325 [DOI] [PubMed] [Google Scholar]
- 36. Ihemelandu E. C. (1981) Acta Anat. 110, 311–317 [DOI] [PubMed] [Google Scholar]
- 37. McClung J. M., Davis J. M., Wilson M. A., Goldsmith E. C., Carson J. A. (2006) J. Appl. Physiol. 100, 2012–2023 [DOI] [PubMed] [Google Scholar]